CCR8 as a Therapeutic Novel Target: Omics-Integrated Comprehensive Analysis for Systematically Prioritizing Indications
Abstract
:1. Introduction
2. Materials and Methods
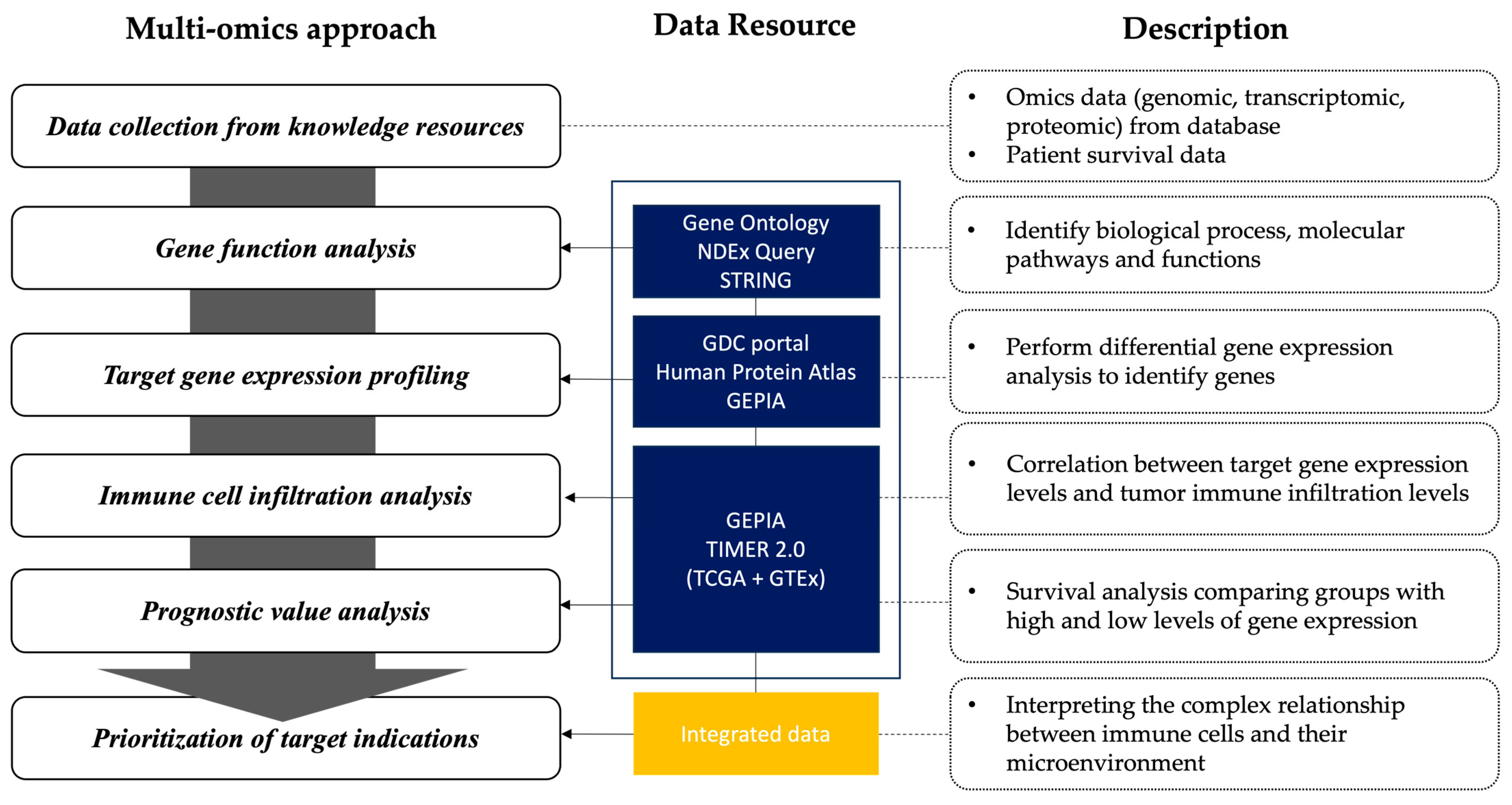
2.1. Framework for Omics-Integrated Analysis
2.2. Gene Function Analysis
2.3. Target Gene Expression Profiling
2.4. Immune Cell Infiltration Analysis
2.5. Prognostic Value Analysis
2.6. Prioritization of Target Indications
3. Results
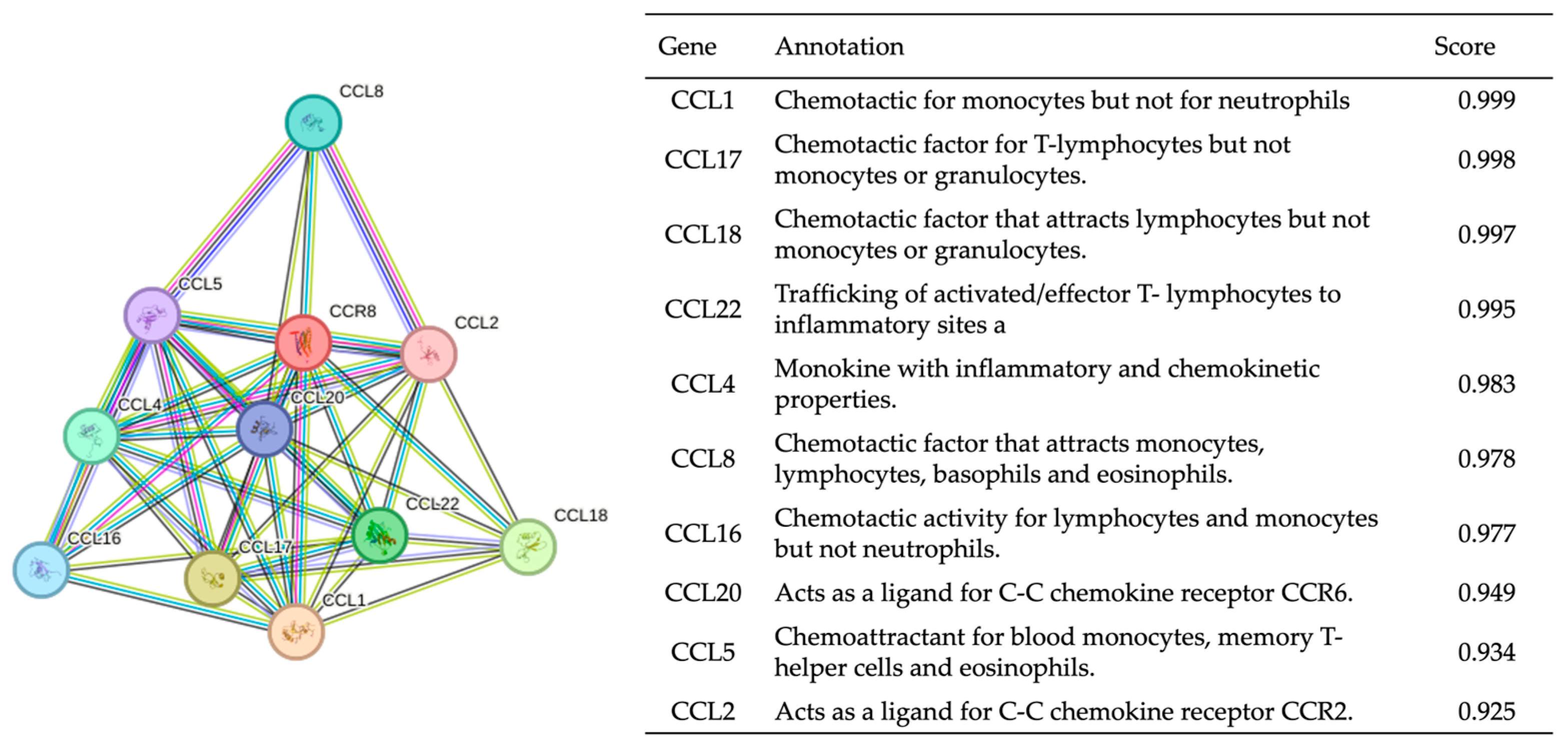
3.1. Gene Function Analysis
3.2. Target Gene Expression Profiling
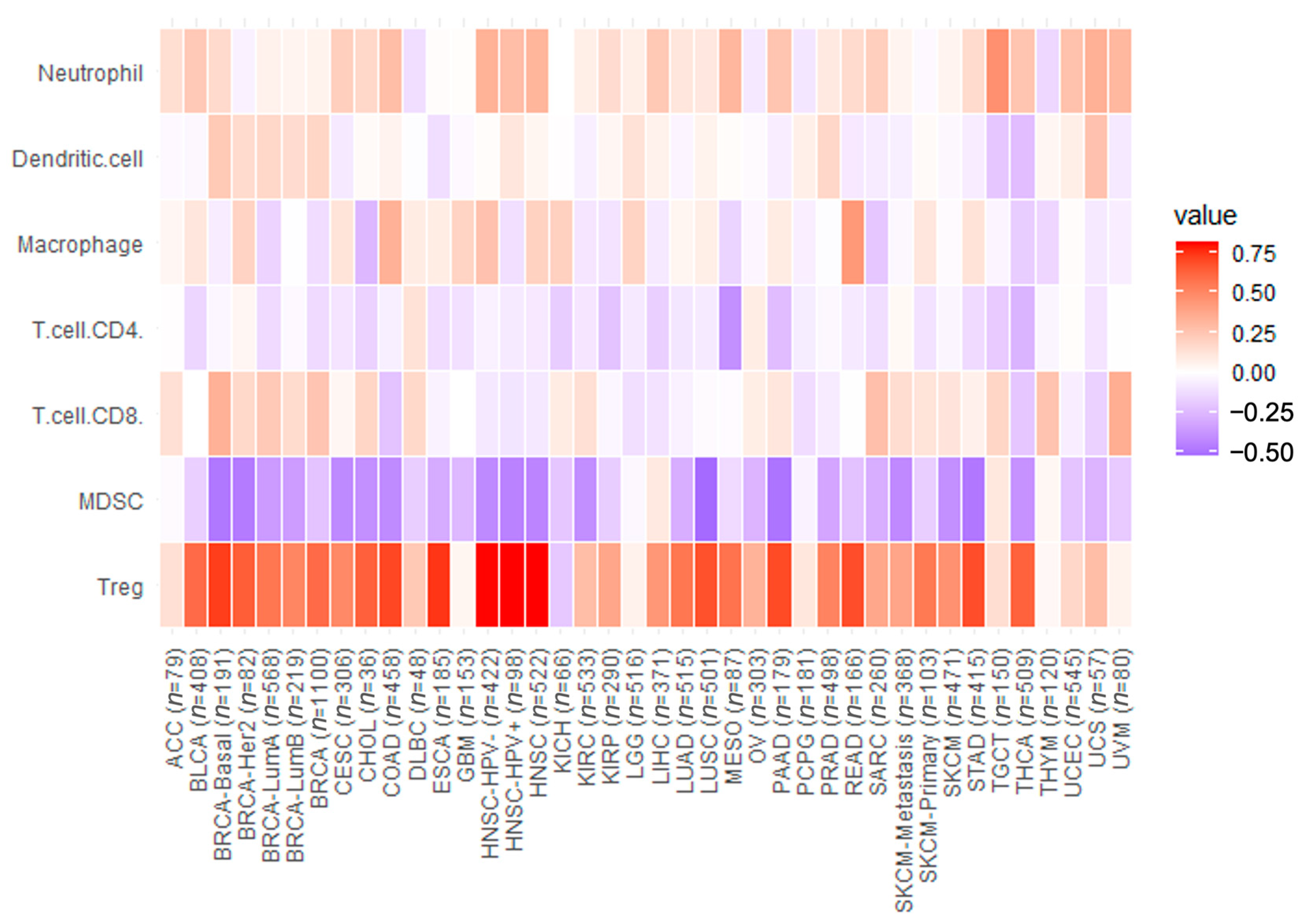
3.3. Immune Cell Infiltration Analysis
3.3.1. Correlation of CCR8 Level with Tumor Immune Cell Infiltration Level
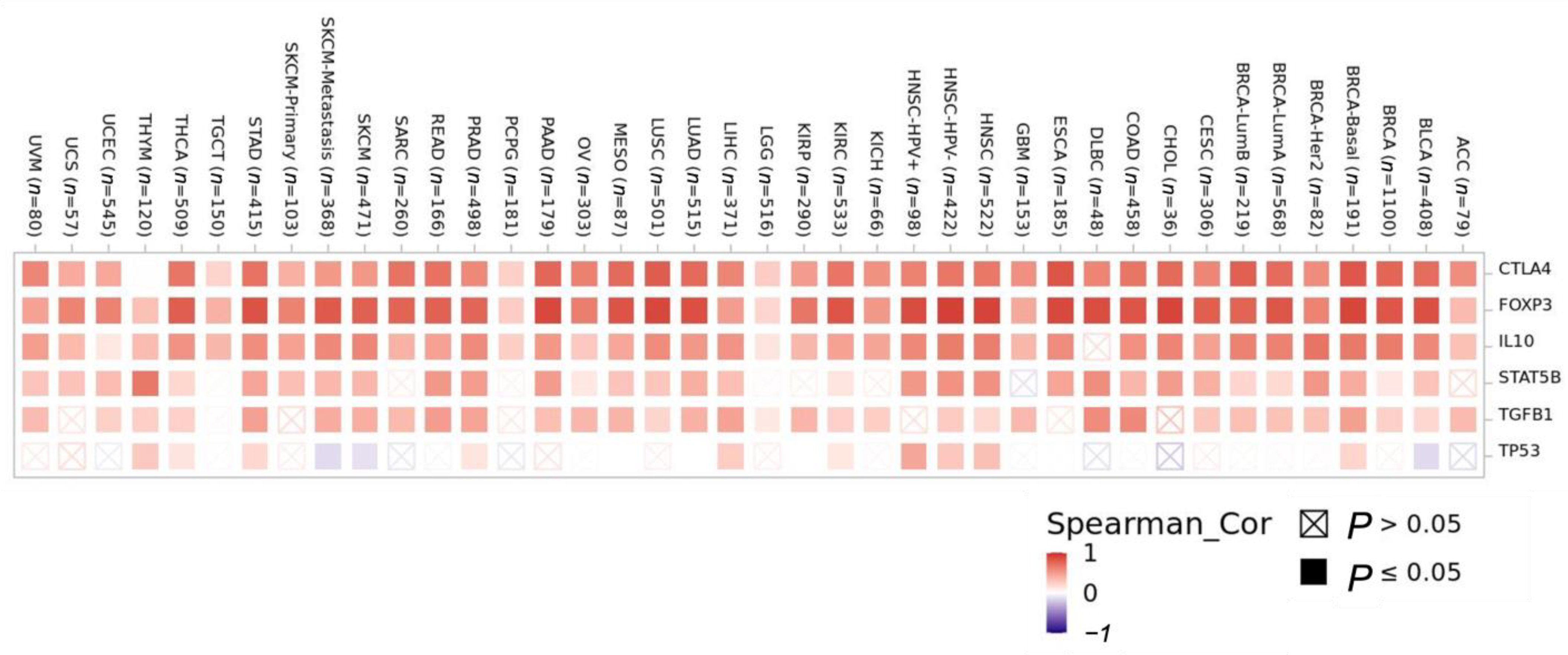
3.3.2. Correlation Analysis of CCR8 Expression Levels with Levels of Tumor-Infiltrating Immune Cell Subgroups
3.3.3. Cancer-Associated Fibroblast (CAF) Level Correlations with CCR8 Expression Level
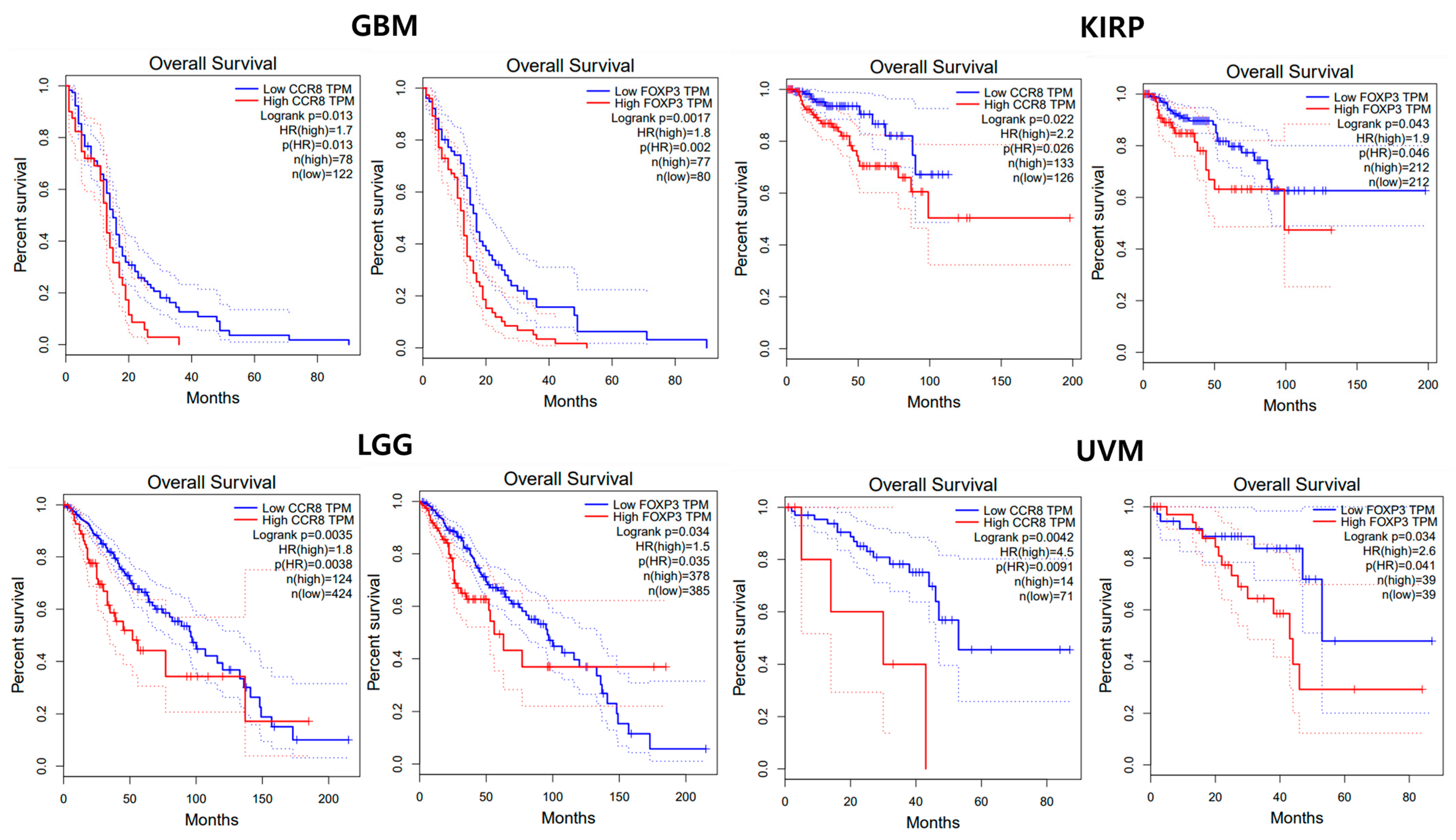
3.4. Prognostic Value Analysis
3.4.1. Univariate Analysis
3.4.2. Multivariate Analysis
3.5. Clinical Trials
3.6. Prioritization of Target Indications
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Qu, C.; Edwards, E.W.; Tacke, F.; Angeli, V.; Llodrá, J.; Sanchez-Schmitz, G.; Garin, A.; Haque, N.S.; Peters, W.; van Rooijen, N.; et al. Role of CCR8 and other chemokine pathways in the migration of monocyte-derived dendritic cells to lymph nodes. J. Exp. Med. 2004, 200, 1231–1241. [Google Scholar] [CrossRef] [PubMed]
- Kidani, Y.; Nogami, W.; Yasumizu, Y.; Kawashima, A.; Tanaka, A.; Sonoda, Y.; Tona, Y.; Nashiki, K.; Matsumoto, R.; Hagiwara, M.; et al. CCR8-targeted specific depletion of clonally expanded Treg cells in tumor tissues evokes potent tumor immunity with long-lasting memory. Proc. Natl. Acad. Sci. USA 2022, 119, e2114282119. [Google Scholar] [PubMed]
- Moser, B. Chemokine Receptor-Targeted Therapies: Special Case for CCR8. Cancers 2022, 14, 511. [Google Scholar] [PubMed]
- Whiteside, S.K.; Grant, F.M.; Gyori, D.S.; Conti, A.G.; Imianowski, C.J.; Kuo, P.; Nasrallah, R.; Sadiyah, F.; Lira, S.A.; Tacke, F.; et al. CCR8 marks highly suppressive Treg cells within tumours but is dispensable for their accumulation and suppressive function. Immunology 2021, 163, 512–520. [Google Scholar] [PubMed]
- Dominguez-Villar, M.; Hafler, D.A. Regulatory T cells in autoimmune disease. Nat. Immunol. 2018, 19, 665–673. [Google Scholar]
- Zheng, D.; Wang, X.; Cheng, L.; Qin, L.; Jiang, Z.; Zhao, R.; Li, Y.; Shi, J.; Wu, Q.; Long, Y.; et al. The Chemokine Receptor CCR8 Is a Target of Chimeric Antigen T Cells for Treating T Cell Malignancies. Front. Immunol. 2022, 13, 808347. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Nanamiya, R.; Takei, J.; Nakamura, T.; Yanaka, M.; Hosono, H.; Sano, M.; Asano, T.; Kaneko, M.K.; Kato, Y. Development of Anti-Mouse CC Chemokine Receptor 8 Monoclonal Antibodies for Flow Cytometry. Monoclon. Antib. Immunodiagn. Immunother. 2021, 40, 65–70. [Google Scholar] [CrossRef]
