gp130 Activates Mitochondrial Dynamics for Hepatocyte Survival in a Model of Steatohepatitis
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Preparation
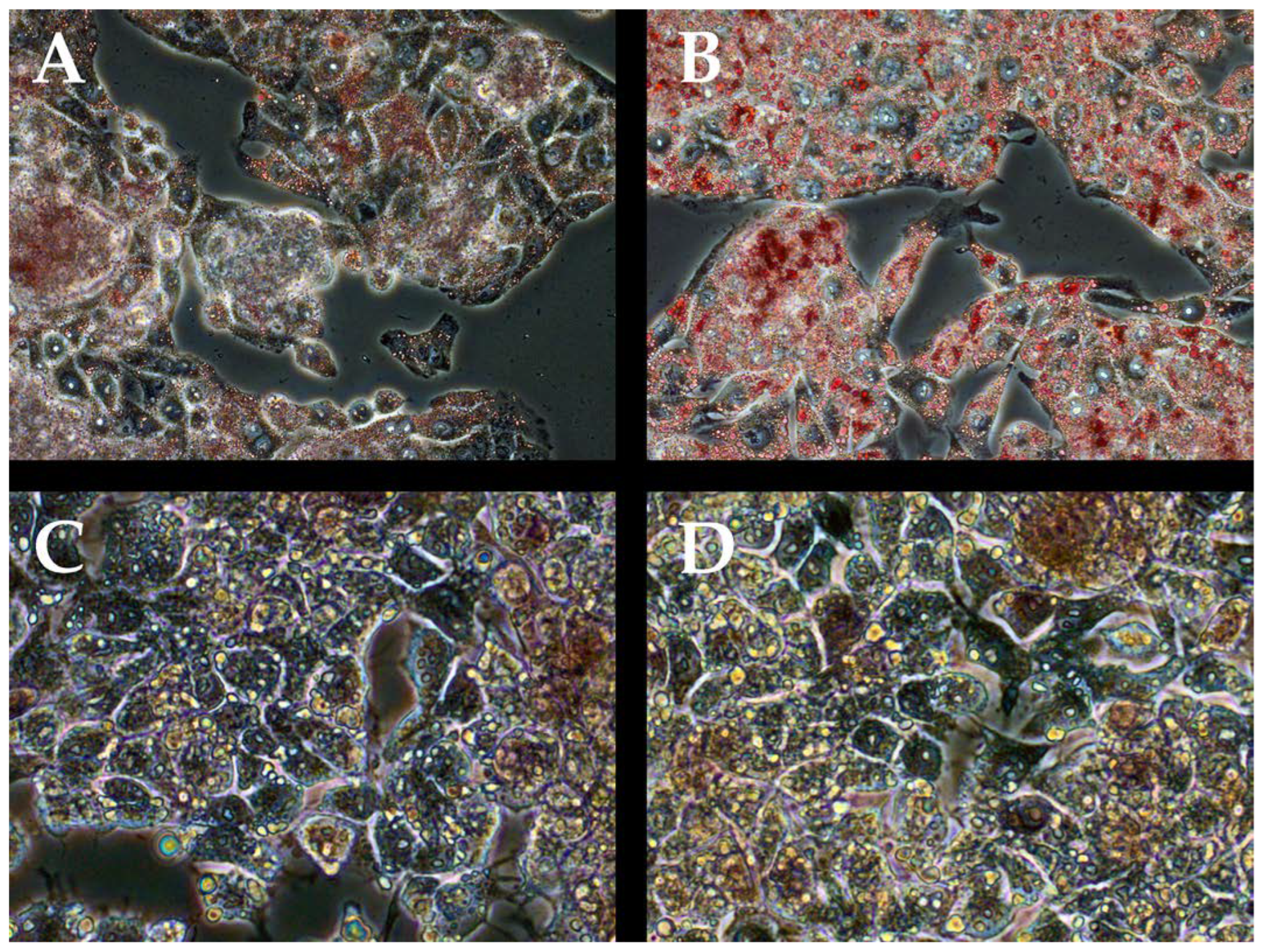
2.2. Oil Red Staining
2.3. Evaluation of the Viability of Cellular Systems
2.4. Analysis of gene expression
2.5. Western Blotting Analysis
2.6. Statistical Analysis
3. Results
3.1. Steatohepatitis Cell Cultures
3.2. IL-6 in Cell Model tBHP/Oleic
3.3. Co-addition of IL-6 and gp130 in Cell Models with tBHP/Oleic
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- CDC. Effects of Overweight and Obesity. Available online: https://www.cdc.gov/healthyweight/effects/index.html (accessed on 29 September 2022).
