Age-Dependent Alterations in Semen Parameters and Human Sperm MicroRNA Profile
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval and Patients’ Recruitment
2.2. Semen Analysis and Sample Preparation
2.3. Total RNA Isolation
2.4. Small RNA Sequencing and Data Analysis
2.5. Bioinformatic Analysis
2.6. Statistical Analysis
3. Results
3.1. Impact of Men’s Age on Conventional Seminal Parameters
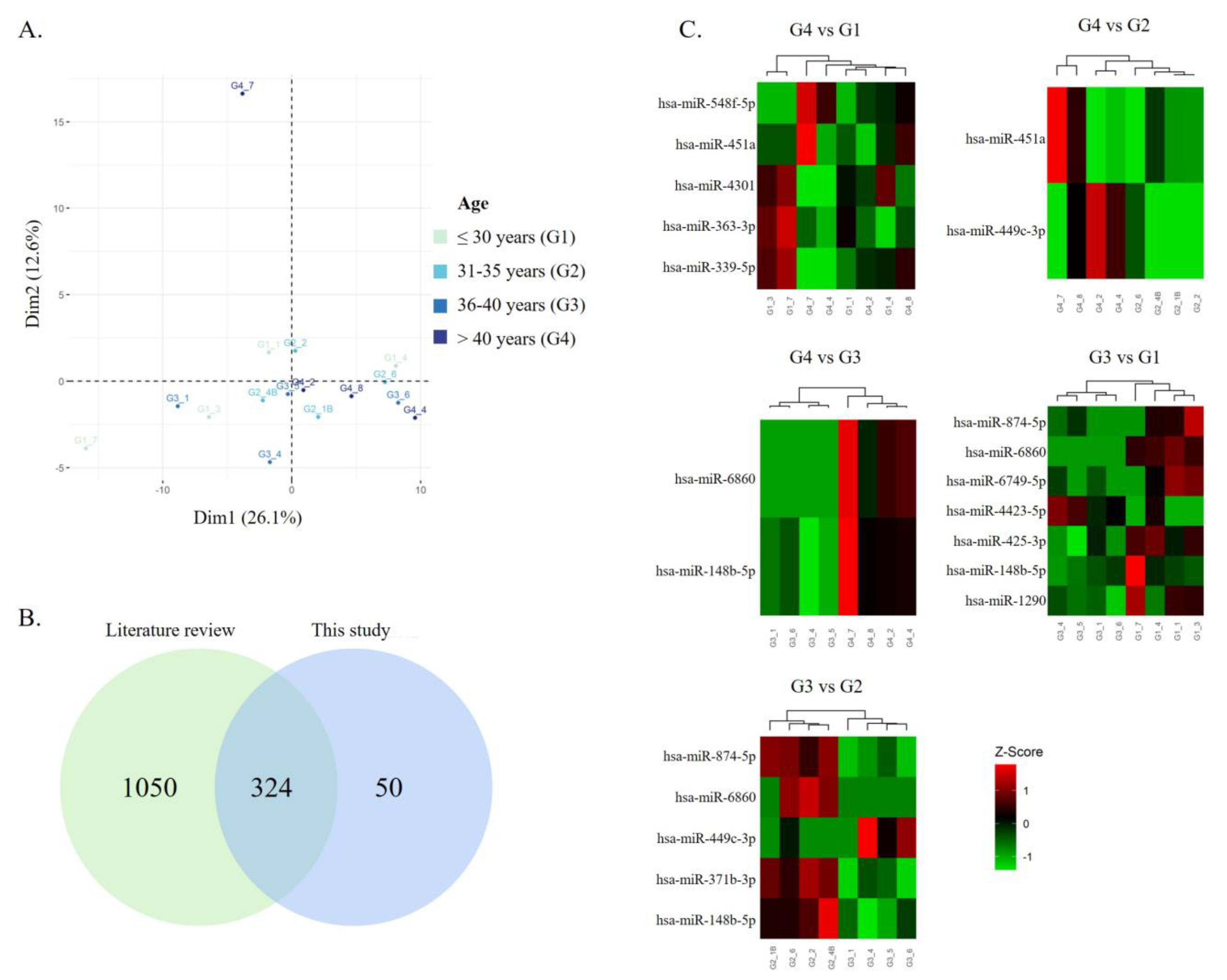
3.2. Impact of Men’s Age on Sperm miRNA Content
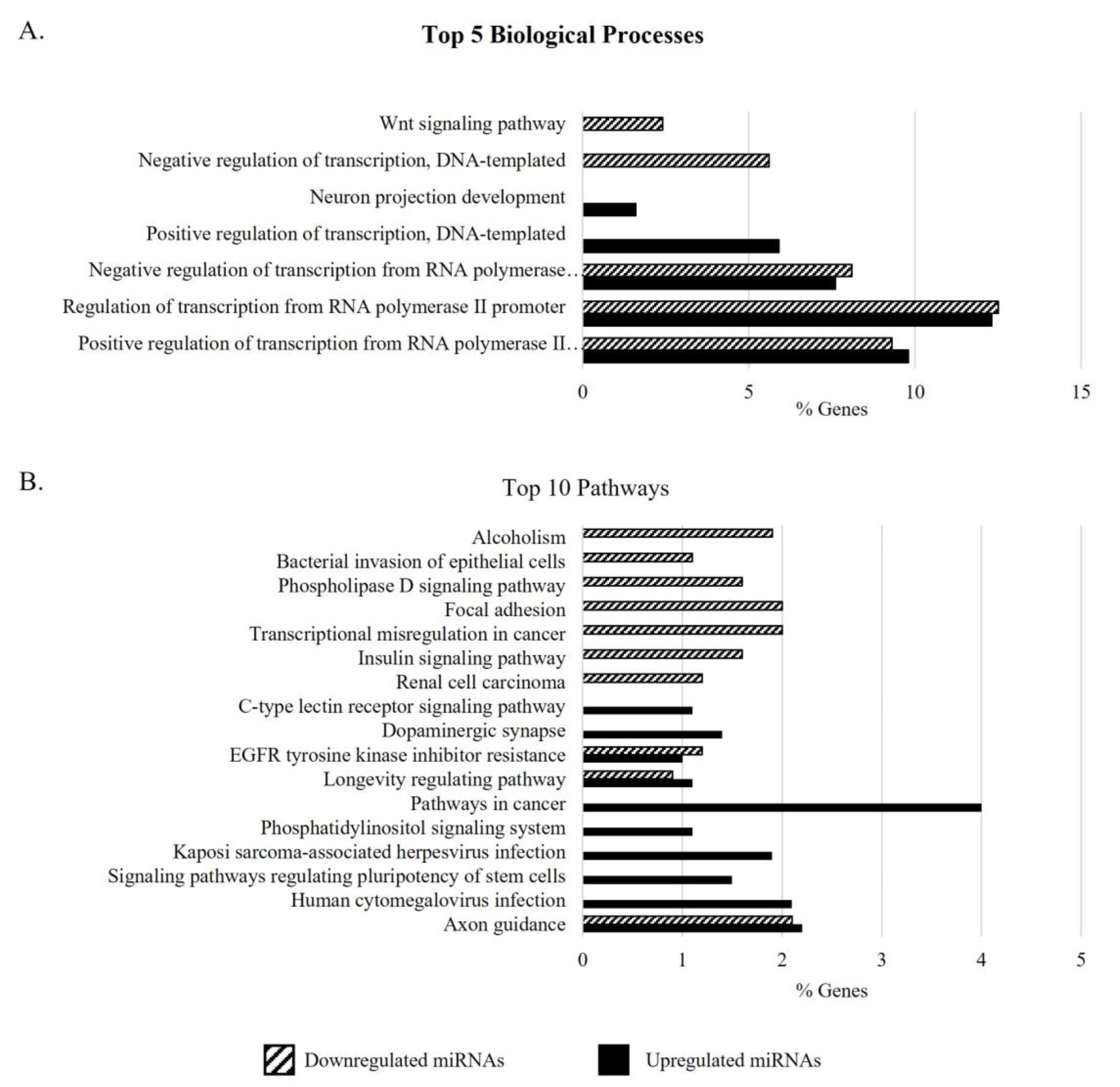
3.3. Gene Ontology Analysis of Target Genes of DEMs in the Sperm of Men with APA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Skakkebæk, N.E.; Lindahl-Jacobsen, R.; Levine, H.; Andersson, A.-M.; Jørgensen, N.; Main, K.M.; Lidegaard, Ø.; Priskorn, L.; Holmboe, S.A.; Bräuner, E.V.; et al. Environmental Factors in Declining Human Fertility. Nat. Rev. Endocrinol. 2022, 18, 139–157. [Google Scholar] [CrossRef] [PubMed]
- WHO. Infertility Prevalence Estimates, 1990–2021; WHO: Geneva, Switzerland, 2023.