- Campbell, J.R.; McDonald, B.R.; Mesko, P.B.; Siemers, N.O.; Singh, P.B.; Selby, M.; Sproul, T.W.; Vlach, L.M.; Houser, J.; Sambanthamoorthy, S.; et al. Fc-Optimized Anti-CCR8 Antibody Depletes Regulatory T Cells in Human Tumor Models. Cancer Res. 2021, 81, 2983–2994. [Google Scholar] [CrossRef]
- Paananen, J.; Fortino, V. An omics perspective on drug target discovery platforms. Brief. Bioinform. 2020, 21, 1937–1953. [Google Scholar]
- Lindsay, M.A. Target discovery. Nat. Rev. Drug Discov. 2003, 2, 831–838. [Google Scholar]
- He, Y.; Dai, X.; Chen, Y.; Huang, S. Comprehensive Analysis of Genomic and Expression Data Identified Potential Markers for Predicting Prognosis and Immune Response in CRC. Genet. Res. 2022, 2022, 1831211. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, W.; Zhang, P.; Sun, W.; Han, Y.; Li, L. A comprehensive analysis of copy number variations in diverse apple populations. BMC Genom. 2023, 24, 256. [Google Scholar] [CrossRef] [PubMed]
- Chinnappan, J.; Ramu, A.; V, V.R.; S, A.K. Integrative Bioinformatics approaches to therapeutic gene target selection in various cancers for Nitroglycerin. Sci. Rep. 2021, 11, 22036. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Xi, J.; Li, Y.; Li, Z.; Zhang, Y.; Wang, J.; Fan, G.-H. Discovery of a Potent and Selective CCR8 Small Molecular Antagonist IPG7236 for the Treatment of Cancer. J. Med. Chem. 2023, 66, 4548–4564. [Google Scholar] [CrossRef]
- Panina-Bordignon, P.; Papi, A.; Mariani, M.; Di Lucia, P.; Casoni, G.; Bellettato, C.; Buonsanti, C.; Miotto, D.; Mapp, C.; Villa, A.; et al. The C-C chemokine receptors CCR4 and CCR8 identify airway T cells of allergen-challenged atopic asthmatics. J. Clin. Investig. 2001, 107, 1357–1364. [Google Scholar] [CrossRef]
- Nagira, Y.; Nagira, M.; Nagai, R.; Nogami, W.; Hirata, M.; Ueyama, A.; Yoshida, T.; Yoshikawa, M.; Shinonome, S.; Yoshida, H.; et al. S-531011, a Novel Anti-Human CCR8 Antibody, Induces Potent Antitumor Responses through Depletion of Tumor-Infiltrating CCR8-Expressing Regulatory T Cells. Mol. Cancer Ther. 2023, 22, 1063–1072. [Google Scholar] [CrossRef]
- Islam, M.A.; Kibria, M.K.; Hossen, M.B.; Reza, M.S.; Tasmia, S.A.; Tuly, K.F.; Mosharof, P.; Kabir, S.R.; Kabir, H.; Mollah, N.H. Bioinformatics-based investigation on the genetic influence between SARS-CoV-2 infections and idiopathic pulmonary fibrosis (IPF) diseases, and drug repurposing. Sci. Rep. 2023, 13, 4685. [Google Scholar] [CrossRef]
