MiR-506 Promotes Antitumor Immune Response in Pancreatic Cancer by Reprogramming Tumor-Associated Macrophages toward an M1 Phenotype
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical Specimens
2.2. Macrophage Analysis
2.3. Macrophage Polarization
2.4. Phagocytosis Assay
2.5. Nitric Oxide (NO) Production
2.6. Arg1 Activity
2.7. VEGF, IL-10, and TGFβ Secretion
2.8. RNA Isolation and qRT-PCR
2.9. Western Blotting
2.10. Reagents and Antibodies
2.11. Orthotopic PDAC Mouse Model
2.12. Statistical Analysis
3. Results
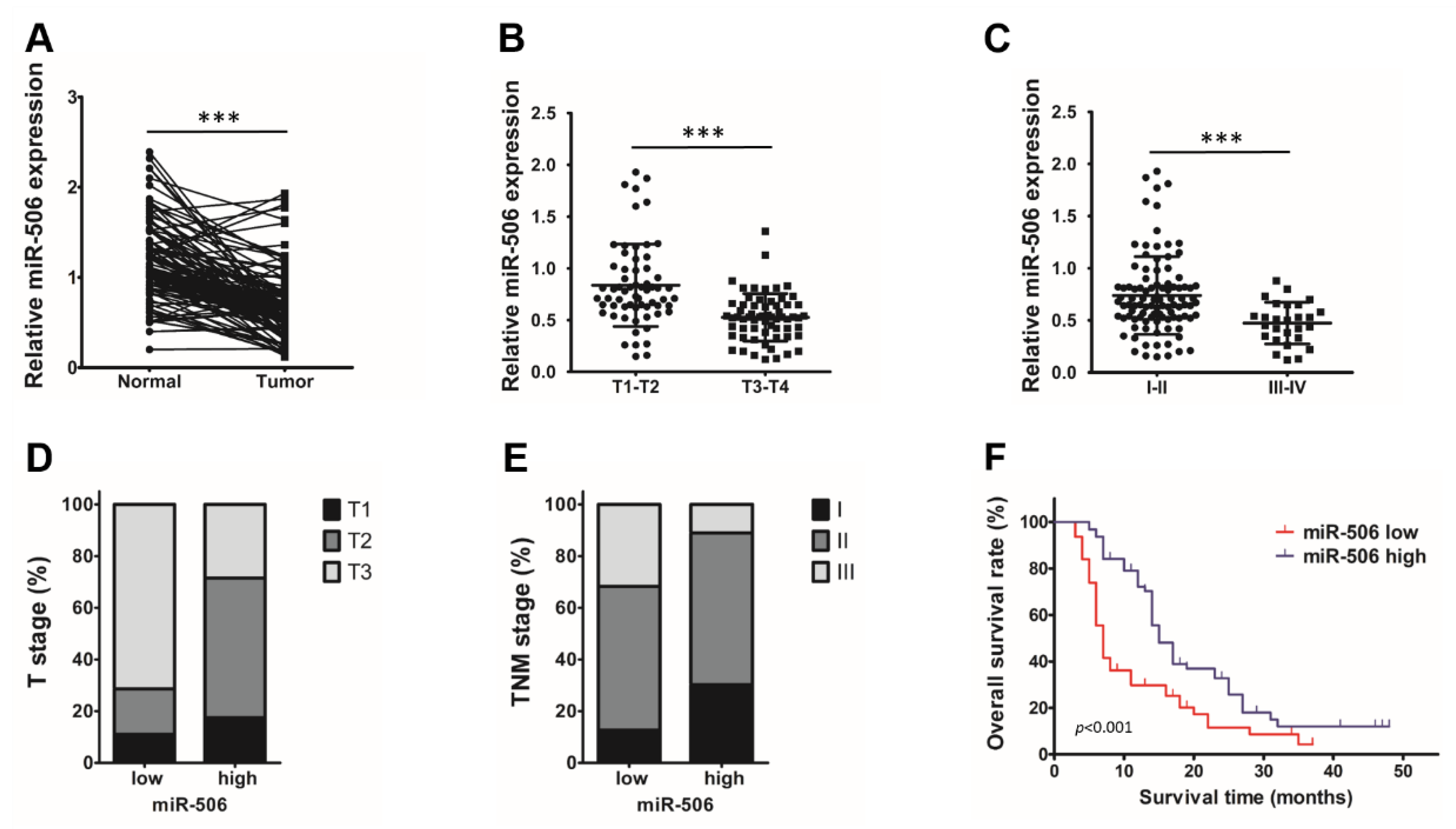
3.1. Low Endogenous miR-506 Levels Are Associated with Poor Outcomes in PDAC Patients
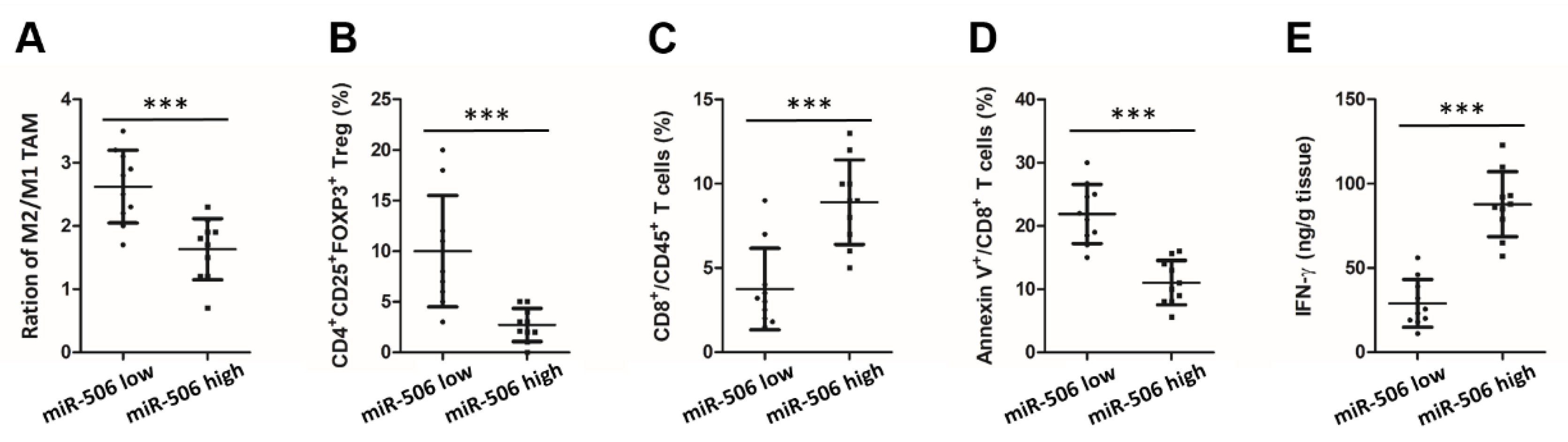
3.2. Low Endogenous miR-506 Levels Are Associated with Immunosuppressive Microenvironment in Human PDAC
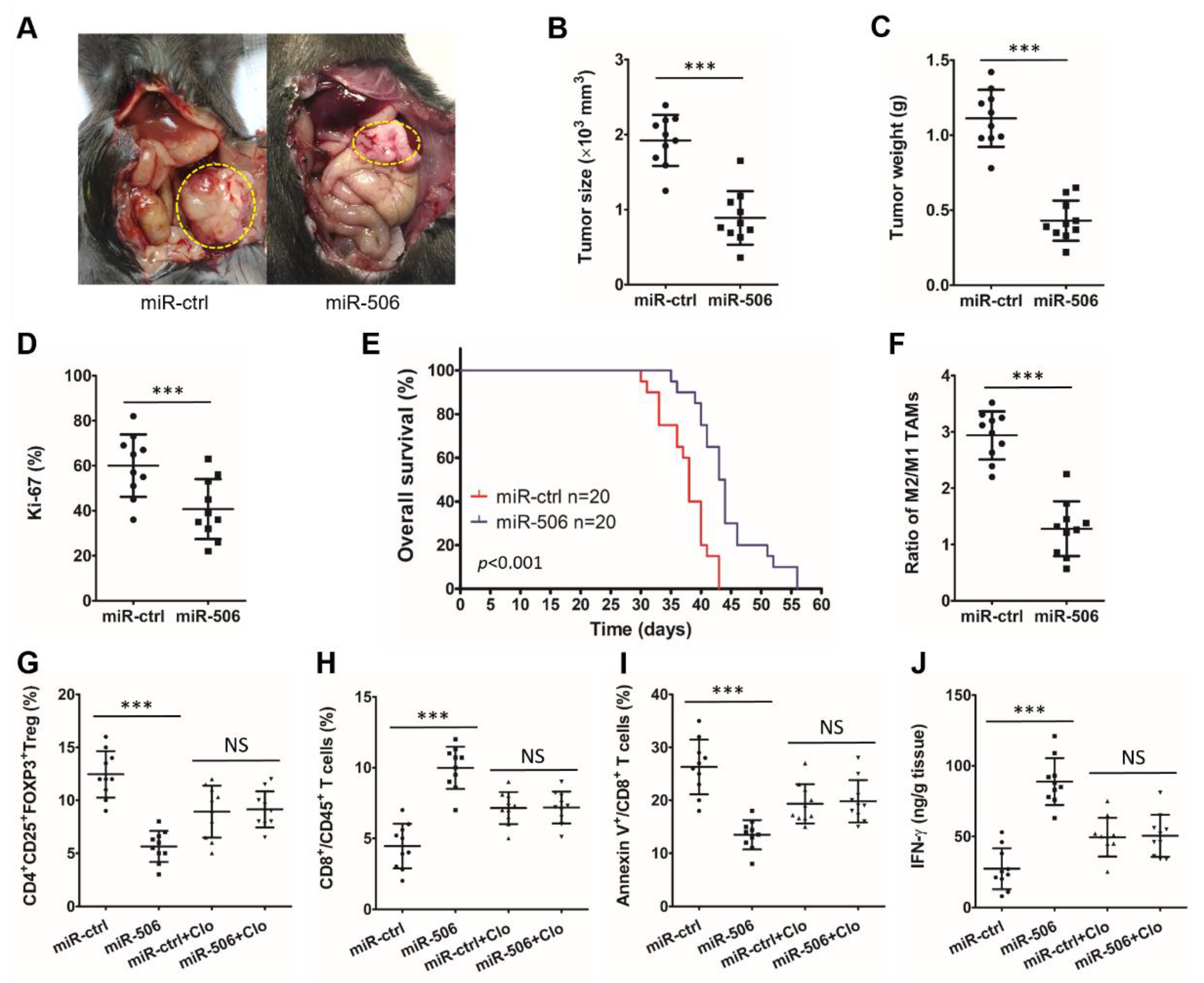
3.3. MiR-506 Inhibits the PDAC Progression and Reverses Its Immunosuppressive Microenvironment in a Macrophage-Dependent Manner
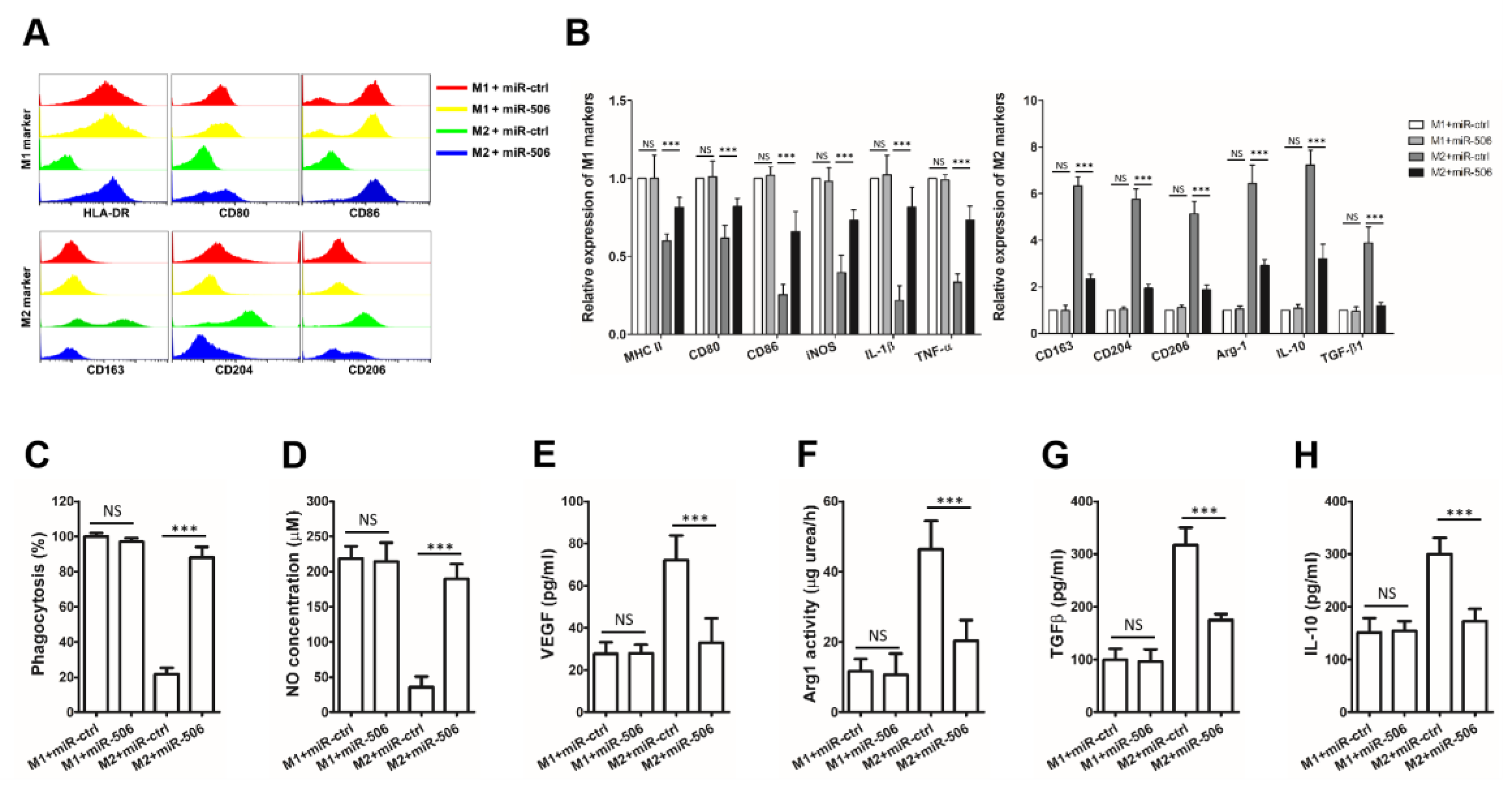
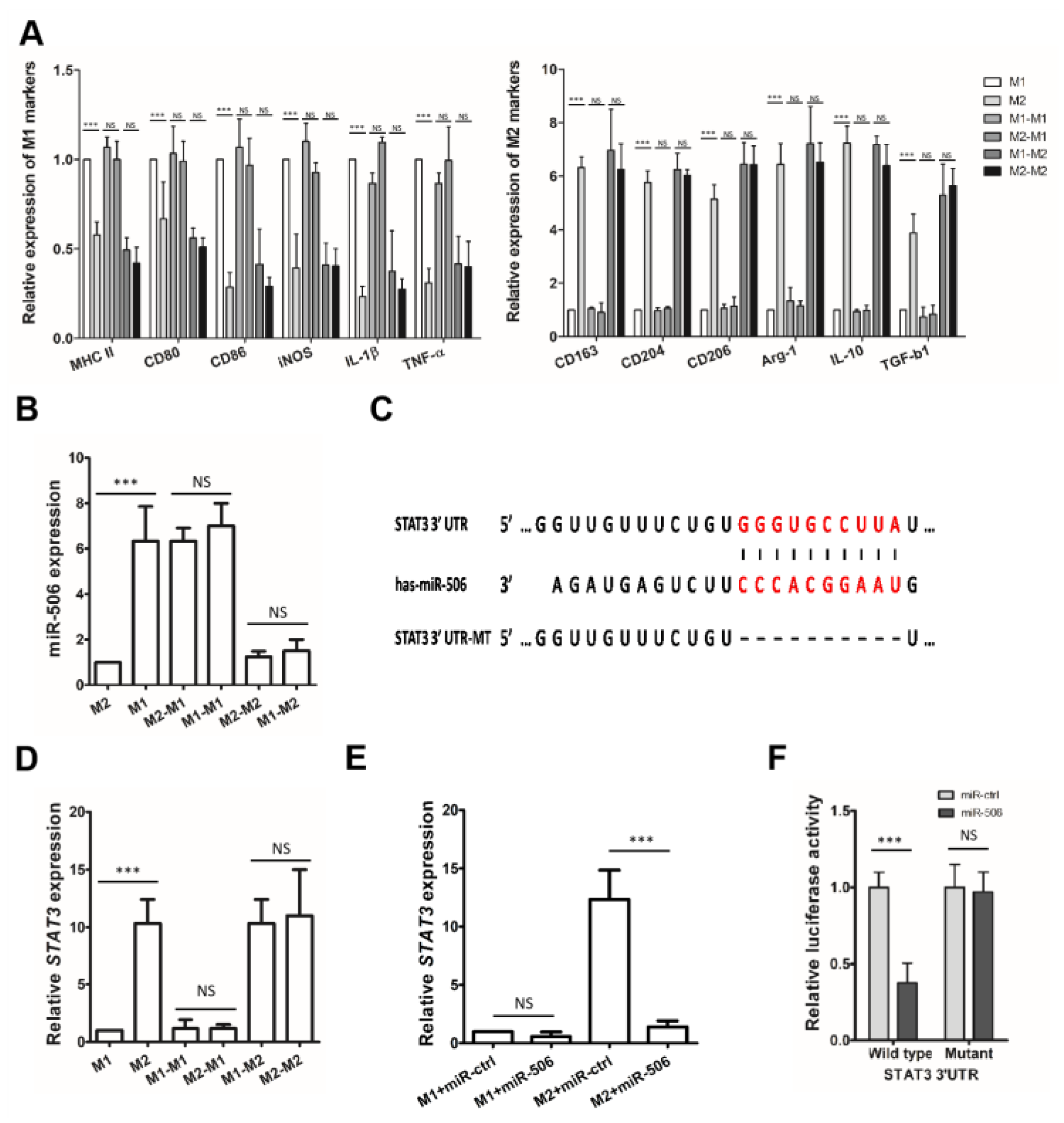
3.4. MiR-506 Reprograms the Polarization of M2 Macrophages
3.5. STAT3 Is a Direct Target of miR-506 Involved in Macrophage Polarization
3.6. MiR-506 Reprograms the Polarization of M2 Macrophage through the STAT3 Signaling Pathway
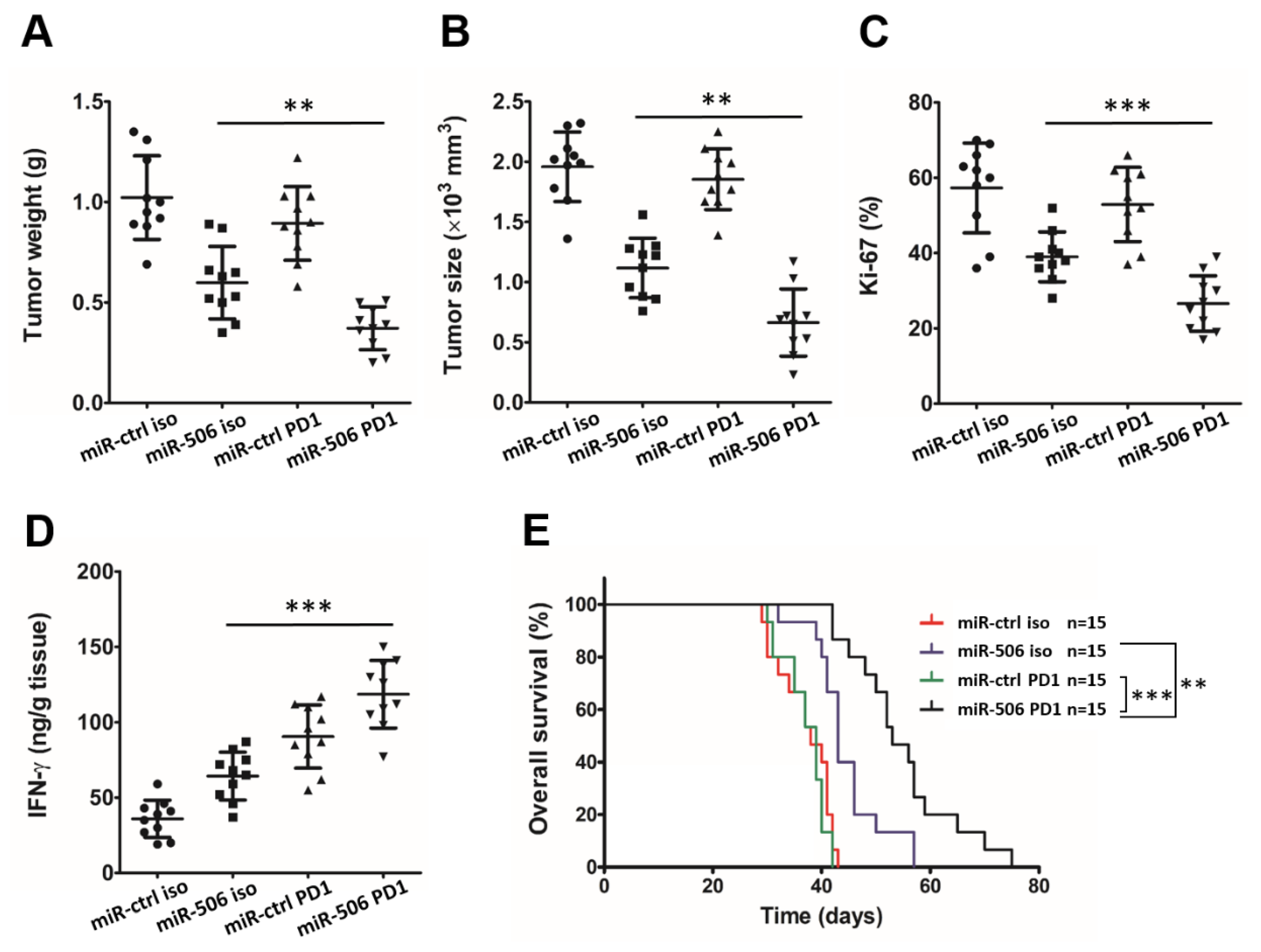
3.7. MiR-506-Induced Repolarization of Macrophages Sensitizes Them to Anti-PD-1 Immunotherapy
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chandana, S.; Babiker, H.M.; Mahadevan, D. Therapeutic trends in pancreatic ductal adenocarcinoma (PDAC). Expert Opin. Investig. Drugs 2019, 28, 161–177. [Google Scholar] [CrossRef]
