VEGFA Status as a Predictive Marker of Therapy Outcome in Metastatic Gastric Cancer Patients Following Ramucirumab-Based Treatment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Samples
2.2. FISH Analysis
FISH Quantification
2.3. IHC Analysis
2.3.1. Scoring of VEGFA Staining
2.3.2. Micro-Vessel Density (MVD) Quantification
2.4. Statistical Analysis
3. Results
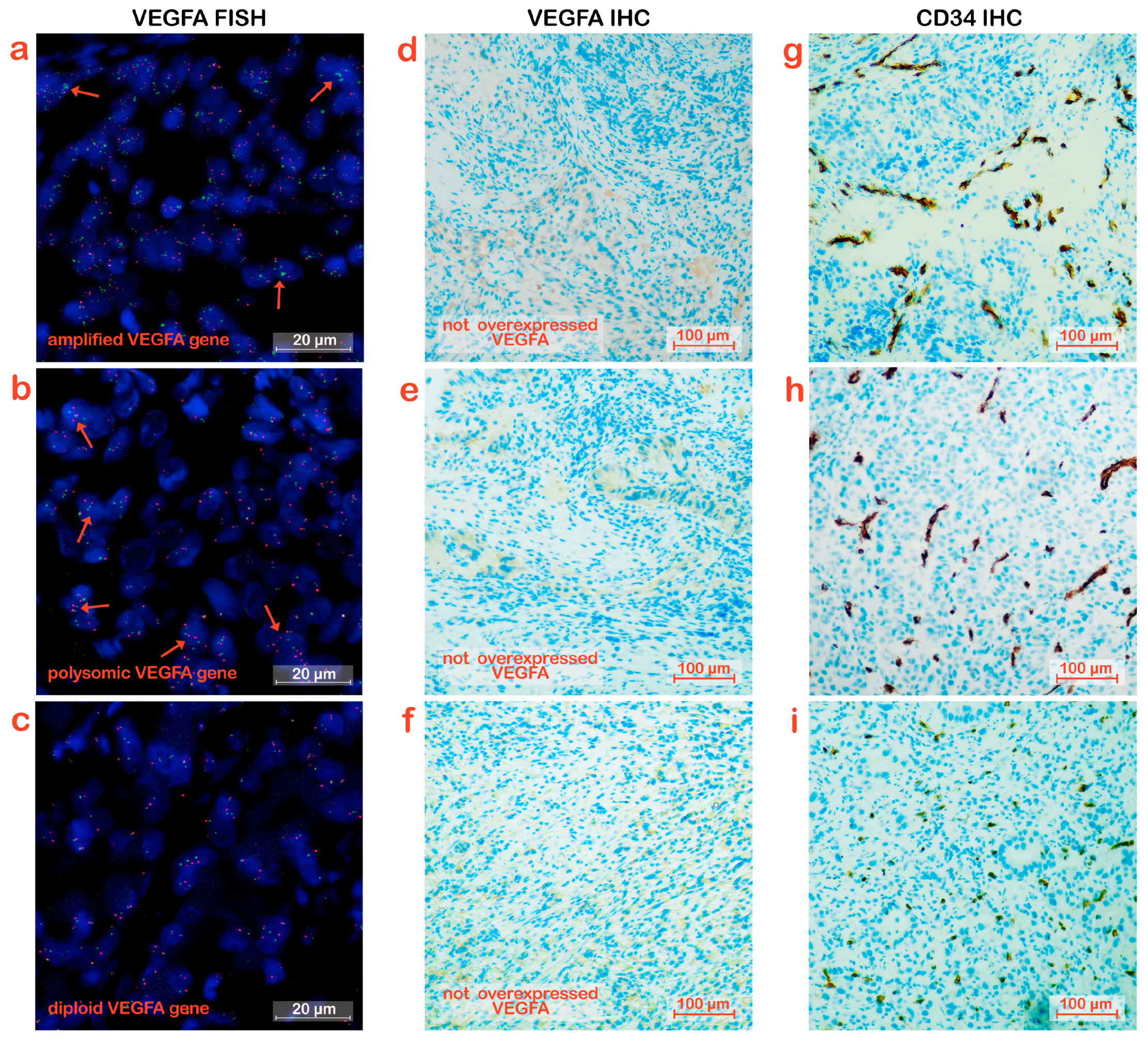
3.1. Gene Amplification of VEGFA in Tumor Tissue
3.2. Overexpression of VEGFA in Tumor Tissues
3.3. Micro-Vessel Density in Tumor Tissues
3.4. Comparative Analysis between VEGFA and CD34 Expression
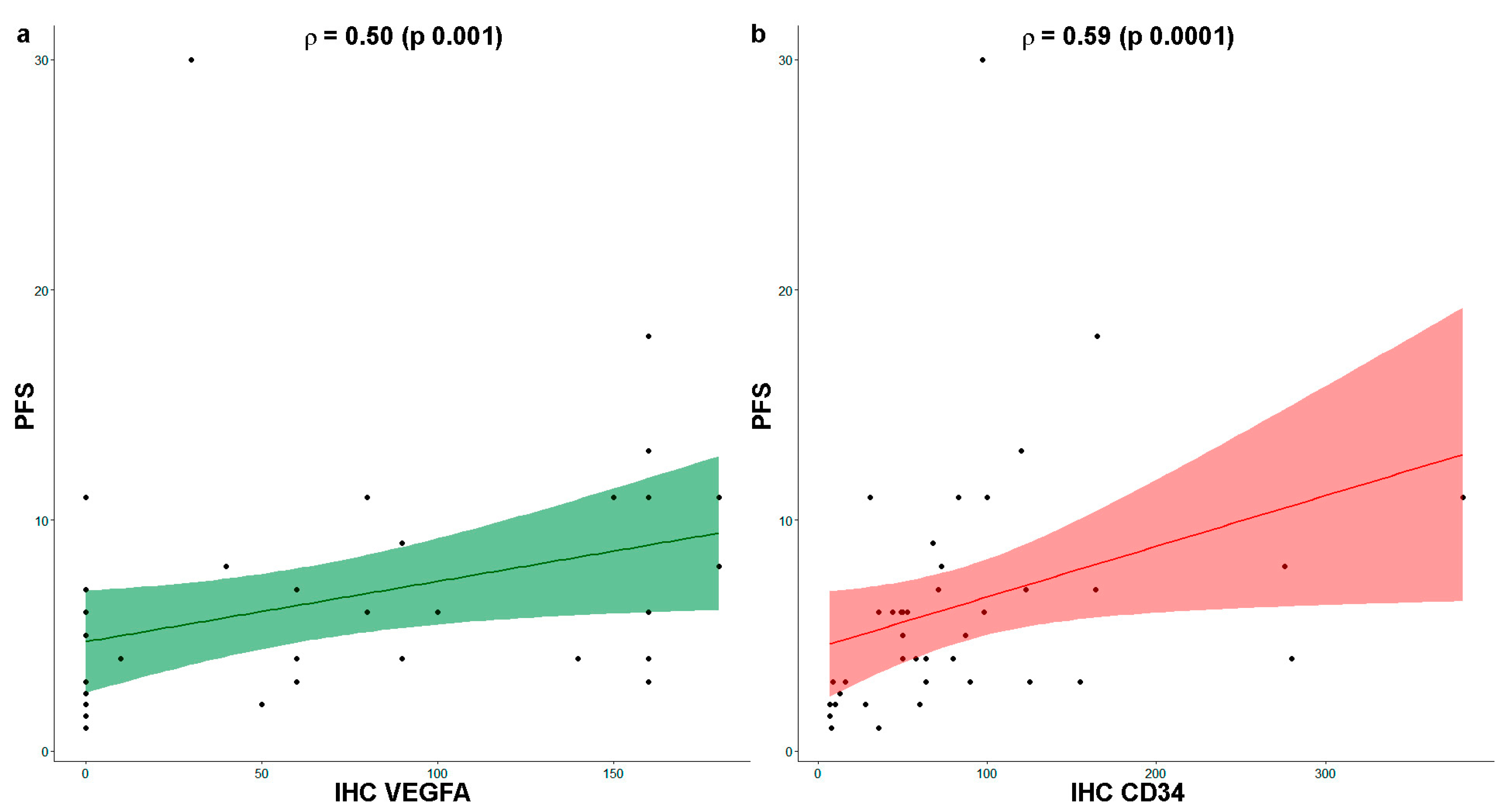
3.5. VEGFA Expression and Micro-Vessel Density Correlate with Therapy Outcome
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.D.; Syn, N.L.; Moehler, M.; Grothe, W.; Yong, W.P.; Tai, B.C.; Ho, J.; Unverzagt, S. Chemotherapy for advanced gastric cancer. Cochrane Database Syst. Rev. 2017, 8, CD004064. [Google Scholar] [PubMed]
- Smyth, E.C.; Nilsson, M.; Grabsch, H.I.; van Grieken, N.C.; Lordick, F. Gastric cancer. Lancet 2020, 396, 635–648. [Google Scholar] [PubMed]
- Fuchs, C.S.; Tomasek, J.; Yong, C.J.; Dumitru, F.; Passalacqua, R.; Goswami, C.; Safran, H.; Dos Santos, L.V.; Aprile, G.; Ferry, D.R.; et al. Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): An international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet 2014, 383, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Wilke, H.; Muro, K.; Van Cutsem, E.; Oh, S.C.; Bodoky, G.; Shimada, Y.; Hironaka, S.; Sugimoto, N.; Lipatov, O.; Kim, T.Y.; et al. Ramucirumab plus paclitaxel versus placebo plus paclitaxel in patients with previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (RAINBOW): A double-blind, randomised phase 3 trial. Lancet Oncol. 2014, 15, 1224–1235. [Google Scholar] [CrossRef] [PubMed]
- Hicklin, D.J.; Ellis, L.M. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2005, 23, 1011–1027. [Google Scholar] [CrossRef]
- Claesson-Welsh, L.; Welsh, M. VEGFA and tumour angiogenesis. J. Intern. Med. 2013, 273, 114–127. [Google Scholar]
