Transient Expression in HEK-293 Cells in Suspension Culture as a Rapid and Powerful Tool: SARS-CoV-2 N and Chimeric SARS-CoV-2N-CD154 Proteins as a Case Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biological Reagents
2.2. Mammalian Cell Lines and Cell Culture Conditions
2.3. SARS-CoV-2 N Plasmid Construction and Lentivirus Production
2.4. Transduction of HEK-293 Cells and Generation of SARS-CoV-2 N-Protein-Expressing Cell Pools
2.5. Assessment of SARS-CoV-2 N-Protein Expression and Degradation in the Cell Culture Supernatant of Recombinant HEK-293 Cells Cultured in Adherent Conditions
2.6. Production of the SARS-CoV-2 N and N-CD Proteins in Cell Suspension Culture
2.7. Cell Cloning of the HEK-293-N-CD-50 and HEK-293-N-CD-100 Cell Pools via Limiting Dilution
2.8. Batch Culture of the N-CD-Expressing Cell Clones Cultured in Suspension Culture and Serum-Free Medium
2.9. Simulation of Perfusion Culture of the N-CD-Expressing Cell Clones Cultured in Suspension Culture and Serum-Free Medium
2.10. Protein Purification
2.11. Protein Quantification with ELISA
2.12. SDS-PAGE and Western Blot
2.13. Statistical Analysis
3. Results
3.1. Production of the SARS-CoV-2 N Protein in Adherent HEK-293 Cells
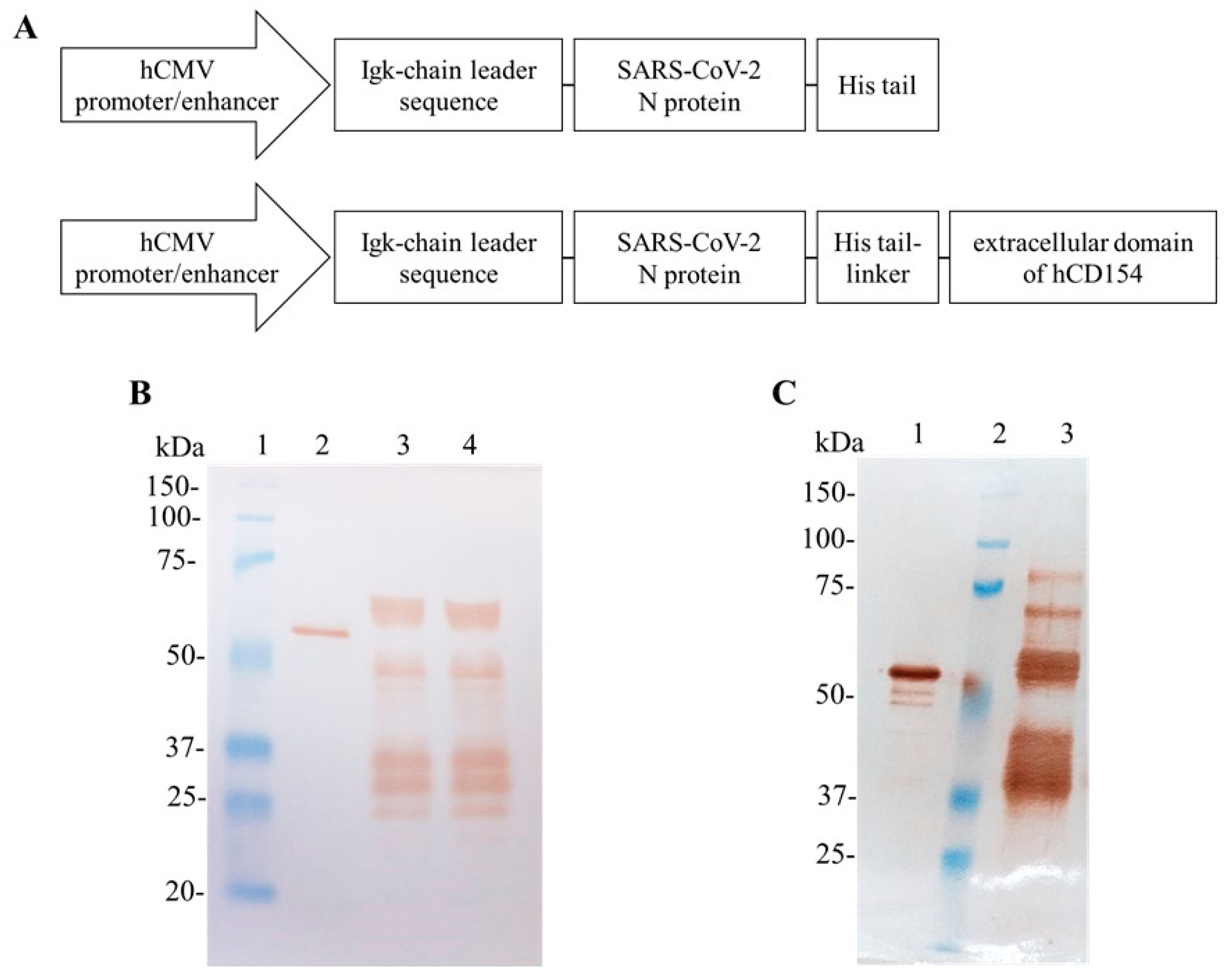
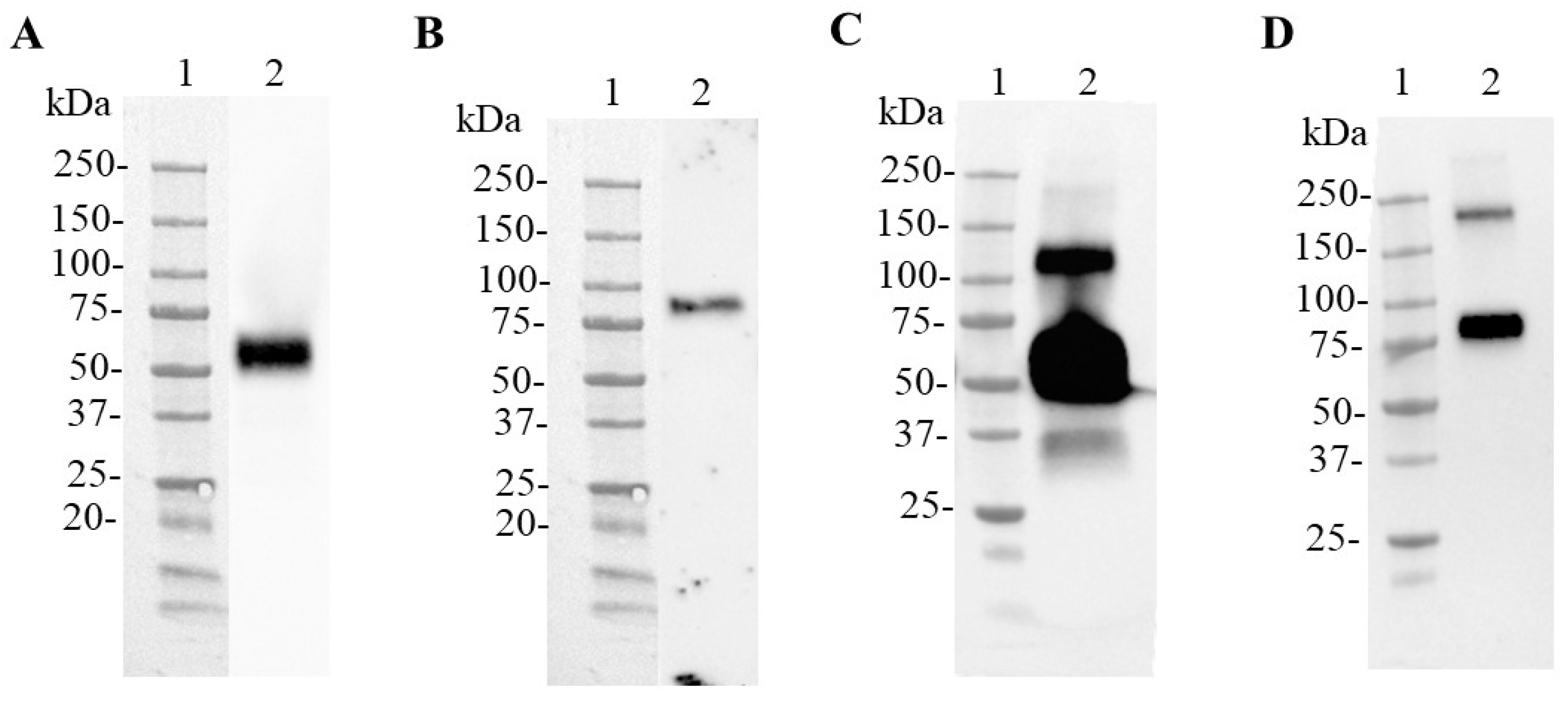
3.2. Production of SARS-CoV-2 N and N-CD Proteins through Transient Expression in Suspension Conditions and Serum-Free Medium Using Shake Flasks
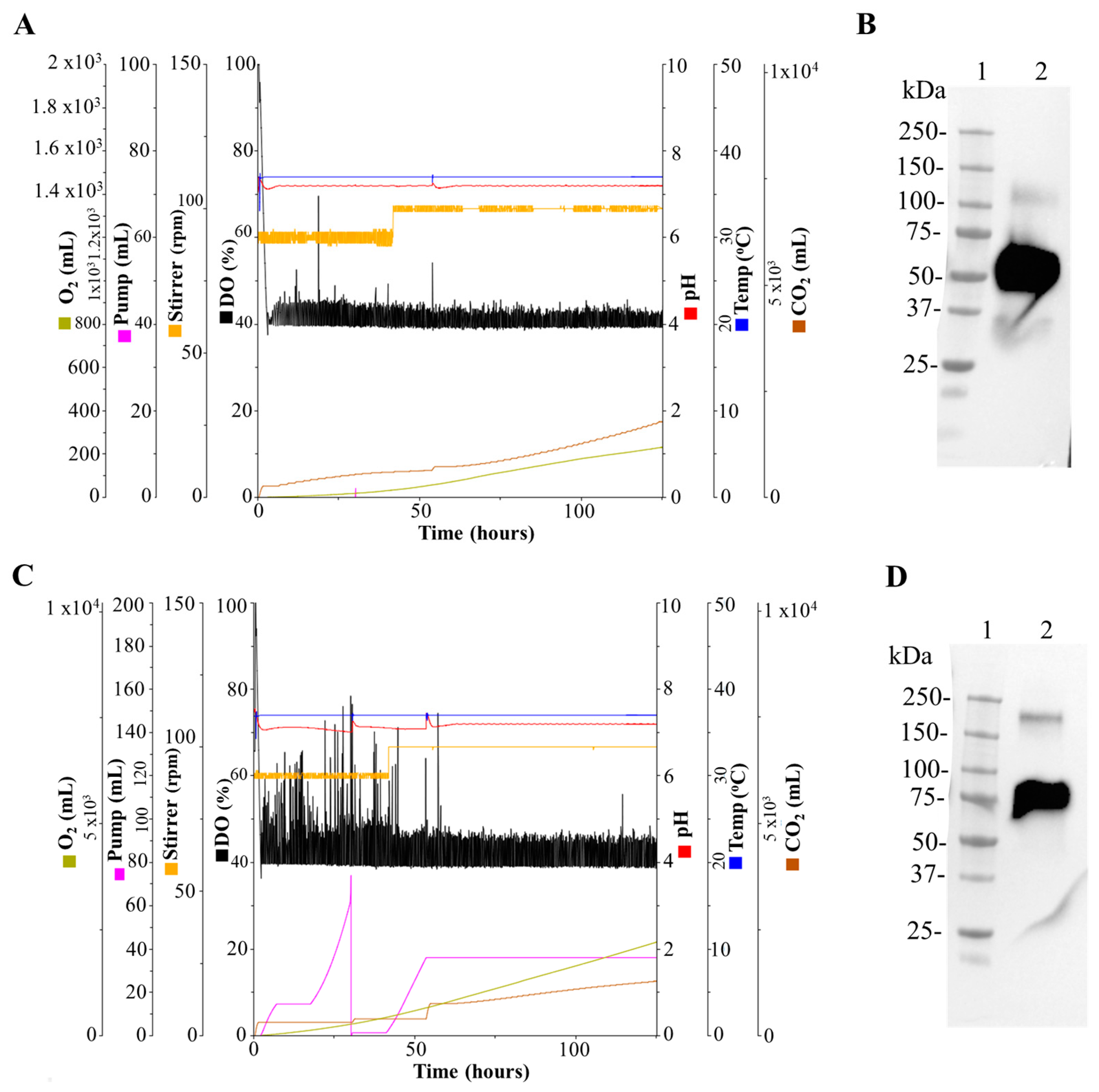
3.3. Production of the SARS-CoV-2 N and N-CD Proteins in 1 L and 3 L Stirred-Tank Bioreactors
3.4. Purification of the SARS-CoV-2 N and N-CD Proteins Produced in Suspension Culture and Serum-Free Medium
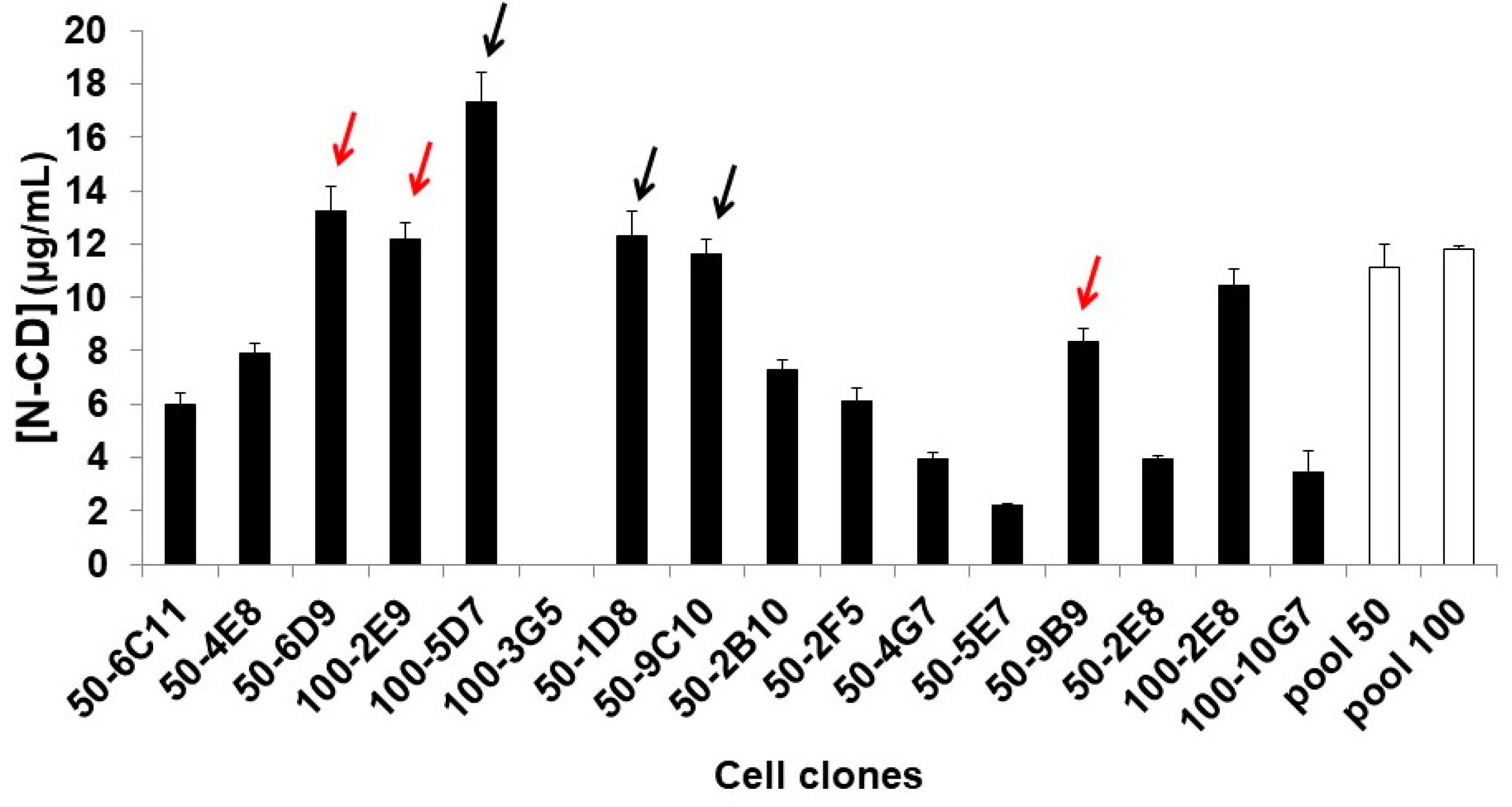
3.5. Obtaining N-CD-Expressing HEK-293 Cell Clones and Adaptation to Serum-Free Medium and Suspension Culture Conditions
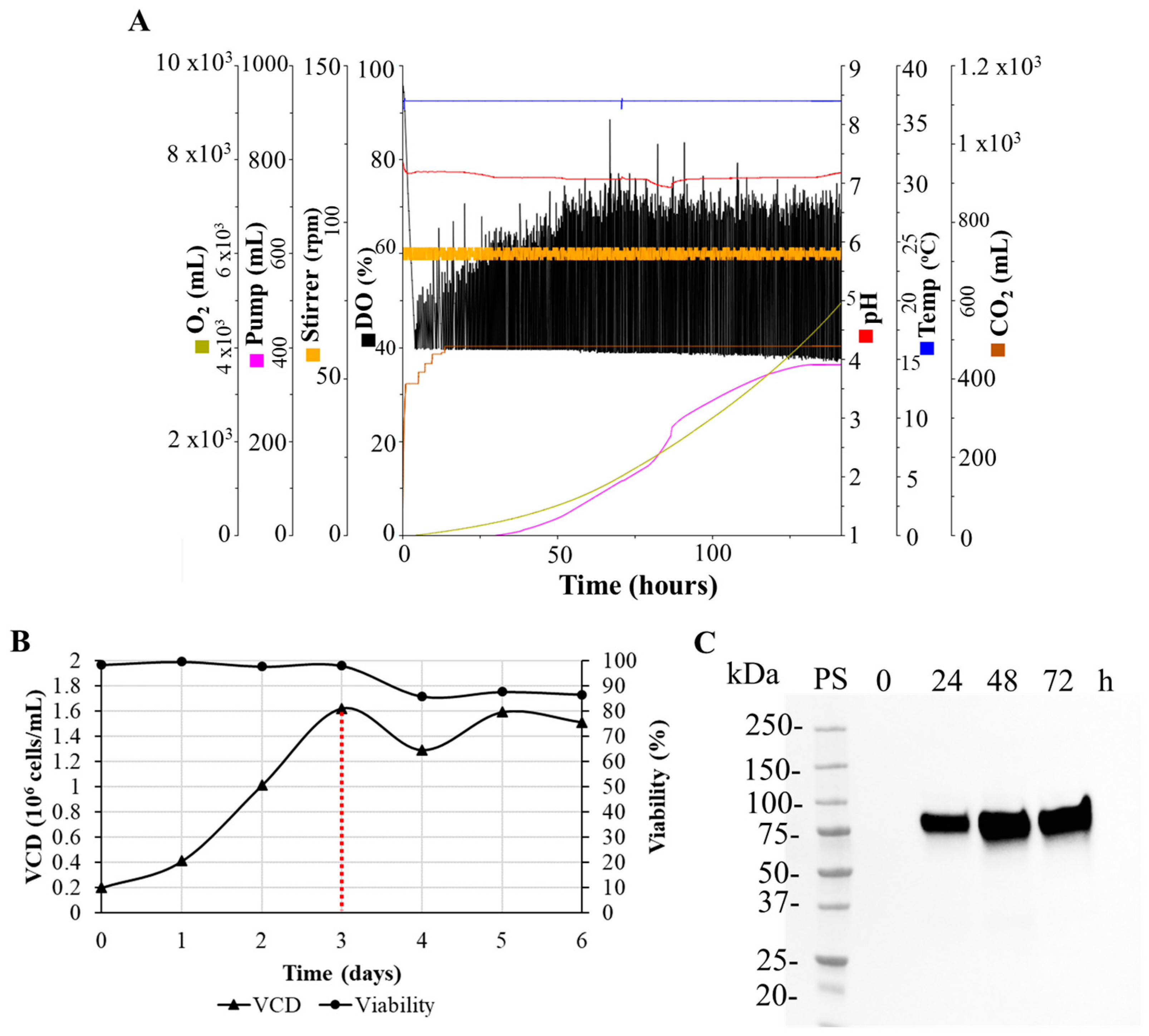
3.6. Batch Culture of the N-CD-Expressing HEK-293 Cell Clone 100-2E9 and Simulation of Perfusion Conditions
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McLean, G.; Kamil, J.; Lee, B.; Moore, P.; Schulz, T.F.; Muik, A.; Sahin, U.; Türeci, Ö.; Pather, S. The impact of evolving SARS-CoV-2 mutations and variants on COVID-19 vaccines. MBio 2022, 13, e02979-21. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; Yang, Y.; Du, L. Advances in SARS-CoV-2 receptor-binding domain-based COVID-19 vaccines. Expert Rev. Vaccines 2023, 22, 422–439. [Google Scholar] [CrossRef]
- Ahlén, G.; Frelin, L.; Nikouyan, N.; Weber, F.; Höglund, U.; Larsson, O.; Westman, M.; Tuvesson, O.; Gidlund, E.-K.; Cadossi, M. The SARS-CoV-2 N protein is a good component in a vaccine. J. Virol. 2020, 94, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Dutta, N.K.; Mazumdar, K.; Gordy, J.T. The nucleocapsid protein of SARS–CoV-2: A target for vaccine development. J. Virol. 2020, 94, 10–1128. [Google Scholar] [CrossRef]
- Bai, Z.; Cao, Y.; Liu, W.; Li, J. The SARS-CoV-2 nucleocapsid protein and its role in viral structure, biological functions, and a potential target for drug or vaccine mitigation. Viruses 2021, 13, 1115. [Google Scholar] [CrossRef] [PubMed]
- Harris, P.E.; Brasel, T.; Massey, C.; Herst, C.; Burkholz, S.; Lloyd, P.; Blankenberg, T.; Bey, T.M.; Carback, R.; Hodge, T. A synthetic peptide CTL vaccine targeting nucleocapsid confers protection from SARS-CoV-2 challenge in rhesus macaques. Vaccines 2021, 9, 520. [Google Scholar] [CrossRef]
- He, J.; Huang, J.R.; Zhang, Y.L.; Zhang, J. SARS-CoV-2 nucleocapsid protein intranasal inoculation induces local and systemic T cell responses in mice. J. Med. Virol. 2021, 93, 1923–1925. [Google Scholar] [CrossRef]