- Bala, R.; Singh, V.; Rajender, S.; Singh, K. Environment, Lifestyle, and Female Infertility. Reprod. Sci. 2021, 28, 617–638. [Google Scholar] [CrossRef] [PubMed]
- Eurostat Mean Age of Women at Childbirth and at Birth of First Child. Available online: https://ec.europa.eu/eurostat/databrowser/view/tps00017/default/table?lang=en (accessed on 16 March 2023).
- Ashapkin, V.; Suvorov, A.; Pilsner, J.R.; Krawetz, S.A.; Sergeyev, O. Age-Associated Epigenetic Changes in Mammalian Sperm: Implications for Offspring Health and Development. Hum. Reprod. Update 2023, 29, 24–44. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.; Mavrelos, D.; Odia, R.; Viñals Gonzalez, X.; Cawood, S.; Yasmin, E.; Saab, W.; Serhal, P.; Seshadri, S. Paternal Age over 50 Years Decreases Assisted Reproductive Technology (ART) Success: A Single UK Center Retrospective Analysis. Acta Obstet. Gynecol. Scand. 2021, 100, 1858–1867. [Google Scholar] [CrossRef] [PubMed]
- Khandwala, Y.S.; Zhang, C.A.; Lu, Y.; Eisenberg, M.L. The Age of Fathers in the USA Is Rising: An Analysis of 168 867 480 Births from 1972 to 2015. Hum. Reprod. 2017, 32, 2110–2116. [Google Scholar] [CrossRef] [PubMed]
- Frick, A.P. Advanced Maternal Age and Adverse Pregnancy Outcomes. Best Pract. Res. Clin. Obstet. Gynaecol. 2021, 70, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.; Silva, J.V.; Alves, M.G.; Oliveira, P.F.; Fardilha, M. Testicular Aging: An Overview of Ultrastructural, Cellular, and Molecular Alterations. J. Gerontol. Ser. A 2019, 74, 860–871. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.L.; Dunleavy, J.; Gemmell, N.J.; Nakagawa, S. Consistent Age-Dependent Declines in Human Semen Quality: A Systematic Review and Meta-Analysis. Ageing Res. Rev. 2015, 19, 22–33. [Google Scholar] [CrossRef]
- Gonzalez, D.C.; Ory, J.; Blachman-Braun, R.; Nackeeran, S.; Best, J.C.; Ramasamy, R. Advanced Paternal Age and Sperm DNA Fragmentation: A Systematic Review. World J. Mens. Health 2022, 40, 1–12. [Google Scholar] [CrossRef]
- Wang, J.J.; Wang, S.X.; Tehmina; Feng, Y.; Zhang, R.F.; Li, X.Y.; Sun, Q.; Ding, J. Age-Related Decline of Male Fertility: Mitochondrial Dysfunction and the Antioxidant Interventions. Pharmaceuticals 2022, 15, 519. [Google Scholar] [CrossRef]
- Nguyen-Powanda, P.; Robaire, B. Oxidative Stress and Reproductive Function in the Aging Male. Biology 2020, 9, 282. [Google Scholar] [CrossRef] [PubMed]
- Paoli, D.; Pecora, G.; Pallotti, F.; Faja, F.; Pelloni, M.; Lenzi, A.; Lombardo, F. Cytological and Molecular Aspects of the Ageing Sperm. Hum. Reprod. 2019, 34, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Goldmann, J.M.; Veltman, J.A.; Gilissen, C. De Novo Mutations Reflect Development and Aging of the Human Germline. Trends Genet. 2019, 35, 828–839. [Google Scholar] [CrossRef] [PubMed]
- Wood, K.A.; Goriely, A. The Impact of Paternal Age on New Mutations and Disease in the next Generation. Fertil. Steril. 2022, 118, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Bertoncelli Tanaka, M.; Agarwal, A.; Esteves, S.C. Paternal Age and Assisted Reproductive Technology: Problem Solver or Trouble Maker? Panminerva Med. 2019, 61, 138–151. [Google Scholar] [CrossRef]
- Halvaei, I.; Litzky, J.; Esfandiari, N. Advanced Paternal Age: Effects on Sperm Parameters, Assisted Reproduction Outcomes and Offspring Health. Reprod. Biol. Endocrinol. 2020, 18, 110. [Google Scholar] [CrossRef] [PubMed]