- Ellis, L.M.; Hicklin, D.J. VEGF-targeted therapy: Mechanisms of anti-tumour activity. Nat. Rev. Cancer 2008, 8, 579–591. [Google Scholar]
- Niu, G.; Chen, X. Vascular endothelial growth factor as an anti-angiogenic target for cancer therapy. Curr. Drug Targets 2010, 11, 1000–1017. [Google Scholar] [CrossRef]
- Goel, H.L.; Mercurio, A.M. VEGF targets the tumour cell. Nat. Rev. Cancer 2013, 13, 871–882. [Google Scholar]
- Refolo, M.G.; Lotesoriere, C.; Messa, C.; Caruso, M.G.; D’Alessandro, R. Integrated immune gene expression signature and molecularclassification in gastriccancer: New insights. J. Leukoc. Biol. 2020, 108, 633–646. [Google Scholar] [CrossRef] [PubMed]
- Andreozzi, M.; Quagliata, L.; Gsponer, J.R.; Ruiz, C.; Vuaroqueaux, V.; Eppenberger-Castori, S.; Tornillo, L.; Terracciano, L.M. VEGFA gene locus analysis across 80 human tumour types reveals gene amplification in several neoplastic entities. Angiogenesis 2014, 17, 519–527. [Google Scholar]
- Raimondi, A.; Gasparini, P.; Lonardi, S.; Corallo, S.; Fornaro, L.; Laterza, M.M.; Salvatore, M.D.; Giommoni, E.; Lotesoriere, C.; Murgioni, S.; et al. Vascular endothelial growth factor A (VEGF-A) amplification and long-termresponse to ramucirumab (ram) in metastaticgastriccancer (mGC): The VERA study. J. Clin. Oncol. 2019, 37 (Suppl. 15), 3143. [Google Scholar] [CrossRef]
- Gravalos, C.; Jimeno, A. HER2 in gastric cancer: A new prognostic factor and a novel therapeutic target. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2008, 19, 1523–1529. [Google Scholar] [CrossRef] [PubMed]
- Hanna, W.; Nofech-Mozes, S.; Kahn, H.J. Intratumoral heterogeneity of HER2/neu in breast cancer—A rare event. Breast J. 2007, 13, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Ruschoff, J.; Dietel, M.; Baretton, G.; Arbogast, S.; Walch, A.; Monges, G.; Chenard, M.P.; Penault-Llorca, F.; Nagelmeier, I.; Schlake, W.; et al. HER2 diagnostics in gastric cancer-guideline validation and development of standardized immunohistochemical testing. Virchows Arch. Int. J. Pathol. 2010, 457, 299–307. [Google Scholar] [CrossRef]