- Dangi, T.; Sanchez, S.; Class, J.; Richner, M.; Visvabharathy, L.; Chung, Y.R.; Bentley, K.; Stanton, R.J.; Koralnik, I.J.; Richner, J.M. Improved control of SARS-CoV-2 by treatment with a nucleocapsid-specific monoclonal antibody. J. Clin. Investig. 2022, 132, e162282. [Google Scholar] [CrossRef]
- López-Muñoz, A.D.; Kosik, I.; Holly, J.; Yewdell, J.W. Cell surface SARS-CoV-2 nucleocapsid protein modulates innate and adaptive immunity. Sci. Adv. 2022, 8, eabp9770. [Google Scholar] [CrossRef]
- Díez, J.M.; Romero, C.; Cruz, M.; Vandeberg, P.; Merritt, W.K.; Pradenas, E.; Trinité, B.; Blanco, J.; Clotet, B.; Willis, T. Anti-severe acute respiratory syndrome coronavirus 2 hyperimmune immunoglobulin demonstrates potent neutralization and antibody-dependent cellular cytotoxicity and phagocytosis through N and S proteins. J. Infect. Dis. 2022, 225, 938–946. [Google Scholar] [CrossRef]
- Lao, T.; Avalos, I.; Rodríguez, E.M.; Zamora, Y.; Rodriguez, A.; Ramón, A.; Alvarez, Y.; Cabrales, A.; Andújar, I.; González, L.J.; et al. Production and characterization of a chimeric antigen, based on nucleocapsid of SARS-CoV-2 fused to the extracellular domain of human CD154 in HEK-293 cells as a vaccine candidate against COVID-19. PLoS ONE 2023, 18, e0288006. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.L.; Goh, J.B.; Srinivasan, H.; Liu, K.I.; Gowher, A.; Shanmugam, R.; Lim, H.L.; Choo, M.; Tang, W.Q.; Tan, A.H.-M. A human expression system based on HEK293 for the stable production of recombinant erythropoietin. Sci. Rep. 2019, 9, 16768. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.; Pete, E.S.; Bruheim, P. The impact of serum-free culture on HEK293 cells: From the establishment of suspension and adherent serum-free adaptation cultures to the investigation of growth and metabolic profiles. Front. Bioeng. Biotechnol. 2022, 10, 964397. [Google Scholar] [CrossRef] [PubMed]
- Shimomura, T.; Ochiai, M.; Kondo, J.; Morimoto, Y. A novel protease obtained from FBS-containing culture supernatant, that processes single chain form hepatocyte growth factor to two chain form in serum-free culture. Cytotechnology 1992, 8, 219–229. [Google Scholar] [CrossRef]
- Qiu, W.Q.; Borth, W.; Ye, Z.; Haass, C.; Teplow, D.B.; Selkoe, D.J. Degradation of amyloid β-protein by a serine protease-α2-macroglobulin complex. J. Biol. Chem. 1996, 271, 8443–8451. [Google Scholar] [CrossRef]
- LeFloch, F.; Tessier, B.; Chenuet, S.; Guillaume, J.-M.; Cans, P.; Goergen, J.-L.; Marc, A. Related effects of cell adaptation to serum-free conditions on murine EPO production and glycosylation by CHO cells. Cytotechnology 2006, 52, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Hua, J.; Wei, Y.; Zhang, Y.; Xu, H.; Ge, J.; Liu, M.; Wang, Y.; Shi, Y.; Hou, L.; Jiang, H. Purification, Adaptation process of engineered cell line FCHO/IL-24 stably secreted rhIL-24 in serum-free suspension culture. Protein Expr. Purif. 2022, 199, 106154. [Google Scholar] [CrossRef]
- Farnós, O.; Venereo-Sánchez, A.; Xu, X.; Chan, C.; Dash, S.; Chaabane, H.; Sauvageau, J.; Brahimi, F.; Saragovi, U.; Leclerc, D. Rapid high-yield production of functional sars-cov-2 receptor binding domain by viral and non-viral transient expression for pre-clinical evaluation. Vaccines 2020, 8, 654. [Google Scholar] [CrossRef]
- Hartley, G.E.; Edwards, E.S.; Aui, P.M.; Varese, N.; Stojanovic, S.; McMahon, J.; Peleg, A.Y.; Boo, I.; Drummer, H.E.; Hogarth, P.M. Rapid generation of durable B cell memory to SARS-CoV-2 spike and nucleocapsid proteins in COVID-19 and convalescence. Sci. Immunol. 2020, 5, eabf8891. [Google Scholar] [CrossRef]