- Marsidi, A.M.; Kipling, L.M.; Kawwass, J.F.; Mehta, A. Influence of Paternal Age on Assisted Reproductive Technology Cycles and Perinatal Outcomes. Fertil. Steril. 2021, 116, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Kidera, N.; Ishikawa, T.; Kawamura, T.; Miyasaka, N. Impact of Paternal Age on IVF and Pregnancy Outcomes with Only Normal Sperm Parameters. Taiwan. J. Obstet. Gynecol. 2022, 61, 1015–1020. [Google Scholar] [CrossRef]
- Murugesu, S.; Kasaven, L.S.; Petrie, A.; Vaseekaran, A.; Jones, B.P.; Bracewell-Milnes, T.; Barcroft, J.F.; Grewal, K.J.; Getreu, N.; Galazis, N.; et al. Does Advanced Paternal Age Affect Outcomes Following Assisted Reproductive Technology? A Systematic Review and Meta-Analysis. Reprod. Biomed. Online 2022, 45, 283–331. [Google Scholar] [CrossRef]
- Ostermeier, G.C.; Goodrich, R.J.; Moldenhauer, J.S.; Diamond, M.P.; Krawetz, S.A. A Suite of Novel Human Spermatozoal RNAs. J. Androl. 2005, 26, 70–74. [Google Scholar] [CrossRef]
- Santiago, J.; Silva, J.V.; Howl, J.; Santos, M.A.S.; Fardilha, M. All You Need to Know about Sperm RNAs. Hum. Reprod. Update 2021, 28, 67–91. [Google Scholar] [CrossRef] [PubMed]
- Marczylo, E.L.; Amoako, A.A.; Konje, J.C.; Gant, T.W.; Marczylo, T.H. Smoking Induces Differential MiRNA Expression in Human Spermatozoa: A Potential Transgenerational Epigenetic Concern? Epigenetics 2012, 7, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Metzler-Guillemain, C.; Victorero, G.; Lepoivre, C.; Bergon, A.; Yammine, M.; Perrin, J.; Sari-Minodier, I.; Boulanger, N.; Rihet, P.; Nguyen, C. Sperm MRNAs and MicroRNAs as Candidate Markers for the Impact of Toxicants on Human Spermatogenesis: An Application to Tobacco Smoking. Syst. Biol. Reprod. Med. 2015, 61, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Nazmara, Z.; Najafi, M.; Movahedin, M.; Zandiyeh, Z.; Shirinbayan, P.; Asgari, H.R.; Roshanpajouh, M.; Maki, C.B.; Bashiri, Z.; Koruji, M. Correlation Between Protamine-2 and MiRNA-122 in Sperm from Heroin-Addicted Men: A Case-Control Study. Urol. J. 2020, 17, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, M.; Liu, Y.; Song, G.; Liu, N. A Microarray for MicroRNA Profiling in Spermatozoa from Adult Men Living in an Environmentally Polluted Site. Bull. Environ. Contam. Toxicol. 2012, 89, 1111–1114. [Google Scholar] [CrossRef] [PubMed]
- Salas-Huetos, A.; Blanco, J.; Vidal, F.; Godo, A.; Grossmann, M.; Pons, M.C.; F-Fernández, S.; Garrido, N.; Anton, E. Spermatozoa from Patients with Seminal Alterations Exhibit a Differential Micro-Ribonucleic Acid Profile. Fertil. Steril. 2015, 104, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, E.; Boekelheide, K.; Sigman, M.; Braun, J.M.; Eliot, M.; Hall, S.J.; Dere, E.; Hwang, K. Spermatozoal Large RNA Content Is Associated with Semen Characteristics, Sociodemographic and Lifestyle Factors. PLoS ONE 2019, 14, e0216584. [Google Scholar] [CrossRef] [PubMed]
- Ostermeier, G.C.; Miller, D.; Huntriss, J.D.; Diamond, M.P.; Krawetz, S.A. Delivering Spermatozoan RNA to the Oocyte. Nature 2004, 429, 154. [Google Scholar] [CrossRef] [PubMed]
- Estill, M.S.; Hauser, R.; Krawetz, S.A. RNA Element Discovery from Germ Cell to Blastocyst. Nucleic Acids Res. 2019, 47, 2263–2275. [Google Scholar] [CrossRef]
- Ntostis, P.; Carter, D.; Iles, D.; Huntriss, J.; Tzetis, M.; Miller, D. Potential Sperm Contributions to the Murine Zygote Predicted by in Silico Analysis. Reproduction 2017, 154, 777–788. [Google Scholar] [CrossRef]
- World Health Organization. WHO Laboratory Manual for the Examination and Processing of Human Semen; World Health Organization: Geneva, Switzerland, 2010; ISBN 9789241547789.