- Lin, G.; Xinhe, Z.; Haoyu, T.; Xing, J.; Dan, L.; Ningning, W.; Jing, S.; Xue, W.; Zilu, Z.; Yiling, L. Epidemiology and Lifestyle Survey of Non-Alcoholic Fatty Liver Disease in School-Age Children and Adolescents in Shenyang, Liaoning. BMC Pediatr. 2022, 22, 286. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global Epidemiology of Nonalcoholic Fatty Liver Disease-Meta-Analytic Assessment of Prevalence, Incidence, and Outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Spiers, J.; Brindley, J.H.; Li, W.; Alazawi, W. What’s New in Non-Alcoholic Fatty Liver Disease? Frontline Gastroenterol. 2022, 13, e102–e108. [Google Scholar] [CrossRef]
- Raza, S.; Rajak, S.; Upadhyay, A.; Tewari, A.; Anthony Sinha, R. Current Treatment Paradigms and Emerging Therapies for NAFLD/NASH. Front. Biosci. 2021, 26, 206–237. [Google Scholar] [CrossRef] [PubMed]
- Lima, K.G.; Schneider Levorse, V.G.; Rosa Garcia, M.C.; de Souza Basso, B.; Pasqualotto Costa, B.; Antunes, G.L.; Luft, C.; Haute, G.V.; Leal Xavier, L.; Donadio, M.V.F.; et al. Octyl Gallate Induces Hepatic Steatosis in HepG2 Cells through the Regulation of SREBP-1c and PPAR-Gamma Gene Expression. EXCLI J. 2020, 19, 962–971. [Google Scholar] [CrossRef]
- Kanda, T.; Goto, T.; Hirotsu, Y.; Masuzaki, R.; Moriyama, M.; Omata, M. Molecular Mechanisms: Connections between Nonalcoholic Fatty Liver Disease, Steatohepatitis and Hepatocellular Carcinoma. Int. J. Mol. Sci. 2020, 21, 1525. [Google Scholar] [CrossRef]
- Caturano, A.; Acierno, C.; Nevola, R.; Pafundi, P.C.; Galiero, R.; Rinaldi, L.; Salvatore, T.; Adinolfi, L.E.; Sasso, F.C. Non-Alcoholic Fatty Liver Disease: From Pathogenesis to Clinical Impact. Processes 2021, 9, 135. [Google Scholar] [CrossRef]
- Masarone, M.; Rosato, V.; Dallio, M.; Gravina, A.G.; Aglitti, A.; Loguercio, C.; Federico, A.; Persico, M. Role of Oxidative Stress in Pathophysiology of Nonalcoholic Fatty Liver Disease. Oxid. Med. Cell Longev. 2018, 2018, 9547613. [Google Scholar] [CrossRef]
- Cheng, Z.; Guo, S.; Copps, K.; Dong, X.; Kollipara, R.; Rodgers, J.T.; Depinho, R.A.; Puigserver, P.; White, M.F. Foxo1 Integrates Insulin Signaling with Mitochondrial Function in the Liver. Nat. Med. 2009, 15, 1307–1311. [Google Scholar] [CrossRef]
- Baker, M.J.; Tatsuta, T.; Langer, T. Quality Control of Mitochondrial Proteostasis. Cold Spring Harb. Perspect. Biol. 2011, 3, a007559. [Google Scholar] [CrossRef]
- Tilokani, L.; Nagashima, S.; Paupe, V.; Prudent, J. Mitochondrial Dynamics: Overview of Molecular Mechanisms. Essays Biochem. 2018, 62, 341–360. [Google Scholar] [CrossRef] [PubMed]
- Madan, S.; Uttekar, B.; Chowdhary, S.; Rikhy, R. Mitochondria Lead the Way: Mitochondrial Dynamics and Function in Cellular Movements in Development and Disease. Front. Cell Dev. Biol. 2021, 9, 781933. [Google Scholar] [CrossRef] [PubMed]
- Mitochondria, Fusion, Division |Learn Science at Scitable. Available online: https://www.nature.com/scitable/topicpage/mitochondrial-fusion-and-division-14264007/ (accessed on 11 October 2022).