- Garcia-Cordero, J.; Mendoza-Ramirez, J.; Fernandez-Benavides, D.; Roa-Velazquez, D.; Filisola-Villasenor, J.; Martinez-Frias, S.P.; Sanchez-Salguero, E.S.; Miguel-Rodriguez, C.E.; Maravillas Montero, J.L.; Torres-Ruiz, J.J. Recombinant protein expression and purification of N, S1, and RBD of SARS-CoV-2 from mammalian cells and their potential applications. Diagnostics 2021, 11, 1808. [Google Scholar] [CrossRef]
- Green, E.A.; Hamaker, N.K.; Lee, K.H. Comparison of vector elements and process conditions in transient and stable suspension HEK293 platforms using SARS-CoV-2 receptor binding domain as a model protein. BMC Biotechnol. 2023, 23, 7. [Google Scholar] [CrossRef] [PubMed]
- Bláha, J.; Pachl, P.; Novák, P.; Vaněk, O. Expression and purification of soluble and stable ectodomain of natural killer cell receptor LLT1 through high-density transfection of suspension adapted HEK293S GnTI− cells. Protein Expr. Purif. 2015, 109, 7–13. [Google Scholar] [CrossRef]
- Liu, H.; Zou, X.; Li, T.; Wang, X.; Yuan, W.; Chen, Y.; Han, W. Enhanced production of secretory glycoprotein VSTM1-v2 with mouse IgGκ signal peptide in optimized HEK293F transient transfection. J. Biosci. Bioeng. 2016, 121, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Chen, W.; Ge, L.; Jiang, W.; Tang, B.; Zhang, Q.; Xu, X.; Wang, C.; Cao, L.; Guo, H. Transient expression of Fc-fused human glycoprotein 130 in Expi293F suspension cells. Protein Expr. Purif. 2016, 124, 41–47. [Google Scholar] [CrossRef]
- Fang, X.T.; Sehlin, D.; Lannfelt, L.; Syvänen, S.; Hultqvist, G. Efficient and inexpensive transient expression of multispecific multivalent antibodies in Expi293 cells. Biol. Proced. Online 2017, 19, 11. [Google Scholar] [CrossRef] [PubMed]
- Côté, J.; Garnier, A.; Massie, B.; Kamen, A. Serum-free production of recombinant proteins and adenoviral vectors by 293SF-3F6 cells. Biotechnol. Bioeng. 1998, 59, 567–575. [Google Scholar] [CrossRef]
- Lao González, T.; Ávalos Olivera, I.; Rodríguez-Mallon, A.J.V.D.M.; Protocols, V.V. Mammalian Cell Culture as a Platform for Veterinary Vaccines. In Vaccine Design; Springer: Berlin/Heidelberg, Germany, 2022; pp. 37–62. [Google Scholar]
- Ávalos, I.; Lao, T.; Rodríguez, E.M.; Zamora, Y.; Rodríguez, A.; Ramón, A.; Lemos, G.; Cabrales, A.; Bequet-Romero, M.; Casillas, D. Chimeric antigen by the fusion of SARS-CoV-2 receptor binding domain with the extracellular domain of human CD154: A promising improved vaccine candidate. Vaccines 2022, 10, 897. [Google Scholar] [CrossRef]
- Bae, D.H.; Marino, M.; Iaffaldano, B.; Fenstermaker, S.; Afione, S.; Argaw, T.; McCright, J.; Kwilas, A.; Chiorini, J.A.; Timmons, A.E.; et al. Design and testing of vector-producing HEK293T cells bearing a genomic deletion of the SV40 T antigen coding region. Mol. Ther.-Methods Clin. Dev. 2020, 18, 631–638. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Bruni, M.; Cecatiello, V.; Diaz-Basabe, A.; Lattanzi, G.; Mileti, E.; Monzani, S.; Pirovano, L.; Rizzelli, F.; Visintin, C.; Bonizzi, G. Persistence of anti-SARS-CoV-2 antibodies in non-hospitalized COVID-19 convalescent health care workers. J. Clin. Med. 2020, 9, 3188. [Google Scholar] [CrossRef]
- Hoste, A.C.; Venteo, A.; Fresco-Taboada, A.; Tapia, I.; Monedero, A.; López, L.; Jebbink, M.F.; Pérez-Ramírez, E.; Jimenez-Clavero, M.A.; Almonacid, M. Two serological approaches for detection of antibodies to SARS-CoV-2 in different scenarios: A screening tool and a point-of-care test. Diagn. Microbiol. Infect. Dis. 2020, 98, 115167. [Google Scholar] [CrossRef]