- Kang, W.; Eldfjell, Y.; Fromm, B.; Estivill, X.; Biryukova, I.; Friedländer, M.R. MiRTrace Reveals the Organismal Origins of MicroRNA Sequencing Data. Genome Biol. 2018, 19, 213. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Zhao, S.; Gordon, W.; Du, S.; Zhang, C.; He, W.; Xi, L.; Mathur, S.; Agostino, M.; Paradis, T.; von Schack, D.; et al. QuickMIRSeq: A Pipeline for Quick and Accurate Quantification of Both Known MiRNAs and IsomiRs by Jointly Processing Multiple Samples from MicroRNA Sequencing. BMC Bioinformat. 2017, 18, 180. [Google Scholar] [CrossRef] [PubMed]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The Biochemical Basis of MicroRNA Targeting Efficacy. Science 2019, 366, 6472. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. MiRDB: An Online Database for Prediction of Functional MicroRNA Targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Liu, W.; Wang, X. Prediction of Functional MicroRNA Targets by Integrative Modeling of MicroRNA Binding and Target Expression Data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates That Thousands of Human Genes Are MicroRNA Targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021 Update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Pantano, L.; Jodar, M.; Bak, M.; Ballescà, J.L.; Tommerup, N.; Oliva, R.; Vavouri, T. The Small RNA Content of Human Sperm Reveals Pseudogene-Derived PiRNAs Complementary to Protein-Coding Genes. RNA 2015, 21, 1085–1095. [Google Scholar] [CrossRef]
- Ingerslev, L.R.; Donkin, I.; Fabre, O.; Versteyhe, S.; Mechta, M.; Pattamaprapanont, P.; Mortensen, B.; Krarup, N.T.; Barrès, R. Endurance Training Remodels Sperm-Borne Small RNA Expression and Methylation at Neurological Gene Hotspots. Clin. Epigenet. 2018, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, X.; Wang, Z.; Li, J.; Xu, Z.; Miao, M.; Chen, G.; Lei, X.; Wu, J.; Shi, H.; et al. MicroRNA Expression Profile Analysis in Sperm Reveals Hsa-Mir-191 as an Auspicious Omen of in Vitro Fertilization. BMC Genomics 2020, 21, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Nätt, D.; Kugelberg, U.; Casas, E.; Nedstrand, E.; Zalavary, S.; Henriksson, P.; Nijm, C.; Jäderquist, J.; Sandborg, J.; Flinke, E.; et al. Human Sperm Displays Rapid Responses to Diet. PLoS Biol. 2019, 17, e3000559. [Google Scholar] [CrossRef] [PubMed]
- Schuster, A.; Tang, C.; Xie, Y.; Ortogero, N.; Yuan, S.; Yan, W. SpermBase: A Database for Sperm-Borne RNA Contents. Biol. Reprod. 2016, 95, 99. [Google Scholar] [CrossRef] [PubMed]
- Hua, M.; Liu, W.; Chen, Y.; Zhang, F.; Xu, B.; Liu, S.; Chen, G.; Shi, H.; Wu, L. Identification of Small Non-Coding RNAs as Sperm Quality Biomarkers for in Vitro Fertilization. Cell Discov. 2019, 5, 20. [Google Scholar] [CrossRef] [PubMed]
- Iroanya, O.O.; Olutunde, O.T.; Egwuatu, T.F.; Igbokwe, C. Stability of Selected MicroRNAs in Human Blood, Semen and Saliva Samples Exposed to Different Environmental Conditions. Forensic Sci. Int. 2022, 336, 111338. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, P. Reviewing Reports of Semen Volume and Male Aging of Last 33 Years: From 1980 through 2013. Asian Pacific J. Reprod. 2015, 4, 242–246. [Google Scholar] [CrossRef]