- Isaza, C.; Rosas, J.F.; Lorenzo, E.; Marrero, A.; Ortiz, C.; Ortiz, M.R.; Perez, L.; Cabrera-Ríos, M. Biological signaling pathways and potential mathematical network representations: Biological discovery through optimization. Cancer Med. 2018, 7, 1875–1895. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Karimizadeh, E.; Sharifi-Zarchi, A.; Nikaein, H.; Salehi, S.; Salamatian, B.; Elmi, N.; Gharibdoost, F.; Mahmoudi, M. Analysis of gene expression profiles and protein-protein interaction networks in multiple tissues of systemic sclerosis. BMC Med. Genom. 2019, 12, 199. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Chen, T.; Lu, J.; Yan, J.; Zhang, Y.; Qin, P.; Ding, S.; Zhou, Y. Integrated Protein-Protein Interaction and Weighted Gene Co-expression Network Analysis Uncover Three Key Genes in Hepatoblastoma. Front. Cell Dev. Biol. 2021, 9, 631982. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Thul, P.J.; Lindskog, C. The human protein atlas: A spatial map of the human proteome. Protein Sci. 2018, 27, 233–244. [Google Scholar] [CrossRef]
- Leclezio, L.; Jansen, A.; Whittemore, V.H.; de Vries, P.J. Pilot validation of the tuberous sclerosis-associated neuropsychiatric disorders (TAND) checklist. Pediatr. Neurol. 2015, 52, 16–24. [Google Scholar] [CrossRef]
- Gui, T.; Yao, C.; Jia, B.; Shen, K. Identification and analysis of genes associated with epithelial ovarian cancer by integrated bioinformatics methods. PLoS ONE 2021, 16, e0253136. [Google Scholar] [CrossRef]
- Fox, J.M.; Najarro, P.; Smith, G.L.; Struyf, S.; Proost, P.; Pease, J.E. Structure/function relationships of CCR8 agonists and antagonists. Amino-terminal extension of CCL1 by a single amino acid generates a partial agonist. J. Biol. Chem. 2006, 281, 36652–36661. [Google Scholar] [CrossRef]
- Miller, H.E.; Bishop, A.J.R. Correlation AnalyzeR: Functional predictions from gene co-expression correlations. BMC Bioinform. 2021, 22, 206. [Google Scholar] [CrossRef] [PubMed]
- von Mering, C.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef] [PubMed]
- Reimand, J.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C.; et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019, 14, 482–517. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, H.; Dombrecht, B.; Kiss, M.; Roose, H.; Allen, E.; Van Overmeire, E.; Kancheva, D.; Martens, L.; Murgaski, A.; Bardet, P.M.R.; et al. Therapeutic depletion of CCR8(+) tumor-infiltrating regulatory T cells elicits antitumor immunity and synergizes with anti-PD-1 therapy. J. Immunother. Cancer 2021, 9, e001749. [Google Scholar] [CrossRef]
- Zhang, S.-C.; Hu, Z.-Q.; Long, J.-H.; Zhu, G.-M.; Wang, Y.; Jia, Y.; Zhou, J.; Ouyang, Y.; Zeng, Z. Clinical Implications of Tumor-Infiltrating Immune Cells in Breast Cancer. J. Cancer 2019, 10, 6175–6184. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chen, J.; Su, Z. KIF4A is a promising prognostic marker and correlates with immune infiltration in clear cell renal cell carcinoma. Transl. Cancer Res. 2020, 9, 7165–7173. [Google Scholar] [CrossRef]