- Pirrelli, M.; Caruso, M.L.; Di Maggio, M.; Armentano, R.; Valentini, A.M. Are biopsy specimens predictive of HER2 status in gastric cancer patients? Dig. Dis. Sci. 2013, 58, 397–404. [Google Scholar] [CrossRef]
- Yang, J.; Yang, D.; Sun, Y.; Sun, B.; Wang, G.; Trent, J.C.; Araujo, D.M.; Chen, K.; Zhang, W. Genetic amplification of the vascular endothelial growth factor (VEGF) pathway genes, including VEGFA, in human osteosarcoma. Cancer 2011, 117, 4925–4938. [Google Scholar] [CrossRef]
- Vlajnic, T.; Andreozzi, M.C.; Schneider, S.; Tornillo, L.; Karamitopoulou, E.; Lugli, A.; Ruiz, C.; Zlobec, I.; Terracciano, L. VEGFA gene locus (6p12) amplification identifies a small but highly aggressive subgroup of colorectal cancer [corrected] patients. Mod. Pathol. Off. J. USA Can. Acad. Pathol. Inc. 2011, 24, 1404–1412. [Google Scholar]
- Schneider, B.P.; Gray, R.J.; Radovich, M.; Shen, F.; Vance, G.; Li, L.; Jiang, G.; Miller, K.D.; Gralow, J.R.; Dickler, M.N.; et al. Prognostic and predictive value of tumor vascular endothelial growth factor gene amplification in metastatic breast cancer treated with paclitaxel with and without bevacizumab; results from ECOG 2100 trial. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 1281–1289. [Google Scholar]
- Luo, X.; Feng, G.S. VEGFA genomic amplification tailors treatment of HCCs with sorafenib. Cancer Discov. 2014, 4, 640–641. [Google Scholar] [CrossRef] [PubMed]
- Oh, C.R.; Kong, S.Y.; Im, H.S.; Kim, H.J.; Kim, M.K.; Yoon, K.A.; Cho, E.H.; Jang, J.H.; Lee, J.; Kang, J.; et al. Genome-wide copy number alteration and VEGFA amplification of circulating cell-free DNA as a biomarker in advanced hepatocellular carcinoma patients treated with Sorafenib. BMC Cancer 2019, 19, 292. [Google Scholar] [CrossRef] [PubMed]
- Bruix, J.; Cheng, A.L.; Meinhardt, G.; Nakajima, K.; De Sanctis, Y.; Llovet, J. Prognostic factors and predictors of sorafenib benefit in patients with hepatocellular carcinoma: Analysis of two phase III studies. J. Hepatol. 2018, 69, 990–991, Corrigendum to J. Hepatol. 2017, 67, 999–1008. [Google Scholar] [CrossRef]