- Cai, J.; Chen, H.; Lu, M.; Zhang, Y.; Lu, B.; You, L.; Zhang, T.; Dai, M.; Zhao, Y. Advances in the epidemiology of pancreatic cancer: Trends, risk factors, screening, and prognosis. Cancer Lett. 2021, 520, 1–11. [Google Scholar] [CrossRef]
- Schnelldorfer, T.; Adams, D.B.; Warshaw, A.L.; Lillemoe, K.D.; Sarr, M.G. Forgotten pioneers of pancreatic surgery: Beyond the favorite few. Ann. Surg. 2008, 247, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Barugola, G.; Partelli, S.; Marcucci, S.; Sartori, N.; Capelli, P.; Bassi, C.; Pederzoli, P.; Falconi, M. Resectable pancreatic cancer: Who really benefits from resection? Ann. Surg. Oncol. 2009, 16, 3316–3322. [Google Scholar] [CrossRef] [PubMed]
- Semaan, A.; Bernard, V.; Lee, J.J.; Wong, J.W.; Huang, J.; Swartzlander, D.B.; Stephens, B.M.; Monberg, M.E.; Weston, B.R.; Bhutani, M.S.; et al. Defining the Comprehensive Genomic Landscapes of Pancreatic Ductal Adenocarcinoma Using Real-World Endoscopic Aspiration Samples. Clin. Cancer Res. 2021, 27, 1082–1093. [Google Scholar] [CrossRef]
- Apte, M.V.; Park, S.; Phillips, P.A.; Santucci, N.; Goldstein, D.; Kumar, R.K.; Ramm, G.A.; Buchler, M.; Friess, H.; McCarroll, J.A.; et al. Desmoplastic reaction in pancreatic cancer: Role of pancreatic stellate cells. Pancreas 2004, 29, 179–187. [Google Scholar] [CrossRef]
- Neesse, A.; Michl, P.; Frese, K.K.; Feig, C.; Cook, N.; Jacobetz, M.A.; Lolkema, M.P.; Buchholz, M.; Olive, K.P.; Gress, T.M.; et al. Stromal biology and therapy in pancreatic cancer. Gut 2011, 60, 861–868. [Google Scholar] [CrossRef]
- Hosein, A.N.; Brekken, R.A.; Maitra, A. Pancreatic cancer stroma: An update on therapeutic targeting strategies. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 487–505. [Google Scholar] [CrossRef]