- Lou, S.; Zhang, J.; Yin, X.; Zhang, Y.; Fang, T.; Wang, Y.; Xue, Y. Comprehensive Characterization of Tumor Purity and Its Clinical Implications in Gastric Cancer. Front. Cell Dev. Biol. 2022, 9, 782529. [Google Scholar] [CrossRef]
- Li, C.; Jiang, P.; Wei, S.; Xu, X.; Wang, J. Regulatory T cells in tumor microenvironment: New mechanisms, potential therapeutic strategies and future prospects. Mol. Cancer 2020, 19, 116. [Google Scholar] [CrossRef]
- Shang, J.; Song, Q.; Yang, Z.; Sun, X.; Xue, M.; Chen, W.; Yang, J.; Wang, S. Analysis of PD-1 related immune transcriptional profile in different cancer types. Cancer Cell Int. 2018, 18, 218. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, Z.; Ping, Y.; Qin, G.; Zhang, K.; Maimela, N.R.; Huang, L.; Yang, S.; Zhang, Y. Comprehensive Analysis of PD-1 Gene Expression, Immune Characteristics and Prognostic Significance in 1396 Glioma Patients. Cancer Manag. Res. 2020, 12, 4399–4410. [Google Scholar] [CrossRef]
- Walsh, R.J.; Sundar, R.; Lim, J.S.J. Immune checkpoint inhibitor combinations-current and emerging strategies. Br. J. Cancer 2023, 128, 1415–1417. [Google Scholar] [CrossRef] [PubMed]
- Giustiniani, J.; Dobos, G.; Moins-Teisserenc, H.; Eustaquio, T.; Battistella, M.; Ortonne, N.; Ram-Wolff, C.; Bouaziz, J.-D.; Marie-Cardine, A.; Mourah, S.; et al. CCR8 is a new therapeutic target in cutaneous T-cell lymphomas. Blood Adv. 2022, 6, 3507–3512. [Google Scholar] [CrossRef] [PubMed]
- Duvic, M.; Evans, M.; Wang, C. Mogamulizumab for the treatment of cutaneous T-cell lymphoma: Recent advances and clinical potential. Ther. Adv. Hematol. 2016, 7, 171–174. [Google Scholar] [CrossRef] [PubMed]
- Kamada, Y.; Arima, N.; Hayashida, M.; Nakamura, D.; Yoshimitsu, M.; Ishitsuka, K. Prediction of the risk for graft versus host disease after allogeneic hematopoietic stem cell transplantation in patients treated with mogamulizumab. Leuk. Lymphoma 2022, 63, 1701–1707. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.-M.; Hwang, Y.-C.; Liu, I.-J.; Lee, C.-C.; Tsai, H.-Z.; Li, H.-J.; Wu, H.-C. Development of therapeutic antibodies for the treatment of diseases. J. Biomed. Sci. 2020, 27, 1. [Google Scholar] [CrossRef]
- Zhong, L.; Li, Y.; Xiong, L.; Wang, W.; Wu, M.; Yuan, T.; Yang, W.; Tian, C.; Miao, Z.; Wang, T.; et al. Small molecules in targeted cancer therapy: Advances, challenges, and future perspectives. Signal Transduct. Target. Ther. 2021, 6, 201. [Google Scholar] [CrossRef]