- Zeng, W.; Liu, G.; Ma, H.; Zhao, D.; Yang, Y.; Liu, M.; Mohammed, A.; Zhao, C.; Yang, Y.; Xie, J. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020, 527, 618–623. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zheng, B.; Gao, X.; Zhang, L.; Pan, H.; Qiao, Y.; Suo, G.; Zhu, F. Development of patient-derived human monoclonal antibodies against nucleocapsid protein of severe acute respiratory syndrome coronavirus 2 for coronavirus disease 2019 diagnosis. Front. Immunol. 2020, 11, 595970. [Google Scholar] [CrossRef]
- Di, D.; Dileepan, M.; Ahmed, S.; Liang, Y.; Ly, H. Recombinant SARS-CoV-2 nucleocapsid protein: Expression, purification, and its biochemical characterization and utility in serological assay development to assess immunological responses to SARS-CoV-2 infection. Pathogens 2021, 10, 1039. [Google Scholar] [CrossRef] [PubMed]
- Djukic, T.; Mladenovic, M.; Stanic-Vucinic, D.; Radosavljevic, J.; Smiljanic, K.; Sabljic, L.; Devic, M.; Cujic, D.; Vasovic, T.; Simovic, A. Expression, purification and immunological characterization of recombinant nucleocapsid protein fragment from SARS-CoV-2. Virology 2021, 557, 15–22. [Google Scholar] [CrossRef]
- Li, G.; Li, W.; Fang, X.; Song, X.; Teng, S.; Ren, Z.; Hu, D.; Zhou, S.; Wu, G.; Li, K. Expression and purification of recombinant SARS-CoV-2 nucleocapsid protein in inclusion bodies and its application in serological detection. Protein Expr. Purif. 2021, 186, 105908. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.-M.; Ko, S.-H.; Chen, W.-Y.; Chang, Y.-L.; Lin, H.-T.; Wu, H.-C. Monoclonal antibodies against nucleocapsid protein of SARS-CoV-2 variants for detection of COVID-19. Int. J. Mol. Sci. 2021, 22, 12412. [Google Scholar] [CrossRef]
- Tariq, M.; Hur, J.; Seo, J.-W.; Kim, D.Y.; Yun, N.R.; Lee, Y.M.; Bang, M.-S.; Hwang, S.Y.; Kim, C.-M.; Lee, J.-H. Usefulness of ELISA using total antibody against plant-expressed recombinant nucleocapsid protein of SARS-CoV-2. Microbiol. Spectr. 2021, 9, e00672-21. [Google Scholar] [CrossRef]
- Terry, J.S.; Anderson, L.B.; Scherman, M.S.; McAlister, C.E.; Perera, R.; Schountz, T.; Geiss, B.J. Development of a SARS-CoV-2 nucleocapsid specific monoclonal antibody. Virology 2021, 558, 28–37. [Google Scholar] [CrossRef]
- Yang, L.; Xu, Q.; Yang, B.; Li, J.; Dong, R.; Da, J.; Ye, Z.; Xu, Y.; Zhou, H.; Zhang, X. IgG antibody titers against SARS-CoV-2 nucleocapsid protein correlate with the severity of COVID-19 patients. BMC Microbiol. 2021, 21, 351. [Google Scholar] [CrossRef]
- Yue, L.; Cao, H.; Xie, T.; Long, R.; Li, H.; Yang, T.; Yan, M.; Xie, Z. N-terminally truncated nucleocapsid protein of SARS-CoV-2 as a better serological marker than whole nucleocapsid protein in evaluating the immunogenicity of inactivated SARS-CoV-2. J. Med. Virol. 2021, 93, 1732–1738. [Google Scholar] [CrossRef] [PubMed]
- Chura-Chambi, R.M.; Prieto-da-Silva, A.R.D.B.; Di Lela, M.M.; Oliveira, J.E.; Abreu, P.E.A.; Meireles, L.R.; de Andrade Junior, H.F.; Morganti, L. High level SARS-CoV-2 nucleocapsid refolding using mild condition for inclusion bodies solubilization: Application of high pressure at pH 9.0. PLoS ONE 2022, 17, e0262591. [Google Scholar] [CrossRef]
- Feng, W.; Xiang, Y.; Wu, L.; Chen, Z.; Li, Q.; Chen, J.; Guo, Y.; Xia, D.; Chen, N.; Zhang, L. Nucleocapsid protein of SARS-CoV-2 is a potential target for developing new generation of vaccine. J. Clin. Lab. Anal. 2022, 36, e24479. [Google Scholar] [CrossRef] [PubMed]