- Feig, C.; Gopinathan, A.; Neesse, A.; Chan, D.S.; Cook, N.; Tuveson, D.A. The pancreas cancer microenvironment. Clin. Cancer Res. 2012, 18, 4266–4276. [Google Scholar] [CrossRef]
- Hinshaw, D.C.; Shevde, L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Sluimer, J.C.; Wang, Y.; Subramanian, M.; Brown, K.; Pattison, J.S.; Robbins, J.; Martinez, J.; Tabas, I. Macrophage autophagy plays a protective role in advanced atherosclerosis. Cell Metab. 2012, 15, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Lin, K.; Li, X.; Yuan, X.; Xu, P.; Ni, P.; Xu, D. Redefining Tumor-Associated Macrophage Subpopulations and Functions in the Tumor Microenvironment. Front. Immunol. 2020, 11, 1731. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Biswas, S.K.; Galdiero, M.R.; Sica, A.; Locati, M. Macrophage plasticity and polarization in tissue repair and remodelling. J. Pathol. 2013, 229, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, Z.; Chen, W.; Wang, X.; Cao, M.; Han, X.; Zhang, K.; Teng, B.; Cao, J.; Wu, W.; et al. M2 Macrophage-Derived Exosomes Promote Angiogenesis and Growth of Pancreatic Ductal Adenocarcinoma by Targeting E2F2. Mol. Ther. 2021, 29, 1226–1238. [Google Scholar] [CrossRef]
- Lin, E.Y.; Li, J.F.; Gnatovskiy, L.; Deng, Y.; Zhu, L.; Grzesik, D.A.; Qian, H.; Xue, X.N.; Pollard, J.W. Macrophages regulate the angiogenic switch in a mouse model of breast cancer. Cancer Res. 2006, 66, 11238–11246. [Google Scholar] [CrossRef]