- Bonaventura, P.; Shekarian, T.; Alcazer, V.; Valladeau-Guilemond, J.; Valsesia-Wittmann, S.; Amigorena, S.; Caux, C.; Depil, S. Cold Tumors: A Therapeutic Challenge for Immunotherapy. Front. Immunol. 2019, 10, 168. [Google Scholar] [CrossRef]
- Wang, M.; Wang, S.; Desai, J.; Trapani, J.A.; Neeson, P.J. Therapeutic strategies to remodel immunologically cold tumors. Clin. Transl. Immunol. 2020, 9, e1226. [Google Scholar] [CrossRef]
- Karin, N. Chemokines in the Landscape of Cancer Immunotherapy: How They and Their Receptors Can Be Used to Turn Cold Tumors into Hot Ones? Cancers 2021, 13, 6317. [Google Scholar] [CrossRef]




| Abbreviation | Cancer Type |
|---|---|
| LAML | Acute Myeloid Leukemia |
| ACC | Adrenocortical carcinoma |
| BLCA | Bladder Urothelial Carcinoma |
| LGG | Brain Lower Grade Glioma |
| BRCA | Breast invasive carcinoma |
| CESC | Cervical squamous cell carcinoma and endocervical adenocarcinoma |
| CHOL | Cholangiocarcinoma |
| LCML | Chronic Myelogenous Leukemia |
| COAD | Colon adenocarcinoma |
| ESCA | Esophageal carcinoma |
| GBM | Glioblastoma multiforme |
| HNSC | Head and Neck squamous cell carcinoma |
| KICH | Kidney Chromophobe |
| KIRC | Kidney renal clear cell carcinoma |
| KIRP | Kidney renal papillary cell carcinoma |
| LIHC | Liver hepatocellular carcinoma |
| LUAD | Lung adenocarcinoma |
| LUSC | Lung squamous cell carcinoma |
| DLBC | Lymphoid Neoplasm Diffuse Large B-cell Lymphoma |
| MESO | Mesothelioma |
| MISC | Miscellaneous |
| OV | Ovarian serous cystadenocarcinoma |
| PAAD | Pancreatic adenocarcinoma |
| PCPG | Pheochromocytoma and Paraganglioma |
| READ | Rectum adenocarcinoma |
| PRAD | Prostate adenocarcinoma |
| SARC | Sarcoma |
| SKCM | Skin Cutaneous Melanoma |
| STAD | Stomach adenocarcinoma |
| TGCT | Testicular Germ Cell Tumors |
| THYM | Thymoma |
| THCA | Thyroid carcinoma |
| UCS | Uterine Carcinosarcoma |
| UCEC | Uterine Corpus Endometrial Carcinoma |
| UVM | Uveal Melanoma |
| Gene Expression Analysis | Immune Cell Infiltration Analysis | Prognostic Value Analysis | Overall Evaluation | |||
|---|---|---|---|---|---|---|
| TIICs | Treg | CAF | Uni | Multi | ||
| High expression | Strong correlation in two or more items | Correlation of one or more items | Strong | |||
| Strong correlation in two or more items | Not applicable | Moderate | ||||
| Strong correlation in only one items | Not applicable | Low | ||||
| Category Domain | Gene Ontology Term |
|---|---|
| Biological Process | Immune response |
| Cell adhesion | |
| G protein-coupled receptor signaling pathway | |
| Chemokine-mediated signaling pathway | |
| Positive regulation of cytosolic calcium ion concentration | |
| Chemotaxis | |
| Cellular Component | Plasma membrane |
| Molecular Function | Coreceptor activity |
| C-C chemokine receptor activity | |
| Chemokine receptor activity |