- Chen, G.-X.; Li, H.-Y.; Lin, Y.-H.; Huang, Z.-Q.; Huang, P.-Y.; Da, L.-C.; Shi, H.; Yang, L.; Feng, Y.-B.; Zheng, B.-H. The Effect of Age and Abstinence Time on Semen Quality: A Retrospective Study. Asian J. Androl. 2022, 24, 73. [Google Scholar] [CrossRef] [PubMed]
- Kaarouch, I.; Bouamoud, N.; Madkour, A.; Louanjli, N.; Saadani, B.; Assou, S.; Aboulmaouahib, S.; Amzazi, S.; Copin, H.; Benkhalifa, M.; et al. Paternal Age: Negative Impact on Sperm Genome Decays and IVF Outcomes after 40 Years. Mol. Reprod. Dev. 2018, 85, 271–280. [Google Scholar] [CrossRef]
- Gallo, M.; Licata, E.; Meneghini, C.; Dal Lago, A.; Fabiani, C.; Amodei, M.; Antonaci, D.; Miriello, D.; Corno, R.; Liberanome, C.; et al. Impact of Paternal Age on Seminal Parameters and Reproductive Outcome of Intracytoplasmatic Sperm Injection in Infertile Italian Women. Front. Endocrinol. 2019, 10, 35. [Google Scholar] [CrossRef]
- Kasman, A.M.; Li, S.; Zhao, Q.; Behr, B.; Eisenberg, M.L. Relationship between Male Age, Semen Parameters and Assisted Reproductive Technology Outcomes. Andrology 2021, 9, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.; Jarvi, K. Steps in the Investigation and Management of Low Semen Volume in the Infertile Man. Can. Urol. Assoc. J. 2013, 3, 479. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Ryan, D.P.; Pearson, B.L.; Henzel, K.S.; Neff, F.; Vidal, R.O.; Hennion, M.; Lehmann, I.; Schleif, M.; Schröder, S.; et al. Epigenetic Alterations in Longevity Regulators, Reduced Life Span, and Exacerbated Aging-Related Pathology in Old Father Offspring Mice. Proc. Natl. Acad. Sci. USA 2018, 115, E2348–E2357. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Chen, Q.; Wang, S.; Ma, R.; Jing, J.; Yang, Y.; Feng, Y.; Zou, Z.; Zhang, Y.; Ge, X.; et al. Mitochondria-Related MiR-574 Reduces Sperm ATP by Targeting ND5 in Aging Males. Aging 2020, 12, 8321–8338. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, D.A.; Sakkas, D.; Gardner, D.K. Sperm Selection Methods in the 21st Century. Biol. Reprod. 2019, 101, 1076–1082. [Google Scholar] [CrossRef] [PubMed]
- Stone, B.A.; Alex, A.; Werlin, L.B.; Marrs, R.P. Age Thresholds for Changes in Semen Parameters in Men. Fertil. Steril. 2013, 100, 952–958. [Google Scholar] [CrossRef] [PubMed]
- Rassoulzadegan, M.; Grandjean, V.; Gounon, P.; Cuzin, F. Sperm RNA, an “Epigenetic Rheostat” of Gene Expression? Arch. Androl. 2007, 53, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yan, W.; Duan, E. Epigenetic Inheritance of Acquired Traits through Sperm RNAs and Sperm RNA Modifications. Nat. Rev. Genet. 2016, 17, 733–743. [Google Scholar] [CrossRef]
- Dominguez, F.; Moreno-Moya, J.M.; Lozoya, T.; Romero, A.; Martínez, S.; Monterde, M.; Gurrea, M.; Ferri, B.; Núñez, M.J.; Simón, C.; et al. Embryonic MiRNA Profiles of Normal and Ectopic Pregnancies. PLoS ONE 2014, 9, e102185. [Google Scholar] [CrossRef]
- Krupp, D.R.; Xu, P.-T.; Thomas, S.; Dellinger, A.; Etchevers, H.C.; Vekemans, M.; Gilbert, J.R.; Speer, M.C.; Ashley-Koch, A.E.; Gregory, S.G. Transcriptome Profiling of Genes Involved in Neural Tube Closure during Human Embryonic Development Using Long Serial Analysis of Gene Expression (Long-SAGE). Birth Defects Res. Part A Clin. Mol. Teratol. 2012, 94, 683–692. [Google Scholar] [CrossRef]