- Horwitz, E.; Stein, I.; Andreozzi, M.; Nemeth, J.; Shoham, A.; Pappo, O.; Schweitzer, N.; Tornillo, L.; Kanarek, N.; Quagliata, L.; et al. Human and mouse VEGFA-amplified hepatocellular carcinomas are highly sensitive to sorafenib treatment. Cancer Discov. 2014, 4, 730–743. [Google Scholar] [PubMed]
- Saponaro, C.; Fanizzi, A.; Sonnessa, M.; Mondelli, P.; Vergara, D.; Loisi, D.; Massafra, R.; Latorre, A.; Zito, F.A.; Schirosi, L. Downstream Signaling of Inflammasome Pathway Affects Patients’ Outcome in the Context of Distinct Molecular Breast Cancer Subtypes. Pharmaceuticals 2022, 15, 651. [Google Scholar] [CrossRef]
- Wolff, A.C.; Hammond, M.E.H.; Allison, K.H.; Harvey, B.E.; Mangu, P.B.; Bartlett, J.M.S.; Bilous, M.; Ellis, I.O.; Fitzgibbons, P.; Hanna, W.; et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2018, 36, 2105–2122. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, C.S.; Dong, X.Y.; Hu, Y.; Duan, B.J.; Bai, J.; Wu, Y.Y.; Fan, L.; Liao, X.H.; Kang, Y.; et al. Claudin 18.2 is a potential therapeutic target for zolbetuximab in pancreatic ductal adenocarcinoma. World J. Gastrointest. Oncol. 2022, 14, 1252–1264. [Google Scholar] [CrossRef]
- Apte, R.S.; Chen, D.S.; Ferrara, N. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell 2019, 176, 1248–1264. [Google Scholar]
- Van Cutsem, E.; Muro, K.; Cunningham, D.; Bodoky, G.; Sobrero, A.; Cascinu, S.; Ajani, J.; Oh, S.C.; Al-Batran, S.E.; Wainberg, Z.A.; et al. Biomarker analyses of second-line ramucirumab in patients with advanced gastric cancer from RAINBOW, a global, randomized, double-blind, phase 3 study. Eur. J. Cancer 2020, 127, 150–157. [Google Scholar] [CrossRef]
- Fuchs, C.S.; Tabernero, J.; Tomasek, J.; Chau, I.; Melichar, B.; Safran, H.; Tehfe, M.A.; Filip, D.; Topuzov, E.; Schlittler, L.; et al. Biomarker analyses in REGARD gastric/GEJ carcinoma patients treated with VEGFR2-targeted antibody ramucirumab. Br. J. Cancer 2016, 115, 974–982. [Google Scholar] [CrossRef]
- Bais, C.; Mueller, B.; Brady, M.F.; Mannel, R.S.; Burger, R.A.; Wei, W.; Marien, K.M.; Kockx, M.M.; Husain, A.; Birrer, M.J. Tumor Microvessel Density as a Potential Predictive Marker for Bevacizumab Benefit: GOG-0218 Biomarker Analyses. J. Natl. Cancer Inst. 2017, 109, djx066. [Google Scholar] [CrossRef] [PubMed]