| Description | Tregs | Description | Tregs | ||
|---|---|---|---|---|---|
| Rho | p-Value | Rho | p-Value | ||
| ACC (n = 79) | 0.14 | 0.30 | LIHC (n = 371) | 0.44 *** | <0.001 |
| BLCA (n = 408) | 0.60 *** | <0.001 | LUAD (n = 515) | 0.56 *** | <0.001 |
| BRCA (n = 1100) | 0.60 *** | <0.001 | LUSC (n = 501) | 0.68 *** | <0.001 |
| BRCA-Basal (n = 191) | 0.72 *** | <0.001 | MESO (n = 87) | 0.57 *** | <0.001 |
| BRCA-Her2 (n = 82) | 0.64 *** | <0.001 | OV (n = 303) | 0.33 ** | <0.001 |
| BRCA-LumA (n = 568) | 0.56 *** | <0.001 | PAAD (n = 179) | 0.69 *** | <0.001 |
| BRCA-LumB (n = 219) | 0.50 *** | <0.001 | PCPG (n = 181) | 0.11 | 0.22 |
| CESC (n = 306) | 0.49 *** | <0.001 | PRAD (n = 498) | 0.51 *** | <0.001 |
| CHOL (n = 36) | 0.63 *** | <0.001 | READ (n = 166) | 0.68 *** | <0.001 |
| COAD (n = 458) | 0.70 *** | <0.001 | SARC (n = 260) | 0.37 ** | <0.001 |
| DLBC (n = 48) | 0.23 * | 0.19 | SKCM (n = 471) | 0.46 *** | <0.001 |
| ESCA (n = 185) | 0.74 *** | <0.001 | SKCM-Metastasis (n = 368) | 0.38 ** | <0.001 |
| GBM (n = 153) | 0.04 | 0.70 | SKCM-Primary (n = 103) | 0.55 *** | <0.001 |
| HNSC (n = 522) | 0.81 *** | <0.001 | STAD (n = 415) | 0.68 *** | <0.001 |
| HNSC-HPV− (n = 422) | 0.81 *** | <0.001 | TGCT (n = 150) | 0.14 | 0.12 |
| HNSC-HPV+ (n = 98) | 0.81 *** | <0.001 | THCA (n = 509) | 0.62 *** | <0.001 |
| KICH (n = 66) | −0.19 | 0.17 | THYM (n = 120) | 0.03 | 0.77 |
| KIRC (n = 533) | 0.28 * | <0.001 | UCEC (n = 545) | 0.17 | 0.15 |
| KIRP (n = 290) | 0.38 ** | <0.001 | UCS (n = 57) | 0.28 | 0.07 |
| LGG (n = 516) | 0.06 | 0.25 | UVM (n = 80) | 0.05 | 0.70 |
| Cancer Type | HR (High vs. Low) | p-Value | No. of Patients (High/Low) |
|---|---|---|---|
| ACC | 0.90 | 0.850 | 19/64 |
| BLCA | 0.85 | 0.430 | 325/341 |
| BRCA | 0.95 | 0.740 | 532/533 |
| CESC | 0.79 | 0.320 | 146/142 |
| CHOL | 0.58 | 0.330 | 16/16 |
| COAD | 0.80 | 0.370 | 134/135 |
| DLBC | 1.50 | 0.590 | 22/21 |
| ESCA | 1.20 | 0.430 | 91/91 |
| GBM | 1.70 | 0.013 * | 78/122 |
| HNSC | 0.68 | 0.019 | 389/389 |
| KICH | 0.89 | 0.860 | 29/31 |
| KIRC | 1.20 | 0.230 | 251/256 |
| KIRP | 2.20 | 0.026 * | 133/126 |
| LAML | 1.00 | 0.970 | 52/50 |
| LGG | 1.80 | 0.004 ** | 124/424 |
| LIHC | 0.83 | 0.560 | 244/327 |
| LAUD | 0.80 | 0.150 | 237/239 |
| LUSC | 1.20 | 0.210 | 240/236 |
| MESO | 1.10 | 0.810 | 39/41 |
| OV | 0.81 | 0.095 | 211/210 |
| PADD | 0.84 | 0.400 | 87/88 |
| PCPG | 2.90 | 0.240 | 71/124 |
| PRAD | 1.00 | 1.000 | 240/233 |
| READ | 0.70 | 0.460 | 46/46 |
| SARC | 0.95 | 0.810 | 122/127 |
| SKCM | 0.55 | 1.2 × 10−5 | 225/228 |
| STAD | 1.00 | 0.820 | 192/192 |
| TGCT | 5.63 × 108 | 1.000 | 68/67 |
| THCA | 1.10 | 0.840 | 254/250 |
| THYM | 1.30 | 0.750 | 58/53 |
| UCEC | 1.30 | 0.480 | 83/75 |
| UCS | 0.64 | 0.200 | 25/24 |
| UVM | 4.50 | 0.0091 ** | 14/71 |