- Hanlon, M.M.; Rakovich, T.; Cunningham, C.C.; Ansboro, S.; Veale, D.J.; Fearon, U.; McGarry, T. STAT3 Mediates the Differential Effects of Oncostatin M and TNFα on RA Synovial Fibroblast and Endothelial Cell Function. Front. Immunol. 2019, 10, 2056. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, J.S.; Rosler, K.M.; Harrison, D.A. The JAK/STAT signaling pathway. J. Cell Sci. 2004, 117 Pt 8, 1281–1283. [Google Scholar] [CrossRef]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef]
- Wang, T.; Niu, G.; Kortylewski, M.; Burdelya, L.; Shain, K.; Zhang, S.; Bhattacharya, R.; Gabrilovich, D.; Heller, R.; Coppola, D.; et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat. Med. 2004, 10, 48–54. [Google Scholar] [CrossRef]
- Zou, S.; Tong, Q.; Liu, B.; Huang, W.; Tian, Y.; Fu, X. Targeting STAT3 in Cancer Immunotherapy. Mol. Cancer 2020, 19, 145. [Google Scholar] [CrossRef] [PubMed]
- Nefedova, Y.; Huang, M.; Kusmartsev, S.; Bhattacharya, R.; Cheng, P.; Salup, R.; Jove, R.; Gabrilovich, D. Hyperactivation of STAT3 is involved in abnormal differentiation of dendritic cells in cancer. J. Immunol. 2004, 172, 464–474. [Google Scholar] [CrossRef] [PubMed]
- Zorn, E.; Nelson, E.A.; Mohseni, M.; Porcheray, F.; Kim, H.; Litsa, D.; Bellucci, R.; Raderschall, E.; Canning, C.; Soiffer, R.J.; et al. IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo. Blood 2006, 108, 1571–1579. [Google Scholar] [CrossRef]
- Aveta, A.; Cilio, S.; Contieri, R.; Spena, G.; Napolitano, L.; Manfredi, C.; Franco, A.; Crocerossa, F.; Cerrato, C.; Ferro, M.; et al. Urinary MicroRNAs as Biomarkers of Urological Cancers: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 10846. [Google Scholar] [CrossRef] [PubMed]