- Iglewski, M.; Hill, J.A.; Lavandero, S.; Rothermel, B.A. Mitochondrial Fission and Autophagy in the Normal and Diseased Heart. Curr. Hypertens. Rep. 2010, 12, 418–425. [Google Scholar] [CrossRef]
- Longo, M.; Meroni, M.; Paolini, E.; Macchi, C.; Dongiovanni, P. Mitochondrial Dynamics and Nonalcoholic Fatty Liver Disease (NAFLD): New Perspectives for a Fairy-Tale Ending? Metabolism 2021, 117, 154708. [Google Scholar] [CrossRef] [PubMed]
- White, J.P.; Puppa, M.J.; Sato, S.; Gao, S.; Price, R.L.; Baynes, J.W.; Kostek, M.C.; Matesic, L.E.; Carson, J.A. IL-6 Regulation on Skeletal Muscle Mitochondrial Remodeling during Cancer Cachexia in the ApcMin/+mouse. Skelet. Muscle 2012, 2, 14. [Google Scholar] [CrossRef]
- Sánchez-Garrido, M.A.; Chico, Y.; González, R.; Ranchal, I.; González-Rubio, S.; Hidalgo, A.B.; Díaz-López, C.; Costán, G.; Padillo, F.J.; De la Mata, M.; et al. Interleukin-6 Is Associated with Liver Lipid Homeostasis but Not with Cell Death in Experimental Hepatic Steatosis. Innate Immun. 2009, 15, 337–349. [Google Scholar] [CrossRef]
- Kishimoto, T. Interleukin-6: From Basic Science to Medicine—40 Years in Immunology. Annu. Rev. Immunol. 2005, 23, 1–21. [Google Scholar] [CrossRef]
- Novick, D.; Engelmann, H.; Wallach, D.; Rubinstein, M. Soluble Cytokine Receptors Are Present in Normal Human Urine. J. Exp. Med. 1989, 170, 1409–1414. [Google Scholar] [CrossRef]
- Rose-John, S. IL-6 Trans-Signaling via the Soluble IL-6 Receptor: Importance for the pro-Inflammatory Activities of IL-6. Int. J. Biol. Sci. 2012, 8, 1237–1247. [Google Scholar] [CrossRef]
- Skuratovskaia, D.; Komar, A.; Vulf, M.; Quang, H.V.; Shunkin, E.; Volkova, L.; Gazatova, N.; Zatolokin, P.; Litvinova, L. IL-6 Reduces Mitochondrial Replication, and IL-6 Receptors Reduce Chronic Inflammation in NAFLD and Type 2 Diabetes. Int. J. Mol. Sci. 2021, 22, 1774. [Google Scholar] [CrossRef]
- Fernandes, R.O.; De Castro, A.L.; Bonetto, J.H.P.; Ortiz, V.D.; Müller, D.D.; Campos-Carraro, C.; Barbosa, S.; Neves, L.T.; Xavier, L.L.; Schenkel, P.C.; et al. Sulforaphane Effects on Postinfarction Cardiac Remodeling in Rats: Modulation of Redox-Sensitive Prosurvival and Proapoptotic Proteins. J. Nutr. Biochem. 2016, 34, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, D.-W.; Yan, H.-Y.; Wang, Z.-Y.; Zhao, S.-H.; Wang, B. Obesity Is an Independent Risk Factor for Non-Alcoholic Fatty Liver Disease: Evidence from a Meta-Analysis of 21 Cohort Studies. Obes. Rev 2016, 17, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Lambert, J.E.; Ramos-Roman, M.A.; Browning, J.D.; Parks, E.J. Increased de Novo Lipogenesis Is a Distinct Characteristic of Individuals with Nonalcoholic Fatty Liver Disease. Gastroenterology 2014, 146, 726–735. [Google Scholar] [CrossRef]
- Schmidt-Arras, D.; Rose-John, S. IL-6 Pathway in the Liver: From Physiopathology to Therapy. J. Hepatol. 2016, 64, 1403–1415. [Google Scholar] [CrossRef]