| Covariate | HR (95% CIs) | p-Value | Covariate | HR (95% CIs) | p-Value |
|---|---|---|---|---|---|
| BRCA | COAD | ||||
| CCR8 expression | 1.29 (1.01–1.64) | 0.037 * | CCR8 expression | 0.39 (0.17–0.91) | 0.029 * |
| B-cell | 0.48 (0.00–34.34) | 0.739 | B-cell | 1.27 (0.01–143.46) | 0.920 |
| CD8+ T-cell | 0.36 (0.03–3.87) | 0.399 | CD8+ T-cell | 0.02 (0.00–0.98) | 0.049 * |
| CD4+ T-cell | 1.19 (0.02–49.38) | 0.924 | CD4+ T-cell | 0.53 (0.00–79.10) | 0.804 |
| Macrophage | 5.91 (0.41–83.68) | 0.188 | Macrophage | 9.61 (0.08–1149.82) | 0.354 |
| Neutrophil | 7.32 (0.03–1487.46) | 0.463 | Neutrophil | 0.02 (0.00–46.09) | 0.343 |
| Dendritic cell | 0.37 (0.04–2.91) | 0.347 | Dendritic cell | 59.39 (2.29–1537.39) | 0.014 * |
| HNSC | Kidney Chromophobe KICH | ||||
| CCR8 expression | 0.62 (0.43–0.88) | 0.009 ** | CCR8 expression | 1.96 (6.10–6.34) | 0.006 ** |
| B-cell | 0.10 (0.00–1.77) | 0.119 | B-cell | 9.93 (3.68–2.67) | 0.000 *** |
| CD8+ T-cell | 0.24 (0.03–1.77) | 0.164 | CD8+ T-cell | 0.00 (0.00–0.00) | 0.000 *** |
| CD4+ T-cell | 0.23 (0.00–6.67) | 0.394 | CD4+ T-cell | 0.00 (0.00–0.00) | 0.000 *** |
| Macrophage | 10.98 (0.53–223.99) | 0.119 | Macrophage | 4.68 (3.51–6.23) | 0.000 *** |
| Neutrophil | 2.75 (0.11–63.68) | 0.528 | Neutrophil | 4.14 (2.59–6.62) | 0.000 *** |
| Dendritic cell | 3.64 (0.74–17.93) | 0.112 | Dendritic cell | 0.00 (0.00–0.00) | 0.000 *** |
| LIHC | MESO | ||||
| CCR8 expression | 0.51 (0.15–1.79) | 0.299 | CCR8 expression | 0.72 (0.22–2.35) | 0.593 |
| B-cell | 0.00 (0.00–8.96) | 0.175 | B-cell | 0.15 (0.00–249.47) | 0.617 |
| CD8+ T-cell | 0.00 (0.00–0.26) | 0.012 * | CD8+ T-cell | 2.81 (0.01–472.12) | 0.692 |
| CD4+ T-cell | 0.03 (0.00–30.76) | 0.329 | CD4+ T-cell | 5.97 (0.01–3583.79) | 0.584 |
| Macrophage | 265.66 (1.17–60,085.23) | 0.044 * | Macrophage | 5681.43 (3.82–8,434,211.64) | 0.020 * |
| Neutrophil | 5.32 (0.00–690,216.97) | 0.781 | Neutrophil | 0.00 (0.00–0.00) | 0.000 *** |
| Dendritic cell | 95.65 (2.40–3801.49) | 0.015 * | Dendritic cell | 389.04 (5.41–27,948.15) | 0.006 ** |
| PAAD | OV | ||||
| CCR8 expression | 0.41 (0.19–8.65) | 0.019 * | CCR8 expression | 0.94 (0.80–1.12) | 0.540 |
| B-cell | 7.72 (0.03–1.93) | 0.46844 | B-cell | 0.16 (0.00–64.42) | 0.549 |
| CD8+ T-cell | 44.50 (0.07–2.84) | 0.250 | CD8+ T-cell | 0.04 (0.00–0.00) | 0.000 *** |
| CD4+ T-cell | 0.00 (0.00–2.14) | 0.079 | CD4+ T-cell | 0.00 (0.00–0.00) | 0.000 *** |
| Macrophage | 0.01 (0.00–3.26) | 0.118 | Macrophage | 10,467.65 (48.64–2,252,393.25) | 0.001 ** |
| Neutrophil | 0.01 (0.00–3.26) | 0.015 * | Neutrophil | 8707.68 (1.58–47,760,091.36) | 0.039 * |
| Dendritic cell | 2.76 (0.05–1.32) | 0.601 | Dendritic cell | 0.44 (0.00–44.78) | 0.731 |
| NCT No. | Drug Name | Combination | Conditions | Types | Status |