- Kotov, A.A.; Olenkina, O.M.; Godneeva, B.K.; Adashev, V.E.; Olenina, L. V Progress in Understanding the Molecular Functions of DDX3Y (DBY) in Male Germ Cell Development and Maintenance. Biosci. Trends 2017, 11, 46–53. [Google Scholar] [CrossRef] [PubMed]
- La Salle, S.; Palmer, K.; O’Brien, M.; Schimenti, J.C.; Eppig, J.; Handel, M.A. Spata22, a Novel Vertebrate-Specific Gene, Is Required for Meiotic Progress in Mouse Germ Cells. Biol. Reprod. 2012, 86, 45. [Google Scholar] [CrossRef] [PubMed]
- Tabibzadeh, S. Signaling Pathways and Effectors of Aging. Front. Biosci. 2021, 26, 4889. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.V.; Cabral, M.; Correia, B.R.; Carvalho, P.; Sousa, M.; Oliveira, P.F.; Fardilha, M. mTOR Signaling Pathway Regulates Sperm Quality in Older Men. Cells 2019, 8, 629. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.V.; Freitas, M.J.; Correia, B.R.; Korrodi-Gregório, L.; Patrício, A.; Pelech, S.; Fardilha, M. Profiling Signaling Proteins in Human Spermatozoa: Biomarker Identification for Sperm Quality Evaluation. Fertil. Steril. 2015, 104, 845–856.e8. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pizzari, T.; Dean, R.; Pacey, A.; Moore, H.; Bonsall, M.B. The Evolutionary Ecology of Pre- and Post-Meiotic Sperm Senescence. Trends Ecol. Evol. 2008, 23, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Darmishonnejad, Z.; Tavalaee, M.; Izadi, T.; Tanhaei, S.; Nasr-Esfahani, M.H. Evaluation of Sperm Telomere Length in Infertile Men with Failed/Low Fertilization after Intracytoplasmic Sperm Injection. Reprod. Biomed. Online 2019, 38, 579–587. [Google Scholar] [CrossRef]
- Muñoz-Espín, D.; Cañamero, M.; Maraver, A.; Gómez-López, G.; Contreras, J.; Murillo-Cuesta, S.; Rodríguez-Baeza, A.; Varela-Nieto, I.; Ruberte, J.; Collado, M.; et al. Programmed Cell Senescence during Mammalian Embryonic Development. Cell 2013, 155, 1104–1118. [Google Scholar] [CrossRef]
- Pan, H.; Rui, X.; Wei, W.; Shao, S.; Zhu, Y. Prognostic Value of MiR-339-5p in Patients with Prostate Cancer and Its Effects on Tumor Progression. Exp. Ther. Med. 2021, 21, 390. [Google Scholar] [CrossRef]
| Parameter | Mean ± SD | Spearman Correlation Test | |
|---|---|---|---|
| r | p-Value | ||
| Age (years) | 35.73 ± 6.74 | - | - |
| Semen parameters | |||
| Semen volume (mL) | 3.07 ± 1.45 | −0.132 | 0.016 * |
| Sperm concentration (106/mL) | 46.74 ± 46.97 | −0.010 | 0.853 |
| Total sperm count (106) | 136.70 ± 127.59 | −0.032 | 0.568 |
| Total motility (%) | 56.94 ± 13.24 | −0.010 | 0.856 |
| Progressive motility (%) | 41.17 ± 13.46 | −0.006 | 0.909 |
| Non-progressive motility (%) | 15.55 ± 5.81 | −0.067 | 0.225 |
| Immobility (%) | 43.3 ± 13.14 | 0.023 | 0.672 |
| Morphological normal sperm (%) | 5.77 ± 2.26 | 0.032 | 0.560 |
| Head defects (%) | 85.79 ± 5.49 | 0.032 | 0.568 |
| Midpiece defects (%) | 49.81 ± 10.21 | −0.063 | 0.262 |
| Principal piece defects (%) | 24.32 ± 7.62 | −0.060 | 0.287 |
| Teratozoospermic index | 1.70 ± 0.16 | −0.062 | 0.267 |
| Parameter/Group | ≤30 (n = 66) | 31–35 (n = 96) | 36–40 (n = 100) | >40 (n = 71) |
|---|---|---|---|---|
| Age (years) | 26.97 ± 3.09 b,c,d * | 33.26 ± 1.42 a,c,d * | 37.68 ± 1.29 a,b,d * | 44.86 ± 4.41 a,b,c * |
| Semen parameters | ||||
| Semen volume (mL) | 3.07 ± 1.64 | 3.48 ± 1.47 | 2.99 ± 1.28 | 2.63 ± 4.41 b* |