- Humbert, M.V.; Opurum, P.C.; Brendish, N.J.; Poole, S.; He, P.; Katis, I.; Quaye, J.; Bediako, Y.; Duriez, P.J.; Eason, R.W. A SARS-CoV-2 nucleocapsid ELISA represents a low-cost alternative to lateral flow testing for community screening in LMI countries. J. Infect. 2022, 84, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Khan, W.H.; Khan, N.; Mishra, A.; Gupta, S.; Bansode, V.; Mehta, D.; Bhambure, R.; Ansari, M.A.; Das, S.; Rathore, A.S. Dimerization of SARS-CoV-2 nucleocapsid protein affects sensitivity of ELISA based diagnostics of COVID-19. Int. J. Biol. Macromol. 2022, 200, 428–437. [Google Scholar] [CrossRef]
- Luo, J.; Brakel, A.; Krizsan, A.; Ludwig, T.; Mötzing, M.; Volke, D.; Lakowa, N.; Grünewald, T.; Lehmann, C.; Wolf, J. Sensitive and specific serological ELISA for the detection of SARS-CoV-2 infections. Virol. J. 2022, 19, 50. [Google Scholar] [CrossRef]
- Xie, C.; Ding, H.; Ding, J.; Xue, Y.; Lu, S.; Lv, H. Preparation of highly specific monoclonal antibodies against SARS-CoV-2 nucleocapsid protein and the preliminary development of antigen detection test strips. J. Med. Virol. 2022, 94, 1633–1640. [Google Scholar] [CrossRef]
- Tapela, K.; Opurum, P.C.; Nuokpem, F.Y.; Tetteh, B.; Siaw, G.K.; Humbert, M.V.; Tawiah-Eshun, S.; Barakisu, A.I.; Asiedu, K.; Arhin, S.K. Development of an Affordable ELISA Targeting the SARS-CoV-2 Nucleocapsid and Its Application to Samples from the Ongoing COVID-19 Epidemic in Ghana. Mol. Diagn. Ther. 2023, 27, 583–592. [Google Scholar] [CrossRef]
- Jack, A.; Ferro, L.S.; Trnka, M.J.; Wehri, E.; Nadgir, A.; Nguyenla, X.; Fox, D.; Costa, K.; Stanley, S.; Schaletzky, J. SARS-CoV-2 nucleocapsid protein forms condensates with viral genomic RNA. PLoS Biol. 2021, 19, e3001425. [Google Scholar] [CrossRef]
- Supekar, N.T.; Shajahan, A.; Gleinich, A.S.; Rouhani, D.S.; Heiss, C.; Chapla, D.G.; Moremen, K.W.; Azadi, P. Variable posttranslational modifications of severe acute respiratory syndrome coronavirus 2 nucleocapsid protein. Glycobiology 2021, 31, 1080–1092. [Google Scholar] [CrossRef]
- Sun, Z.; Zheng, X.; Ji, F.; Zhou, M.; Su, X.; Ren, K.; Li, L. Mass spectrometry analysis of SARS-CoV-2 nucleocapsid protein reveals camouflaging glycans and unique post-translational modifications. Infect. Microbes Dis. 2021, 3, 149. [Google Scholar] [CrossRef]
- Williams, D.M.; Hornsby, H.; Shehata, O.M.; Brown, R.; Zafred, D.; Shun-Shion, A.S.; Hodder, A.J.; Bliss, D.; Metcalfe, A.; Edgar, J.R. A high content microscopy-based platform for detecting antibodies to the nucleocapsid, spike and membrane proteins of SARS-CoV-2. medRxiv 2021. [Google Scholar] [CrossRef]
- Zheng, X.; Sun, Z.; Yu, L.; Shi, D.; Zhu, M.; Yao, H.; Li, L. Interactome analysis of the nucleocapsid protein of SARS-CoV-2 virus. Pathogens 2021, 10, 1155. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.S.; Kim, N.H.; Song, H.; Cha, S.Y.; Hwang, K.H.; Lee, J.E.; Jeong, C.-H.; Song, S.H.; Kim, S.; Cho, E.S. Emergence of glycogen synthase kinase-3 interaction domain enhances phosphorylation of SARS-CoV-2 nucleocapsid protein. bioRxiv 2022. [Google Scholar] [CrossRef]
- Kim, J.; Kim, D.; Baek, K.; Kim, M.; Kang, B.M.; Maharjan, S.; Park, S.; Choi, J.-K.; Kim, S.; Kim, Y.K. Production of a Monoclonal Antibody to the Nucleocapsid Protein of SARS-CoV-2 and Its Application to ELISA-Based Detection Methods with Broad Specificity by Combined Use of Detector Antibodies. Viruses 2022, 15, 28. [Google Scholar] [CrossRef]
- Wen, Y.; Guo, W.; Min, Y.; Zhong, K.; Zhang, X.; Xing, X.; Tong, Y.; Pan, Y.; Hong, W.; Cai, W. Patient-derived monoclonal antibodies to SARS-CoV-2 nucleocapsid protein N-terminal and C-terminal domains cross-react with their counterparts of SARS-CoV, but not other human betacoronaviruses. Front. Immunol. 2023, 14, 1093709. [Google Scholar] [CrossRef]