- Acierno, C.; Caturano, A.; Pafundi, P.C.; Nevola, R.; Adinolfi, L.E.; Sasso, F.C. Nonalcoholic Fatty Liver Disease and Type 2 Diabetes: Pathophysiological Mechanisms Shared between the Two Faces of the Same Coin. Explor. Med. 2020, 1, 287–306. [Google Scholar] [CrossRef]
- Fontes-Cal, T.C.M.; Mattos, R.T.; Medeiros, N.I.; Pinto, B.F.; Belchior-Bezerra, M.; Roque-Souza, B.; Dutra, W.O.; Ferrari, T.C.A.; Vidigal, P.V.T.; Faria, L.C.; et al. Crosstalk Between Plasma Cytokines, Inflammation, and Liver Damage as a New Strategy to Monitoring NAFLD Progression. Front. Immunol. 2021, 12, 708959. [Google Scholar] [CrossRef]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in Inflammation, Immunity, and Disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016295. [Google Scholar] [CrossRef]
- Lorenzen, I.; Shang, W.; Perbandt, M.; Petoukhov, M.V.; Svergun, D.I.; Waetzig, G.H.; Rose-John, S.; Hilgenfeld, R.; Grötzinger, J. The Structure of the Unliganded Extracellular Domain of the Interleukin-6 Signal Transducer Gp130 in Solution. Eur. J. Cell Biol. 2011, 90, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Fleischman, A.; Shoelson, S.E.; Bernier, R.; Goldfine, A.B. Salsalate Improves Glycemia and Inflammatory Parameters in Obese Young Adults. Diabetes Care 2008, 31, 289–294. [Google Scholar] [CrossRef]
- Sun, S.-C. The Non-Canonical NF-ΚB Pathway in Immunity and Inflammation. Nat. Rev. Immunol. 2017, 17, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Sivitz, W.I.; Yorek, M.A. Mitochondrial Dysfunction in Diabetes: From Molecular Mechanisms to Functional Significance and Therapeutic Opportunities. Antioxid. Redox. Signal 2010, 12, 537–577. [Google Scholar] [CrossRef] [PubMed]
- El-Hattab, A.W.; Craigen, W.J.; Scaglia, F. Mitochondrial DNA Maintenance Defects. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 1539–1555. [Google Scholar] [CrossRef] [PubMed]
- Litvinova, L.; Zatolokin, P.; Vulf, M.; Mazunin, I.; Skuratovskaia, D. The Relationship between the MtDNA Copy Number in Insulin-Dependent Tissues and Markers of Endothelial Dysfunction and Inflammation in Obese Patients. BMC Med. Genomics 2019, 12, 41. [Google Scholar] [CrossRef] [PubMed]
- Yoneda, M.; Miyatake, T.; Attardi, G. Complementation of Mutant and Wild-Type Human Mitochondrial DNAs Coexisting since the Mutation Event and Lack of Complementation of DNAs Introduced Separately into a Cell within Distinct Organelles. Mol. Cell Biol. 1994, 14, 2699–2712. [Google Scholar] [CrossRef]
- Schumacher, N.; Yan, K.; Gandraß, M.; Müller, M.; Krisp, C.; Häsler, R.; Carambia, A.; Nofer, J.-R.; Bernardes, J.P.; Khouja, M.; et al. Cell-Autonomous Hepatocyte-Specific GP130 Signaling Is Sufficient to Trigger a Robust Innate Immune Response in Mice. J. Hepatol. 2021, 74, 407–418. [Google Scholar] [CrossRef]