- Fathi, M.; Ghafouri-Fard, S.; Abak, A.; Taheri, M. Emerging roles of miRNAs in the development of pancreatic cancer. Biomed. Pharmacother. 2021, 141, 111914. [Google Scholar] [CrossRef]
- Cheng, R.F.; Wang, J.; Zhang, J.Y.; Sun, L.; Zhao, Y.R.; Qiu, Z.Q.; Sun, B.C.; Sun, Y. MicroRNA-506 is up-regulated in the development of pancreatic ductal adenocarcinoma and is associated with attenuated disease progression. Chin. J. Cancer 2016, 35, 64. [Google Scholar] [CrossRef]
- Li, J.; Wu, H.; Li, W.; Yin, L.; Guo, S.; Xu, X.; Ouyang, Y.; Zhao, Z.; Liu, S.; Tian, Y.; et al. Downregulated miR-506 expression facilitates pancreatic cancer progression and chemoresistance via SPHK1/Akt/NF-κB signaling. Oncogene 2016, 35, 5501–5514. [Google Scholar] [CrossRef]
- Borchardt, H.; Kogel, A.; Kalwa, H.; Weirauch, U.; Aigner, A. Therapeutic miR-506-3p Replacement in Pancreatic Carcinoma Leads to Multiple Effects including Autophagy, Apoptosis, Senescence, and Mitochondrial Alterations In Vitro and In Vivo. Biomedicines 2022, 10, 1692. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, X.; Song, Q.; Liu, L.; Forbes, E.; Tian, W.; Zhang, Z.; Kang, Y.; Wang, H.; Fleming, J.B.; et al. IGFBP2 promotes tumor progression by inducing alternative polarization of macrophages in pancreatic ductal adenocarcinoma through the STAT3 pathway. Cancer Lett. 2021, 500, 132–146. [Google Scholar] [CrossRef]
- Park, W.; Chawla, A.; O’Reilly, E.M. Pancreatic Cancer: A Review. JAMA 2021, 326, 851–862. [Google Scholar] [CrossRef]
- Wang-Gillam, A.; Li, C.P.; Bodoky, G.; Dean, A.; Shan, Y.S.; Jameson, G.; Macarulla, T.; Lee, K.H.; Cunningham, D.; Blanc, J.F.; et al. Nanoliposomal irinotecan with fluorouracil and folinic acid in metastatic pancreatic cancer after previous gemcitabine-based therapy (NAPOLI-1): A global, randomised, open-label, phase 3 trial. Lancet 2016, 387, 545–557. [Google Scholar] [CrossRef]