- Rump, A.; Risti, R.; Kristal, M.-L.; Reut, J.; Syritski, V.; Lookene, A.; Boudinot, S.R. Dual ELISA using SARS-CoV-2 nucleocapsid protein produced in E. coli and CHO cells reveals epitope masking by N-glycosylation. Biochem. Biophys. Res. Commun. 2021, 534, 457–460. [Google Scholar] [CrossRef]
- Colwill, K.; Galipeau, Y.; Stuible, M.; Gervais, C.; Arnold, C.; Rathod, B.; Abe, K.T.; Wang, J.H.; Pasculescu, A.; Maltseva, M. A scalable serology solution for profiling humoral immune responses to SARS-CoV-2 infection and vaccination. Clin. Transl. Immunol. 2022, 11, e1380. [Google Scholar] [CrossRef]
- Mamedov, T.; Yuksel, D.; Ilgın, M.; Gürbüzaslan, I.; Gulec, B.; Mammadova, G.; Ozdarendeli, A.; Yetiskin, H.; Kaplan, B.; Islam Pavel, S.T. Production and characterization of nucleocapsid and RBD cocktail antigens of SARS-CoV-2 in Nicotiana benthamiana plant as a vaccine candidate against COVID-19. Vaccines 2021, 9, 1337. [Google Scholar] [CrossRef]
- Williams, L.; Jurado, S.; Llorente, F.; Romualdo, A.; González, S.; Saconne, A.; Bronchalo, I.; Martínez-Cortes, M.; Pérez-Gómez, B.; Ponz, F. The C-terminal half of SARS-CoV-2 nucleocapsid protein, industrially produced in plants, is valid as antigen in COVID-19 serological tests. Front. Plant Sci. 2021, 12, 699665. [Google Scholar] [CrossRef]
- de Camargo, B.R.; da Silva, L.A.; de Oliveira, A.S.; Ribeiro, B.M. An easy pipeline for one-step purification of SARS-CoV-2 nucleocapsid protein from insect cell suspension culture. J. Virol. Methods 2022, 299, 114341. [Google Scholar] [CrossRef]
- Tian, Y.; Zhang, G.; Liu, H.; Ding, P.; Jia, R.; Zhou, J.; Chen, Y.; Qi, Y.; Du, J.; Liang, C. Screening and identification of B cell epitope of the nucleocapsid protein in SARS-CoV-2 using the monoclonal antibodies. Appl. Microbiol. Biotechnol. 2022, 106, 1151–1164. [Google Scholar] [CrossRef]
- Lutomski, C.A.; El-Baba, T.J.; Bolla, J.R.; Robinson, C.V. Proteoforms of the sars-cov-2 nucleocapsid protein are primed to proliferate the virus and attenuate the antibody response. medRxiv 2020. [Google Scholar] [CrossRef]
- Wilson, N.; Simpson, R.; Cooper-Liddell, C. Introductory glycosylation analysis using SDS-PAGE and peptide mass fingerprinting. Glycom. Methods Protoc. 2009, 534, 205–212. [Google Scholar]
- Scheller, C.; Krebs, F.; Wiesner, R.; Wätzig, H.; Oltmann-Norden, I. A comparative study of CE-SDS, SDS-PAGE, and Simple Western—Precision, repeatability, and apparent molecular mass shifts by glycosylation. Electrophoresis 2021, 42, 1521–1531. [Google Scholar] [CrossRef]
- Mark, J.; Li, X.; Cyr, T.; Fournier, S.; Jaentschke, B.; Hefford, M.A. SARS coronavirus: Unusual lability of the nucleocapsid protein. Biochem. Biophys. Res. Commun. 2008, 377, 429–433. [Google Scholar] [CrossRef]
- Diemer, C.; Schneider, M.; Seebach, J.; Quaas, J.; Frösner, G.; Schätzl, H.M.; Gilch, S. Cell type-specific cleavage of nucleocapsid protein by effector caspases during SARS coronavirus infection. J. Mol. Biol. 2008, 376, 23–34. [Google Scholar] [CrossRef]
- Ying, W.; Hao, Y.; Zhang, Y.; Peng, W.; Qin, E.; Cai, Y.; Wei, K.; Wang, J.; Chang, G.; Sun, W. Proteomic analysis on structural proteins of Severe Acute Respiratory Syndrome coronavirus. Proteomics 2004, 4, 492–504. [Google Scholar] [CrossRef] [PubMed]
- Bielser, J.-M.; Chappuis, L.; Xiao, Y.; Souquet, J.; Broly, H.; Morbidelli, M. Perfusion cell culture for the production of conjugated recombinant fusion proteins reduces clipping and quality heterogeneity compared to batch-mode processes. J. Biotechnol. 2019, 302, 26–31. [Google Scholar] [CrossRef]