- Su, H.; Lei, C.-T.; Zhang, C. Interleukin-6 Signaling Pathway and Its Role in Kidney Disease: An Update. Front. Immunol. 2017, 8, 405. [Google Scholar] [CrossRef]
- Skuratovskaia, D.A.; Sofronova, J.K.; Zatolokin, P.A.; Popadin, K.Y.; Vasilenko, M.A.; Litvinova, L.S.; Mazunin, I.O. Additional Evidence of the Link between MtDNA Copy Number and the Body Mass Index. Mitochondrial DNA Part A 2018, 29, 1240–1244. [Google Scholar] [CrossRef]
- Shunkina Skuratovskaia, D.; Komar, A.; Vulf, M.; Quang, H.V.; Shunkin, E.; Kirienkova, E.; Dakchnevich, A.; Malkov, D.; Zatolokin, P.; Litvinova, L. Tumor Necrosis Receptor Superfamily Interact with Fusion and Fission of Mitochondria of Adipose Tissue in Obese Patients without Type 2 Diabetes. Biomedicines 2021, 9, 1260. [Google Scholar] [CrossRef] [PubMed]
- Kozhukhar, N.; Alexeyev, M.F. Limited Predictive Value of TFAM in Mitochondrial Biogenesis. Mitochondrion 2019, 49, 156–165. [Google Scholar] [CrossRef]
- Koh, J.-H.; Johnson, M.L.; Dasari, S.; LeBrasseur, N.K.; Vuckovic, I.; Henderson, G.C.; Cooper, S.A.; Manjunatha, S.; Ruegsegger, G.N.; Shulman, G.I.; et al. TFAM Enhances Fat Oxidation and Attenuates High-Fat Diet-Induced Insulin Resistance in Skeletal Muscle. Diabetes 2019, 68, 1552–1564. [Google Scholar] [CrossRef]
- Flynn, J.M.; Melov, S. SOD2 in Mitochondrial Dysfunction and Neurodegeneration. Free Radic. Biol. Med. 2013, 62, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Palma, F.R.; He, C.; Danes, J.M.; Paviani, V.; Coelho, D.R.; Gantner, B.N.; Bonini, M.G. Mitochondrial Superoxide Dismutase: What the Established, the Intriguing, and the Novel Reveal About a Key Cellular Redox Switch. Antioxid. Redox. Signal 2020, 32, 701–714. [Google Scholar] [CrossRef] [PubMed]
- Ekoue, D.N.; He, C.; Diamond, A.M.; Bonini, M.G. Manganese Superoxide Dismutase and Glutathione Peroxidase-1 Contribute to the Rise and Fall of Mitochondrial Reactive Oxygen Species Which Drive Oncogenesis. Biochim. Et Biophys. Acta (BBA)-Bioenerg. 2017, 1858, 628–632. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Lu, Y.; Saredy, J.; Wang, X.; Drummer Iv, C.; Shao, Y.; Saaoud, F.; Xu, K.; Liu, M.; Yang, W.Y.; et al. ROS Systems Are a New Integrated Network for Sensing Homeostasis and Alarming Stresses in Organelle Metabolic Processes. Redox. Biol. 2020, 37, 101696. [Google Scholar] [CrossRef]
- Koh, J.-H.; Kim, Y.-W.; Seo, D.-Y.; Sohn, T.-S. Mitochondrial TFAM as a Signaling Regulator between Cellular Organelles: A Perspective on Metabolic Diseases. Diabetes Metab. J. 2021, 45, 853–865. [Google Scholar] [CrossRef]
- Reeg, S.; Jung, T.; Castro, J.P.; Davies, K.J.A.; Henze, A.; Grune, T. The Molecular Chaperone Hsp70 Promotes the Proteolytic Removal of Oxidatively Damaged Proteins by the Proteasome. Free Radic. Biol. Med. 2016, 99, 153–166. [Google Scholar] [CrossRef]








| Gene | Sequence |
|---|---|