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouché, O.; Guimbaud, R.; Bécouarn, Y.; Adenis, A.; Raoul, J.L.; Gourgou-Bourgade, S.; de la Fouchardière, C.; et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef]
- Grant, T.J.; Hua, K.; Singh, A. Molecular Pathogenesis of Pancreatic Cancer. Prog. Mol. Biol. Transl. Sci. 2016, 144, 241–275. [Google Scholar]
- Li, P.; Hao, Z.; Wu, J.; Ma, C.; Xu, Y.; Li, J.; Lan, R.; Zhu, B.; Ren, P.; Fan, D.; et al. Comparative Proteomic Analysis of Polarized Human THP-1 and Mouse RAW264.7 Macrophages. Front. Immunol. 2021, 12, 700009. [Google Scholar] [CrossRef]
- Martinez, F.O.; Gordon, S. The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000Prime Rep. 2014, 6, 13. [Google Scholar] [CrossRef]
- Fong, C.H.; Bebien, M.; Didierlaurent, A.; Nebauer, R.; Hussell, T.; Broide, D.; Karin, M.; Lawrence, T. An antiinflammatory role for IKKbeta through the inhibition of “classical” macrophage activation. J. Exp. Med. 2008, 205, 1269–1276. [Google Scholar] [CrossRef]
- Gordon, S.; Martinez, F.O. Alternative activation of macrophages: Mechanism and functions. Immunity 2010, 32, 593–604. [Google Scholar] [CrossRef]
- Shapouri-Moghaddam, A.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, S.A.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.T.; Sahebkar, A. Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef]
- Xia, Y.; Rao, L.; Yao, H.; Wang, Z.; Ning, P.; Chen, X. Engineering Macrophages for Cancer Immunotherapy and Drug Delivery. Adv. Mater. 2020, 32, e2002054. [Google Scholar] [CrossRef]