|---|---|---|---|---|---|
| NCT05537740 | BAY3375968 | Monotherapy vs. Combination with anti-PD-1 | Advanced Solid Tumors | Antibody | Recruiting |
| NCT05635643 | SRF114 | Monotherapy | Advanced Solid Tumors and Head and Neck Squamous Cell Carcinoma | Antibody | Recruiting |
| NCT05101070 | S-531011 | Monotherapy vs. Combination with anti-PD-1 | Advanced Solid Tumors | Antibody | Recruiting |
| NCT05007782 | GS-1811 | Monotherapy vs. Combination with anti-PD-1 | Advanced Solid Tumors | Antibody | Recruiting |
| Gene Expression Analysis | Immune Cell Infiltration Analysis | Prognostic Value Analysis | Overall Evaluation | ||||
|---|---|---|---|---|---|---|---|
| TIICs | Treg | CAF | Uni | Multi | |||
| ACC | |||||||
| BLCA | ** | *** | *** | ||||
| BRCA | *** | *** | *** | ** | *** | ||
| CESC | ** | *** | |||||
| CHOL | ** | *** | |||||
| COAD | *** | *** | *** | ** | ** | *** | |
| DLBC | |||||||
| ESCA | *** | *** | * | ||||
| GBM | ** | ||||||
| HNSC | *** | *** | *** | ** | ** | *** | |
| KICH | ** | ||||||
| KIRC | *** | ** | |||||
| KIRP | ** | ||||||
| LAML | |||||||
| LGG | ** | ||||||
| LIHC | *** | *** | ** | *** | |||
| LUAD | *** | *** | * | ||||
| LUSC | *** | *** | * | ||||
| MESO | *** | ** | |||||
| OV | ** | ||||||
| PAAD | ** | *** | ** | ||||
| PCPG | |||||||
| PRAD | *** | ||||||
| READ | ** | *** | ** | ** | |||
| SARC | |||||||
| SKCM | *** | *** | |||||
| STAD | *** | *** | ** | ** | |||
| TGCT | |||||||
| THCA | ** | *** | ** | ** | |||
| THYM | |||||||
| UCEC | *** | ||||||
| UCS | |||||||
| UVM | ** | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, N.; Kim, M.-H.; Pyo, J.; Lee, S.-M.; Jang, J.-S.; Lee, D.-W.; Kim, K.W. CCR8 as a Therapeutic Novel Target: Omics-Integrated Comprehensive Analysis for Systematically Prioritizing Indications. Biomedicines 2023, 11, 2910. https://doi.org/10.3390/biomedicines11112910
Kim N, Kim M-H, Pyo J, Lee S-M, Jang J-S, Lee D-W, Kim KW. CCR8 as a Therapeutic Novel Target: Omics-Integrated Comprehensive Analysis for Systematically Prioritizing Indications. Biomedicines. 2023; 11(11):2910. https://doi.org/10.3390/biomedicines11112910
Chicago/Turabian StyleKim, Nari, Mi-Hyun Kim, Junhee Pyo, Soo-Min Lee, Ji-Sung Jang, Do-Wan Lee, and Kyung Won Kim. 2023. "CCR8 as a Therapeutic Novel Target: Omics-Integrated Comprehensive Analysis for Systematically Prioritizing Indications" Biomedicines 11, no. 11: 2910. https://doi.org/10.3390/biomedicines11112910
APA StyleKim, N., Kim, M.-H., Pyo, J., Lee, S.-M., Jang, J.-S., Lee, D.-W., & Kim, K. W. (2023). CCR8 as a Therapeutic Novel Target: Omics-Integrated Comprehensive Analysis for Systematically Prioritizing Indications. Biomedicines, 11(11), 2910. https://doi.org/10.3390/biomedicines11112910