- Kuang, B.; Dhara, V.G.; Hoang, D.; Jenkins, J.; Ladiwala, P.; Tan, Y.; Shaffer, S.A.; Galbraith, S.C.; Betenbaugh, M.J.; Yoon, S. Identification of novel inhibitory metabolites and impact verification on growth and protein synthesis in mammalian cells. Metab. Eng. Commun. 2021, 13, e00182. [Google Scholar] [CrossRef] [PubMed]
- Raymond, C.; Tom, R.; Perret, S.; Moussouami, P.; L’Abbé, D.; St-Laurent, G.; Durocher, Y. A simplified polyethylenimine-mediated transfection process for large-scale and high-throughput applications. Methods 2011, 55, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Granados, S.; Cervera, L.; Kamen, A.A.; Gòdia, F. Advancements in mammalian cell transient gene expression (TGE) technology for accelerated production of biologics. Crit. Rev. Biotechnol. 2018, 38, 918–940. [Google Scholar] [CrossRef] [PubMed]
- Hunter, M.; Yuan, P.; Vavilala, D.; Fox, M. Optimization of protein expression in mammalian cells. Curr. Protoc. Protein Sci. 2019, 95, e77. [Google Scholar] [CrossRef]
- Pham, P.L.; Perret, S.; Doan, H.C.; Cass, B.; St-Laurent, G.; Kamen, A.; Durocher, Y. Large-scale transient transfection of serum-free suspension-growing HEK293 EBNA1 cells: Peptone additives improve cell growth and transfection efficiency. Biotechnol. Bioeng. 2003, 84, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Durocher, Y.; Perret, S.; Kamen, A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002, 30, e9. [Google Scholar] [CrossRef] [PubMed]
- Hacker, D.L.; Kiseljak, D.; Rajendra, Y.; Thurnheer, S.; Baldi, L.; Wurm, F.M. Polyethyleneimine-based transient gene expression processes for suspension-adapted HEK-293E and CHO-DG44 cells. Protein Expr. Purif. 2013, 92, 67–76. [Google Scholar] [CrossRef]
- Tan, E.; Chin, C.S.H.; Lim, Z.F.S.; Ng, S.K. HEK293 cell line as a platform to produce recombinant proteins and viral vectors. Front. Bioeng. Biotechnol. 2021, 9, 796991. [Google Scholar] [CrossRef] [PubMed]
- Backliwal, G.; Hildinger, M.; Chenuet, S.; Wulhfard, S.; De Jesus, M.; Wurm, F.M. Rational vector design and multi-pathway modulation of HEK 293E cells yield recombinant antibody titers exceeding 1 g/l by transient transfection under serum-free conditions. Nucleic Acids Res. 2008, 36, e96. [Google Scholar] [CrossRef]
- Backliwal, G.; Hildinger, M.; Hasija, V.; Wurm, F.M. High-density transfection with HEK-293 cells allows doubling of transient titers and removes need for a priori DNA complex formation with PEI. Biotechnol. Bioeng. 2008, 99, 721–727. [Google Scholar] [CrossRef]
- Arena, T.A.; Chou, B.; Harms, P.D.; Wong, A.W. An anti-apoptotic HEK293 cell line provides a robust and high titer platform for transient protein expression in bioreactors. MAbs 2019, 11, 977–986. [Google Scholar] [CrossRef]
- Chang, C.-K.; Hou, M.-H.; Chang, C.-F.; Hsiao, C.-D.; Huang, T.-H. The SARS coronavirus nucleocapsid protein–forms and functions. Antivir. Res. 2014, 103, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Zeng, R.; von Brunn, A.; Lei, J. Structural characterization of the C-terminal domain of SARS-CoV-2 nucleocapsid protein. Mol. Biomed. 2020, 1, 2. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Du, N.; Lei, Y.; Dorje, S.; Qi, J.; Luo, T.; Gao, G.F.; Song, H. Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design. EMBO J. 2020, 39, e105938. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; West, A.M.; Silletti, S.; Corbett, K.D. Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein. Protein Sci. 2020, 29, 1890–1901. [Google Scholar] [CrossRef]
- van Kooten, C.; Banchereau, J. CD40-CD40 ligand. J. Leukoc. Biol. 2000, 67, 2–17. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Cheng, X.; Truong, B.; Sun, L.; Yang, X.; Wang, H. Molecular basis and therapeutic implications of CD40/CD40L immune checkpoint. Pharmacol. Ther. 2021, 219, 107709. [Google Scholar] [CrossRef]