- Sica, A.; Mantovani, A. Macrophage plasticity and polarization: In vivo veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef]
- Hu, H.; Hang, J.J.; Han, T.; Zhuo, M.; Jiao, F.; Wang, L.W. The M2 phenotype of tumor-associated macrophages in the stroma confers a poor prognosis in pancreatic cancer. Tumour Biol. 2016, 37, 8657–8664. [Google Scholar] [CrossRef]
- Bentwich, I.; Avniel, A.; Karov, Y.; Aharonov, R.; Gilad, S.; Barad, O.; Barzilai, A.; Einat, P.; Einav, U.; Meiri, E.; et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005, 37, 766–770. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, H.; Li, Y.; Wu, J.; Greenlee, A.R.; Yang, C.; Jiang, Y. The role of miR-506 in transformed 16HBE cells induced by anti-benzo[a]pyrene-trans-7,8-dihydrodiol-9,10-epoxide. Toxicol. Lett. 2011, 205, 320–326. [Google Scholar] [CrossRef]
- Yang, D.; Sun, Y.; Hu, L.; Zheng, H.; Ji, P.; Pecot, C.V.; Zhao, Y.; Reynolds, S.; Cheng, H.; Rupaimoole, R.; et al. Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer Cell 2013, 23, 186–199. [Google Scholar] [CrossRef]
- Zhang, Z.; Ma, J.; Luan, G.; Kang, L.; Su, Y.; He, Y.; Luan, F. MiR-506 suppresses tumor proliferation and invasion by targeting FOXQ1 in nasopharyngeal carcinoma. PLoS ONE 2015, 10, e0122851. [Google Scholar] [CrossRef]
- Du, J.; Zheng, X.; Cai, S.; Zhu, Z.; Tan, J.; Hu, B.; Huang, Z.; Jiao, H. MicroRNA-506 participates in pancreatic cancer pathogenesis by targeting PIM3. Mol. Med. Rep. 2015, 12, 5121–5126. [Google Scholar] [CrossRef]