| NF-kB1 | Forward: 5′-CAGGAAGATGTGGTGGACCA-3′ Reverse: 5′-AGGCCCGGCTCTGTCTAGTG-3′ Probe: 5′-FAM-GGCTGGAGGAGGCGGGCGTCTAAA-BHQ1- 3′ |
| HSF1 | Forward: 5′-AGAGAGAGACGACACGGAGTT-3′ Reverse: 5′-CTGTCCTGGCGGATCTTTATGT-3′ Probe: 5′-FAM-AGAGGAAAGTGACCAGTGTGTCCACCCT-BHQ1-3′ |
| SOD2 | Forward: 5′-CGTGGCTGTGGTGGCTTC-3′ Reverse: 5′-CGTGGTGCTTGCTGTGGT-3′ Probe: 5′-FAM-CCTCCCCGACCTGCCCTACGACTA-BHQ1-3′ |
| TFAM | Forward: 5′-CGCTCCCCCTTCAGTTTTGT-3′ Reverse: 5′-TACCTGCCACTCCGCCCTAT-3′ Probe: 5′-FAM-CGAGGTGGTTTTCATCTGTCTTGGCA-BHQ1-3′ |
| HSP70 | Forward: 5′-CTTCGTGGAGGAGTTCAAGAGAAA-3′ Reverse: 5′-TAGAAGTCGATGCCCTCAAACAG-3′ Probe: 5′-FAM-AAGGACATCAGCCAGAACAAGCGAGCC-BHQ1-3′ |
| BAX | Forward: 5′-AGTAACATGGAGCTGCAGAGGA-3′ Reverse: 5′-CCAGTTGAAGTTGCCGTCAGAA-3′ Probe: 5′-FAM-GATTGCCGCCGTGGACACAGACT-BHQ1-3′ |
| MFN2 | Forward: 5′-CCAGCGTCCCATCCCTCT -3′ Reverse: 5′-TCCACACCACTCCTCCAACA-3′ Probe: 5′ ACGGGCTCGCTCACCCAGGAG 3′ |
| DNM1L | Forward: 5′-TCTGGAGGTGGTGGGGTTG-3′ Reverse: 5′-TGGGTTTTGATTTTTCTTCTGCTAAT-3′ Probe: 5′-FAM-ACCAACCACAGGCAACTGGAGAGGA-BHQ1-3′ |
| BCL2L1 | Forward: 5′-CCACCAGGAGAACCACTACA-3′ Reverse: 5′-CAGCCACAAACCCTTCCATA-3′ Probe: 5′-FAM-CACACCTCAGTTCCCTTGGCCTCA-BHQ1-3′ |
| MAP1LC3B | Forward: 5′-TGCCTGTGTTGTTACGGAAAG-3′ Reverse: 5′-AGAAGGGAGTGTGTCTGAATGT-3′ Probe: 5′-FAM-ACTCTGGAGTACAGCGGGAGAAACACA-BHQ1-3′ |
| SQSTM1 | Forward: 5′-CCTGTCCCTGAAAGAGAAGATG-3′ Reverse: 5′-CAGAAAGTGTCAGAACCAGAGG-3′ Probe: 5′-FAM-CATGTGTAAGAACAATGCCAGGGCC-BHQ1-3′ |
| RPLPO | Forward: 5′-GGCGACCTGGAAGTCCAACT-3′ Reverse: 5′-CCATCAGCACCACAGCCTTC-3′ Probe: 5′-Bgl635-ATCTGCTGCATCTGCTTGGAGCCCA-BHQ2-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shunkina, D.; Dakhnevich, A.; Shunkin, E.; Khaziakhmatova, O.; Shupletsova, V.; Vulf, M.; Komar, A.; Kirienkova, E.; Litvinova, L. gp130 Activates Mitochondrial Dynamics for Hepatocyte Survival in a Model of Steatohepatitis. Biomedicines 2023, 11, 396. https://doi.org/10.3390/biomedicines11020396
Shunkina D, Dakhnevich A, Shunkin E, Khaziakhmatova O, Shupletsova V, Vulf M, Komar A, Kirienkova E, Litvinova L. gp130 Activates Mitochondrial Dynamics for Hepatocyte Survival in a Model of Steatohepatitis. Biomedicines. 2023; 11(2):396. https://doi.org/10.3390/biomedicines11020396
Chicago/Turabian StyleShunkina, Daria, Anastasia Dakhnevich, Egor Shunkin, Olga Khaziakhmatova, Valeria Shupletsova, Maria Vulf, Alexandra Komar, Elena Kirienkova, and Larisa Litvinova. 2023. "gp130 Activates Mitochondrial Dynamics for Hepatocyte Survival in a Model of Steatohepatitis" Biomedicines 11, no. 2: 396. https://doi.org/10.3390/biomedicines11020396
APA StyleShunkina, D., Dakhnevich, A., Shunkin, E., Khaziakhmatova, O., Shupletsova, V., Vulf, M., Komar, A., Kirienkova, E., & Litvinova, L. (2023). gp130 Activates Mitochondrial Dynamics for Hepatocyte Survival in a Model of Steatohepatitis. Biomedicines, 11(2), 396. https://doi.org/10.3390/biomedicines11020396



.JPG)