- Santos, G.C.; Zielenska, M.; Prasad, M.; Squire, J.A. Chromosome 6p amplification and cancer progression. J. Clin. Pathol. 2007, 60, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Tse, C.H.; Hwang, H.C.; Goldstein, L.C.; Kandalaft, P.L.; Wiley, J.C.; Kussick, S.J.; Gown, A.M. Determining true HER2 gene status in breast cancers with polysomy by using alternative chromosome 17 reference genes: Implications for anti-HER2 targeted therapy. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2011, 29, 4168–4174. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.H.; Shi, Q.; Sukov, W.R.; Lewis, M.A.; Sattler, C.A.; Wiktor, A.E.; Wu, T.T.; Diasio, R.B.; Jenkins, R.B.; Sinicrope, F.A. Adverse prognostic impact of intratumor heterogeneous HER2 gene amplification in patients with esophageal adenocarcinoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2012, 30, 3932–3938. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network; Analysis Working Group: Asan University; BC Cancer Agency; Brigham and Women’s Hospital; Broad Institute; Brown University; Case Western Reserve University; Dana-Farber Cancer Institute; Duke University; Greater Poland Cancer Centre; et al. Integrated genomic characterization of oesophageal carcinoma. Nature 2017, 541, 169–175. [Google Scholar] [CrossRef]
- Goel, S.; Duda, D.G.; Xu, L.; Munn, L.L.; Boucher, Y.; Fukumura, D.; Jain, R.K. Normalization of the vasculature for treatment of cancer and other diseases. Physiol. Rev. 2011, 91, 1071–1121. [Google Scholar]
- D’Alessandro, R.; Refolo, M.G.; Schirizzi, A.; De Leonardis, G.; Donghia, R.; Guerra, V.; Giannelli, G.; Lolli, I.R.; Laterza, M.M.; De Vita, F.; et al. Corrigendum: Variations in circulating levels of angiopoietin-2 over time are predictive of ramucirumab-paclitaxel therapy outcome in advanced gastric cancer: Results of prospective study. Front. Oncol. 2023, 13, 1187014. [Google Scholar] [CrossRef]