- Yang, F.Q.; Zhang, H.M.; Chen, S.J.; Yan, Y.; Zheng, J.H. MiR-506 is down-regulated in clear cell renal cell carcinoma and inhibits cell growth and metastasis via targeting FLOT1. PLoS ONE 2015, 10, e0120258. [Google Scholar] [CrossRef]
- Li, Z.; Liu, Z.; Dong, S.; Zhang, J.; Tan, J.; Wang, Y.; Ge, C.; Li, R.; Xue, Y.; Li, M.; et al. miR-506 Inhibits Epithelial-to-Mesenchymal Transition and Angiogenesis in Gastric Cancer. Am. J. Pathol. 2015, 185, 2412–2420. [Google Scholar] [CrossRef]
- Li, J.; Ju, J.; Ni, B.; Wang, H. The emerging role of miR-506 in cancer. Oncotarget 2016, 7, 62778–62788. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, T.; Han, Y.; Chen, J.; Liang, X.; Sun, L. MiR-506 Promotes Antitumor Immune Response in Pancreatic Cancer by Reprogramming Tumor-Associated Macrophages toward an M1 Phenotype. Biomedicines 2023, 11, 2874. https://doi.org/10.3390/biomedicines11112874
Yang T, Han Y, Chen J, Liang X, Sun L. MiR-506 Promotes Antitumor Immune Response in Pancreatic Cancer by Reprogramming Tumor-Associated Macrophages toward an M1 Phenotype. Biomedicines. 2023; 11(11):2874. https://doi.org/10.3390/biomedicines11112874
Chicago/Turabian StyleYang, Tiantian, Yitong Han, Junhang Chen, Xiaoyu Liang, and Longhao Sun. 2023. "MiR-506 Promotes Antitumor Immune Response in Pancreatic Cancer by Reprogramming Tumor-Associated Macrophages toward an M1 Phenotype" Biomedicines 11, no. 11: 2874. https://doi.org/10.3390/biomedicines11112874
APA StyleYang, T., Han, Y., Chen, J., Liang, X., & Sun, L. (2023). MiR-506 Promotes Antitumor Immune Response in Pancreatic Cancer by Reprogramming Tumor-Associated Macrophages toward an M1 Phenotype. Biomedicines, 11(11), 2874. https://doi.org/10.3390/biomedicines11112874