- Pietravalle, F.; Lecoanet-Henchoz, S.; Blasey, H.; Aubry, J.P.; Elson, G.; Edgerton, M.D.; Bonnefoy, J.Y.; Gauchat, J.F. Human Native Soluble CD40L Is a Biologically Active Trimer, Processed Inside Microsomes. J. Biol. Chem. 1996, 271, 5965–5967. [Google Scholar] [CrossRef]
- Pullen, S.S.; Labadia, M.E.; Ingraham, R.H.; McWhirter, S.M.; Everdeen, D.S.; Alber, T.; Crute, J.J.; Kehry, M.R. High-affinity interactions of tumor necrosis factor receptor-associated factors (TRAFs) and CD40 require TRAF trimerization and CD40 multimerization. Biochemistry 1999, 38, 10168–10177. [Google Scholar] [CrossRef]
- An, H.J.; Kim, Y.J.; Song, D.H.; Park, B.S.; Kim, H.M.; Lee, J.D.; Paik, S.G.; Lee, J.O.; Lee, H. Crystallographic and mutational analysis of the CD40-CD154 complex and its implications for receptor activation. J. Biol. Chem. 2011, 286, 11226–11235. [Google Scholar] [CrossRef]
- Jiskoot, W.; Randolph, T.W.; Volkin, D.B.; Middaugh, C.R.; Schöneich, C.; Winter, G.; Friess, W.; Crommelin, D.J.A.; Carpenter, J.F. Protein instability and immunogenicity: Roadblocks to clinical application of injectable protein delivery systems for sustained release. J. Pharm. Sci. 2012, 101, 946–954. [Google Scholar] [CrossRef]
- Dorai, H.; Ganguly, S. Mammalian cell-produced therapeutic proteins: Heterogeneity derived from protein degradation. Curr. Opin. Biotechnol. 2014, 30, 198–204. [Google Scholar] [CrossRef] [PubMed]

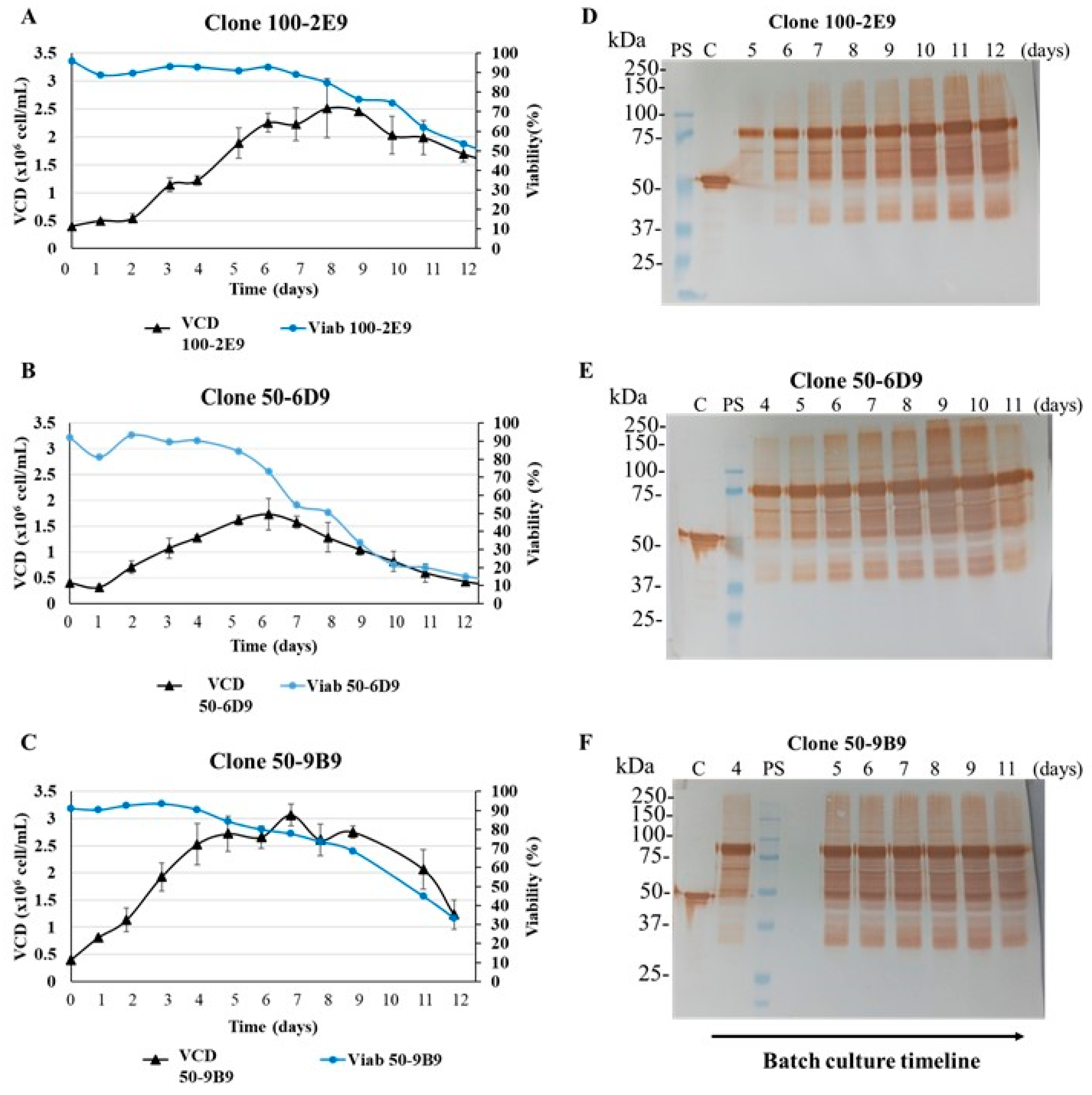
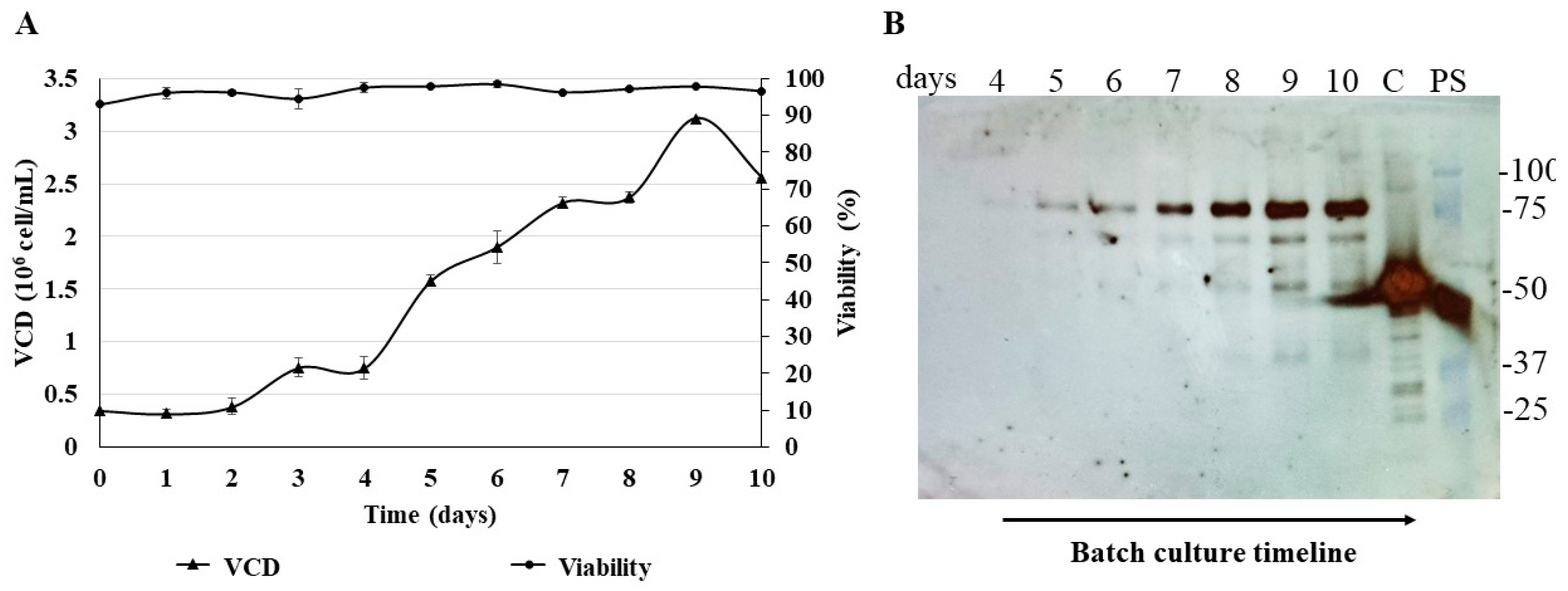
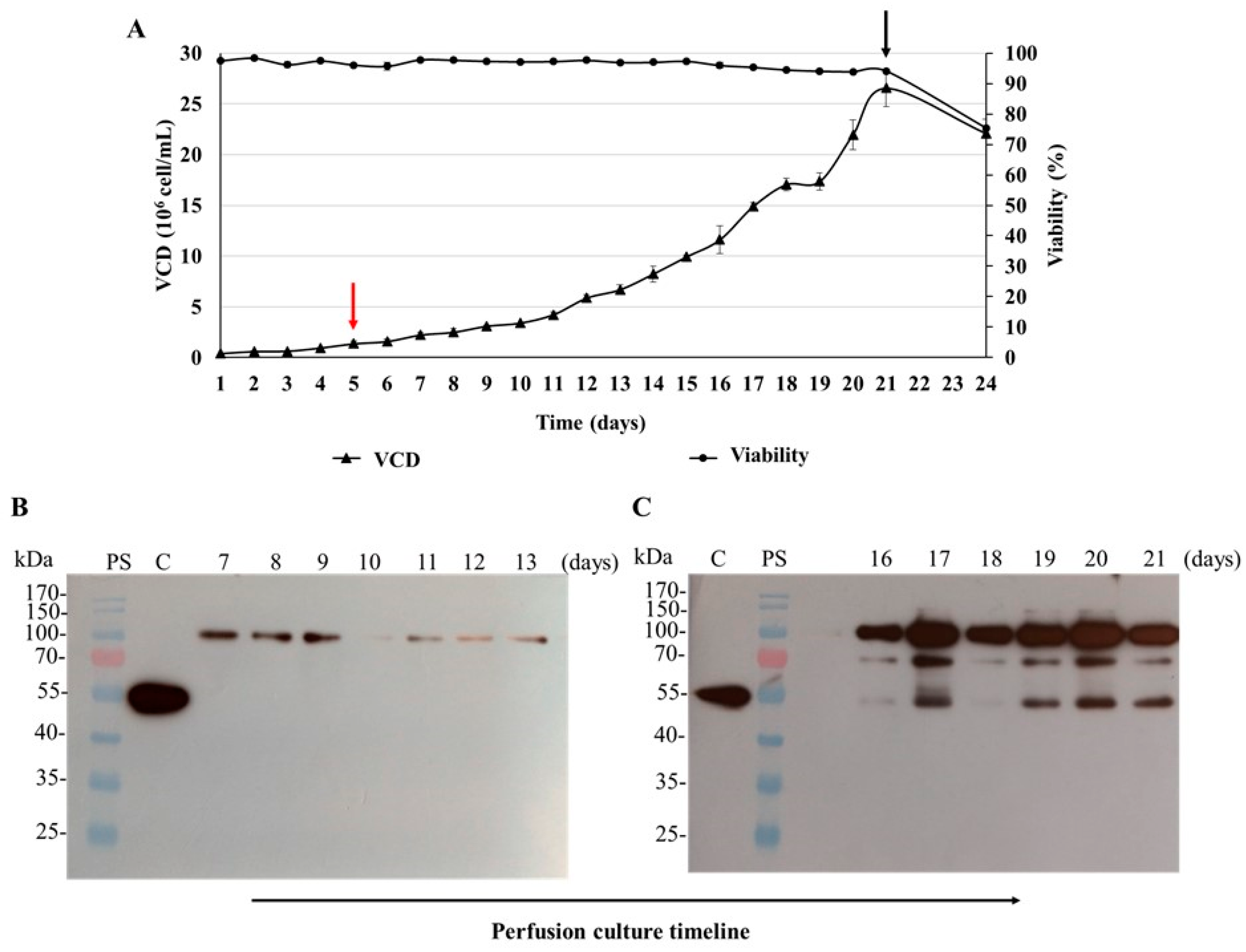
| Cell Clones | Growth Phase Duration § (days) | Experiment Duration (days) | VCDmax (106 cell/mL) | IVCCmax (108 cells × h/mL) | μmax (h−1) |
|---|---|---|---|---|---|
| 50-6D9 * | 4 | 16 | 1.7 | 314.5 | 0.0116 |
| 50-9B9 * | 4 | 13 | 3.1 | 632.8 | 0.0159 |
| 100-2E9 * | 6 | 16 | 2.5 | 592.7 | 0.0105 |
| 100-2E9 ** | 10 | 10 | 3.1 | 354.3 | 0.0101 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lao, T.; Farnos, O.; Bueno, A.; Alvarez, A.; Rodríguez, E.; Palacios, J.; Luz, K.R.d.l.; Kamen, A.; Carpio, Y.; Estrada, M.P. Transient Expression in HEK-293 Cells in Suspension Culture as a Rapid and Powerful Tool: SARS-CoV-2 N and Chimeric SARS-CoV-2N-CD154 Proteins as a Case Study. Biomedicines 2023, 11, 3050. https://doi.org/10.3390/biomedicines11113050
Lao T, Farnos O, Bueno A, Alvarez A, Rodríguez E, Palacios J, Luz KRdl, Kamen A, Carpio Y, Estrada MP. Transient Expression in HEK-293 Cells in Suspension Culture as a Rapid and Powerful Tool: SARS-CoV-2 N and Chimeric SARS-CoV-2N-CD154 Proteins as a Case Study. Biomedicines. 2023; 11(11):3050. https://doi.org/10.3390/biomedicines11113050
Chicago/Turabian StyleLao, Thailin, Omar Farnos, Alexi Bueno, Anays Alvarez, Elsa Rodríguez, Julio Palacios, Kathya Rashida de la Luz, Amine Kamen, Yamila Carpio, and Mario Pablo Estrada. 2023. "Transient Expression in HEK-293 Cells in Suspension Culture as a Rapid and Powerful Tool: SARS-CoV-2 N and Chimeric SARS-CoV-2N-CD154 Proteins as a Case Study" Biomedicines 11, no. 11: 3050. https://doi.org/10.3390/biomedicines11113050
APA StyleLao, T., Farnos, O., Bueno, A., Alvarez, A., Rodríguez, E., Palacios, J., Luz, K. R. d. l., Kamen, A., Carpio, Y., & Estrada, M. P. (2023). Transient Expression in HEK-293 Cells in Suspension Culture as a Rapid and Powerful Tool: SARS-CoV-2 N and Chimeric SARS-CoV-2N-CD154 Proteins as a Case Study. Biomedicines, 11(11), 3050. https://doi.org/10.3390/biomedicines11113050