- Zhao, Y.; Guo, S.; Deng, J.; Shen, J.; Du, F.; Wu, X.; Chen, Y.; Li, M.; Chen, M.; Li, X.; et al. VEGF/VEGFR-Targeted Therapy and Immunotherapy in Non-small Cell Lung Cancer: Targeting the Tumor Microenvironment. Int. J. Biol. Sci. 2022, 18, 3845–3858. [Google Scholar] [CrossRef]
- Solimando, A.G.; Summa, S.; Vacca, A.; Ribatti, D. Cancer-Associated Angiogenesis: The Endothelial Cell as a Checkpoint for Immunological Patrolling. Cancers 2020, 12, 3380. [Google Scholar] [CrossRef]


| Parameters * | Total Cohort (n = 42) | Group | p ^ | |
|---|---|---|---|---|
| Control Disease (n = 28) | Rapid Progression (n = 14) | |||
| Age (yrs) | 64.93 ± 9.25 | 64.36 ± 9.84 | 66.07 ± 8.15 | 0.69 |
| Gender (M) (%) | 31 (73.81) | 20 (71.43) | 11 (78.57) | 0.72 ψ |
| FISH (%) | 0.89 ψ | |||
| Not Amplified | 26 (61.90) | 18 (64.29) | 8 (57.14) | |
| Polysomic | 10 (23.81) | 6 (21.43) | 4 (28.57) | |
| Amplified | 6 (14.29) | 4 (14.29) | 2 (14.29) | |
| PFS (months) | 6.34 ± 5.32 | 8.44 ± 5.46 | 2.28 ± 0.75 | <0.0001 |
| PFS (months) (%) | ||||
| ≤3 | 14 (33.33) | |||
| >3 | 28 (66.67) | |||
| IHC VEGFA score | 62.38 ± 65.44 | 81.78 ± 65.66 | 23.57 ± 46.01 | 0.003 |
| IHC VEGFA score (%) | 0.005 ψ | |||
| 0 (low) | 17 (44.74) | 7 (29.17) | 10 (71.43) | |
| 10–80 (moderate) | 6 (15.79) | 3 (12.50) | 3 (21.43) | |
| 90–180 (high) | 15 (39.47) | 14 (58.33) | 1 (7.14) | |
| IHC VEGFA score (%) | 0.02 ψ | |||
| 0 | 17 (44.74) | 7 (29.17) | 10 (71.43) | |
| 10–180 | 21 (55.26) | 17 (70.83) | 4 (28.57) | |
| CD34 | 84.26 ± 77.51 | 103.96 ± 82.46 | 44.86 ± 48.14 | 0.003 |
| CD34 (count/field) (%) | 0.005 ψ | |||
| 1–50 (low density) | 13 (30.95) | 4 (14.29) | 9 (64.29) | |
| 50–100 (moderate density) | 20 (47.62) | 17 (60.71) | 3 (21.43) | |
| >100 (high density) | 9 (21.43) | 7 (25.00) | 2 (14.29) | |
| Parameters * | Total Cohort (n = 42) | Group | p ^ | |
|---|---|---|---|---|
| Control (n = 28) | RP (n = 14) | |||
| Combo | ||||
| VEGFA IHC and VEGFA FISH (%) | 0.03 | |||
| VEGFA IHC (0) and VEGFA Not Amplified | 11 (28.95) | 6 (25.00) | 5 (35.71) | |
| VEGFA IHC (40–180) and VEGFA Not Amplified | 11 (28.95) | 8 (33.33) | 3 (21.43) | |
| VEGFA IHC (0) and VEGFA Copy Number Variation | 6 (15.79) | 1 (4.17) | 5 (35.71) | |