| Sperm concentration (106/mL) | 49.21 ± 56.78 | 45.50 ± 34.66 | 44.19 ± 43.14 | 49.73 ± 56.42 |
| Total sperm count (106) | 131.4 ± 127.68 | 156.3 ± 142.58 | 127.6 ± 105.19 | 126.76 ± 132.92 |
| Total motility (%) | 55.91 ± 12.46 | 58.52 ± 13.31 | 56.64 ± 13.42 | 56.15 ± 13.68 |
| Progressive motility (%) | 40.02 ± 12.10 | 42.46 ± 13.35 | 41.27 ± 14.49 | 40.34 ± 13.45 |
| Non-progressive motility (%) | 15.72 ± 6.62 | 15.95 ± 4.52 | 15.26 ± 6.33 | 15.22 ± 5.95 |
| Immobility (%) | 44.38 ± 12.75 | 41.59 ± 13.18 | 43.47 ± 13.38 | 44.46 ± 13.16 |
| Morphological normal sperm (%) | 5.60 ± 2.23 | 5.91 ± 2.07 | 5.75 ± 2.40 | 5.75 ± 2.40 |
| Head defects (%) | 85.7 ± 4.82 | 85.67 ± 5.24 | 85.77 ± 5.56 | 86.07 ± 6.34 |
| Midpiece defects (%) | 51.0 ± 9.27 | 48.99 ± 9.86 | 50.25 ± 11.07 | 49.22 ± 10.38 |
| Principal piece defects (%) | 25.11 ± 7.65 | 23.84 ± 6.84 | 24.1 ± 7.75 | 24.56 ± 8.48 |
| Teratozoospermic index | 1.72 ± 0.15 | 1.69 ± 0.16 | 1.70 ± 0.16 | 1.69 ± 0.17 |
| miRNA | Log2 (Fold Change) | p-Value | Previously Identified in Human Sperm |
|---|---|---|---|
| >40 years vs. 36–40 years | |||
| hsa-miR-6860 | 5.650 | 0.0001 | No |
| hsa-miR-148b-5p | 3.884 | 0.0079 | Yes [43,44,45] |
| >40 years vs. 31–35 years | |||
| hsa-miR-451a | 5.540 | 0.0011 | Yes [44,45,46,47,48,49] |
| hsa-miR-499c-3p | 3.884 | 0.0182 | No |
| >40 years vs. ≤30 years | |||
| hsa-miR-451a | 4.809 | 0.0056 | Yes [44,45,46,47,48,49] |
| hsa-miR-4301 | −3.047 | 0.0084 | Yes [47] |
| hsa-miR-548f-3p | 4.091 | 0.0099 | No |
| hsa-miR-363-5p | −2.419 | 0.0245 | Yes [44,45] |
| hsa-miR-339-5p | −2.801 | 0.0462 | Yes [44,45,48] |
| 36–40 years vs. 31–35 years | |||
| hsa-miR-6860 | −4.756 | 0.0019 | No |
| hsa-miR-874-5p | −4.522 | 0.0035 | No |
| hsa-miR-449c-3p | 3.969 | 0.0132 | No |
| hsa-miR-371b-3p | −3.321 | 0.0139 | Yes [44,45] |
| hsa-miR-148b-5p | −3.059 | 0.0439 | Yes [43,44,45] |
| 36–40 years vs. ≤30 years | |||
| hsa-miR-6860 | −5.903 | 6.61 × 10−5 | No |
| hsa-miR-148b-5p | −4.464 | 0.0015 | Yes [43,44,45] |
| hsa-miR-1290 | −2.955 | 0.0223 | Yes [43] |
| hsa-miR-425-3p | −1.948 | 0.0280 | Yes [43,44,45,47,48] |
| hsa-miR-874-5p | −3.541 | 0.0327 | No |
| hsa-miR-4423-5p | 2.757 | 0.0460 | Yes [45] |
| hsa-miR-6749-5p | −3.648 | 0.0498 | Yes [47] |
| 31–35 years vs. ≤30 years | |||
| hsa-miR-449c-3p | −4.702 | 0.0023 | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santiago, J.; Silva, J.V.; Santos, M.A.S.; Fardilha, M. Age-Dependent Alterations in Semen Parameters and Human Sperm MicroRNA Profile. Biomedicines 2023, 11, 2923. https://doi.org/10.3390/biomedicines11112923
Santiago J, Silva JV, Santos MAS, Fardilha M. Age-Dependent Alterations in Semen Parameters and Human Sperm MicroRNA Profile. Biomedicines. 2023; 11(11):2923. https://doi.org/10.3390/biomedicines11112923
Chicago/Turabian StyleSantiago, Joana, Joana V. Silva, Manuel A. S. Santos, and Margarida Fardilha. 2023. "Age-Dependent Alterations in Semen Parameters and Human Sperm MicroRNA Profile" Biomedicines 11, no. 11: 2923. https://doi.org/10.3390/biomedicines11112923
APA StyleSantiago, J., Silva, J. V., Santos, M. A. S., & Fardilha, M. (2023). Age-Dependent Alterations in Semen Parameters and Human Sperm MicroRNA Profile. Biomedicines, 11(11), 2923. https://doi.org/10.3390/biomedicines11112923