| VEGFA IHC (40–180) and VEGFA Copy Number Variation | 10 (26.32) | 9 (37.50) | 1 (7.14) | |
| VEGFA IHC and CD34 IHC (%) | 0.003 | |||
| VEGFA IHC (0) and CD34 IHC (1–50) | 10 (26.32) | 2 (8.33) | 8 (57.14) | |
| VEGFA IHC (40–180) and CD34 IHC (1–50) | 2 (5.26) | 1 (4.17) | 1 (7.14) | |
| VEGFA IHC (0) and CD34 IHC (>50) | 7 (18.42) | 5 (20.83) | 2 (14.29) | |
| VEGFA IHC (40–180) and CD34 IHC (>50) | 19 (50.00) | 16 (66.67) | 3 (21.43) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schirizzi, A.; Arshadi, A.; Tolomeo, D.; Schirosi, L.; Valentini, A.M.; De Leonardis, G.; Refolo, M.G.; Donghia, R.; Storlazzi, C.T.; Zito, A.; et al. VEGFA Status as a Predictive Marker of Therapy Outcome in Metastatic Gastric Cancer Patients Following Ramucirumab-Based Treatment. Biomedicines 2023, 11, 2721. https://doi.org/10.3390/biomedicines11102721
Schirizzi A, Arshadi A, Tolomeo D, Schirosi L, Valentini AM, De Leonardis G, Refolo MG, Donghia R, Storlazzi CT, Zito A, et al. VEGFA Status as a Predictive Marker of Therapy Outcome in Metastatic Gastric Cancer Patients Following Ramucirumab-Based Treatment. Biomedicines. 2023; 11(10):2721. https://doi.org/10.3390/biomedicines11102721
Chicago/Turabian StyleSchirizzi, Annalisa, Aram Arshadi, Doron Tolomeo, Laura Schirosi, Anna Maria Valentini, Giampiero De Leonardis, Maria Grazia Refolo, Rossella Donghia, Clelia Tiziana Storlazzi, Alfredo Zito, and et al. 2023. "VEGFA Status as a Predictive Marker of Therapy Outcome in Metastatic Gastric Cancer Patients Following Ramucirumab-Based Treatment" Biomedicines 11, no. 10: 2721. https://doi.org/10.3390/biomedicines11102721
APA StyleSchirizzi, A., Arshadi, A., Tolomeo, D., Schirosi, L., Valentini, A. M., De Leonardis, G., Refolo, M. G., Donghia, R., Storlazzi, C. T., Zito, A., Ricci, A. D., Vallarelli, S., Ostuni, C., Bencivenga, M., De Manzoni, G., Messa, C., Armentano, R., Giannelli, G., Lotesoriere, C., & D’Alessandro, R. (2023). VEGFA Status as a Predictive Marker of Therapy Outcome in Metastatic Gastric Cancer Patients Following Ramucirumab-Based Treatment. Biomedicines, 11(10), 2721. https://doi.org/10.3390/biomedicines11102721








