Long Non-Coding RNA GAS5 Promotes BAX Expression by Competing with microRNA-128-3p in Response to 5-Fluorouracil
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture, Cloning, and Transfections
2.2. RNA Analysis and RT-qPCR
2.3. Western Blotting
2.4. Cell Viability, Colony Forming Assay, and Flow Cytometric Analysis
2.5. MS2-Trap Assay and Biotin Pull-Down Assay
2.6. Statistical Analysis
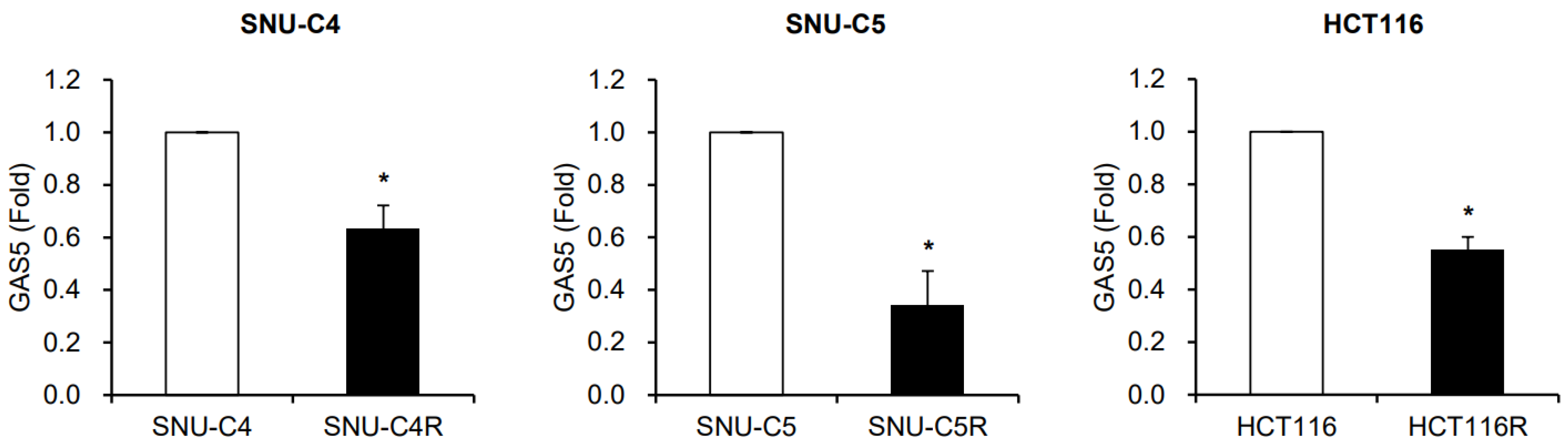
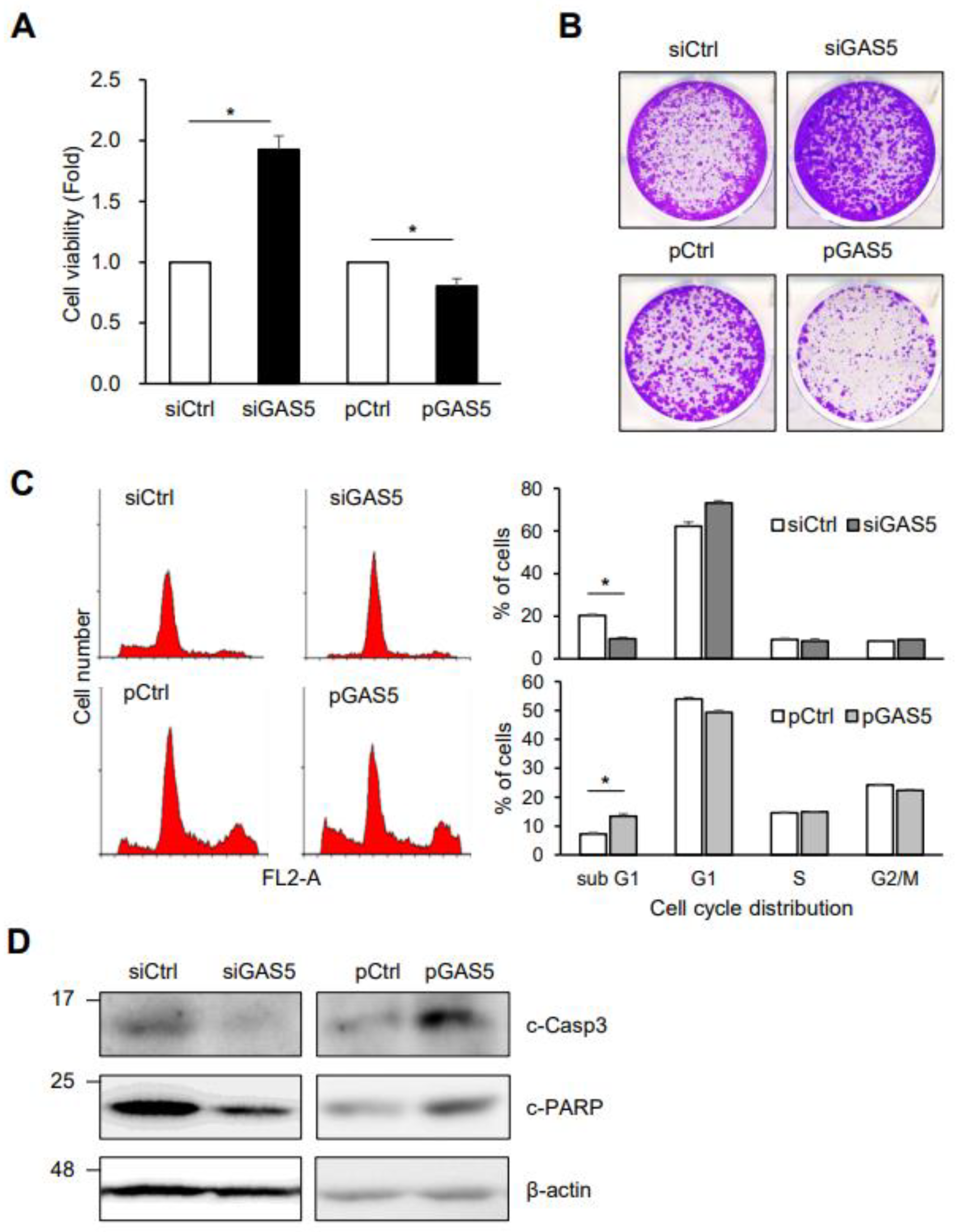
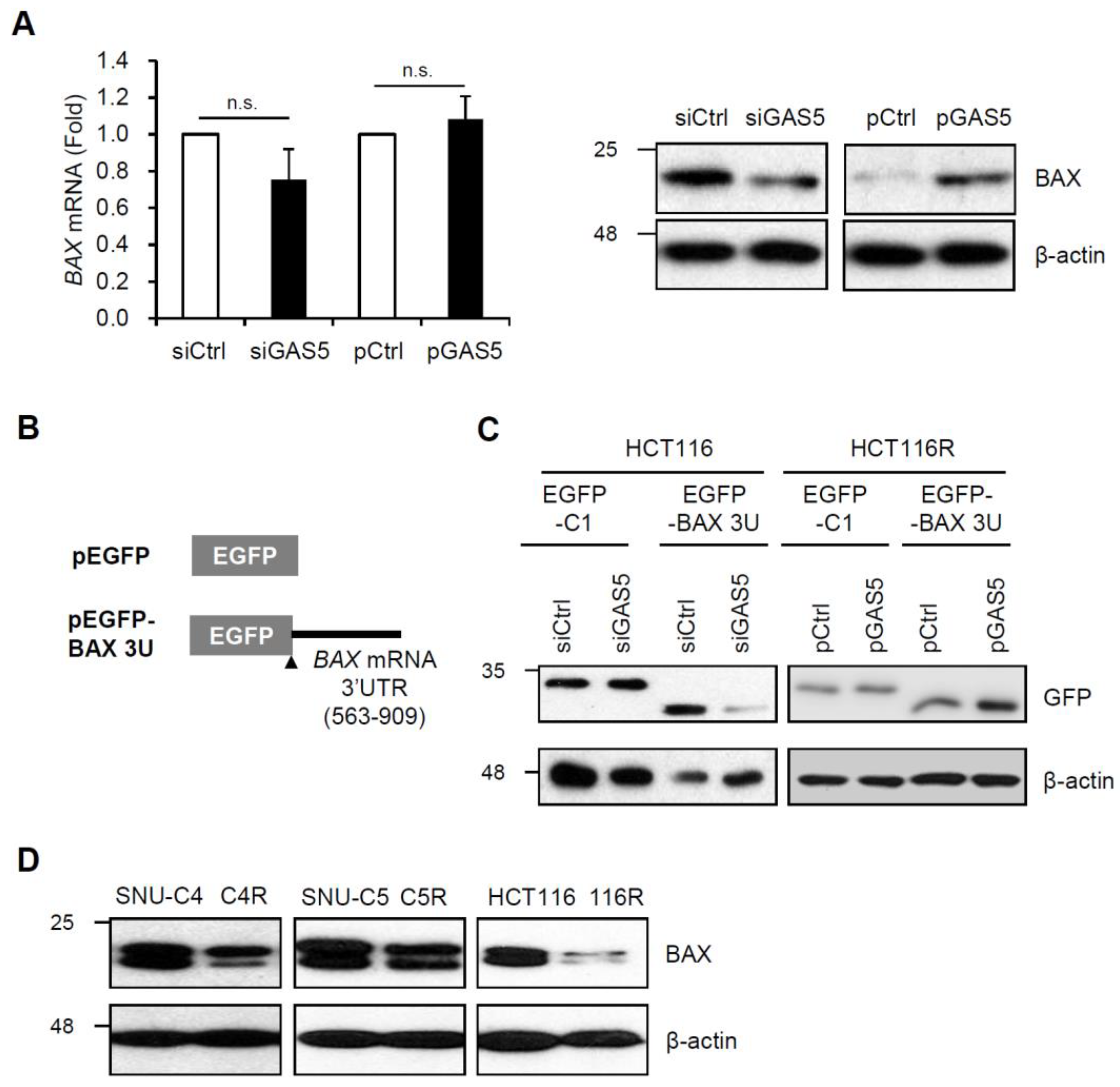
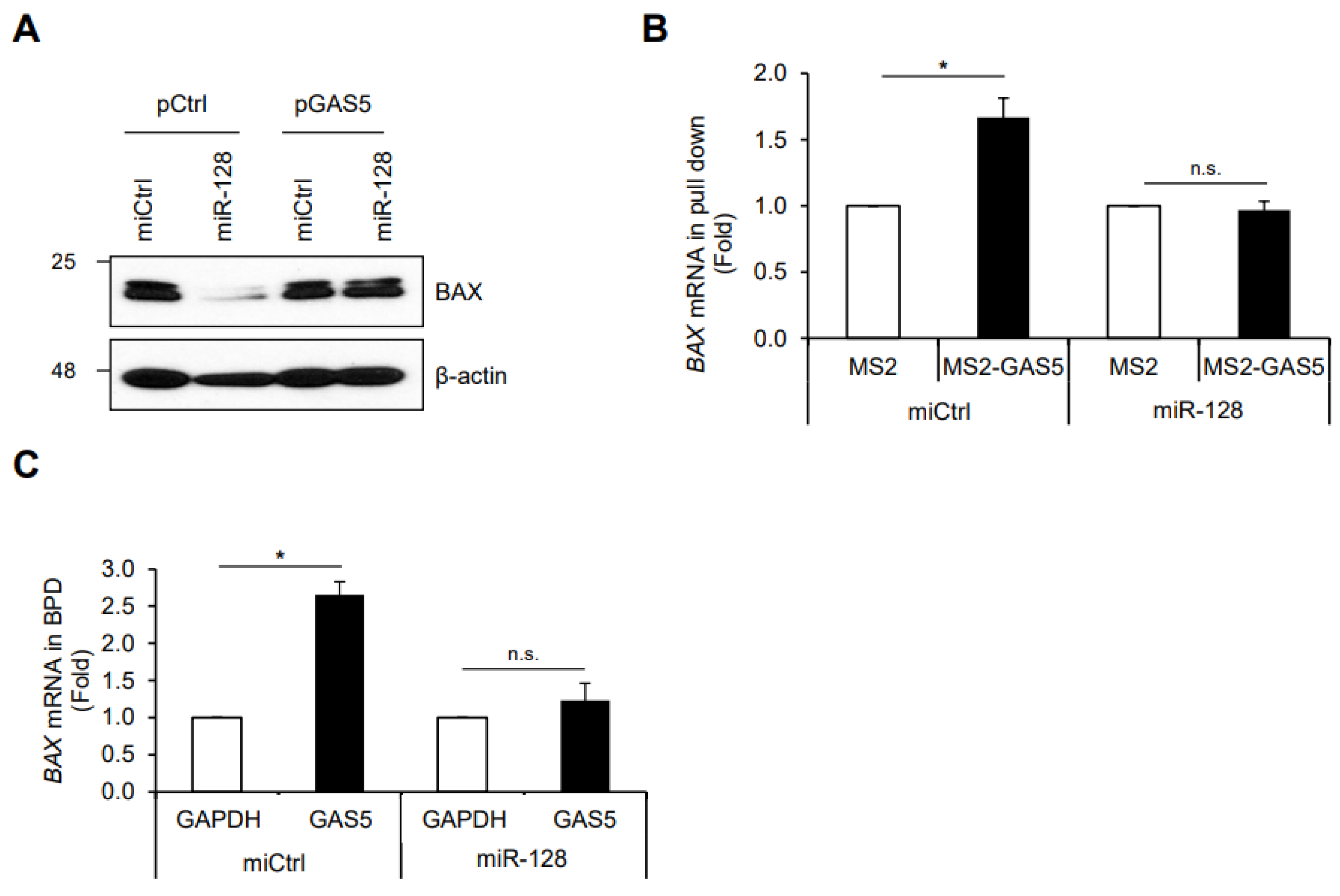
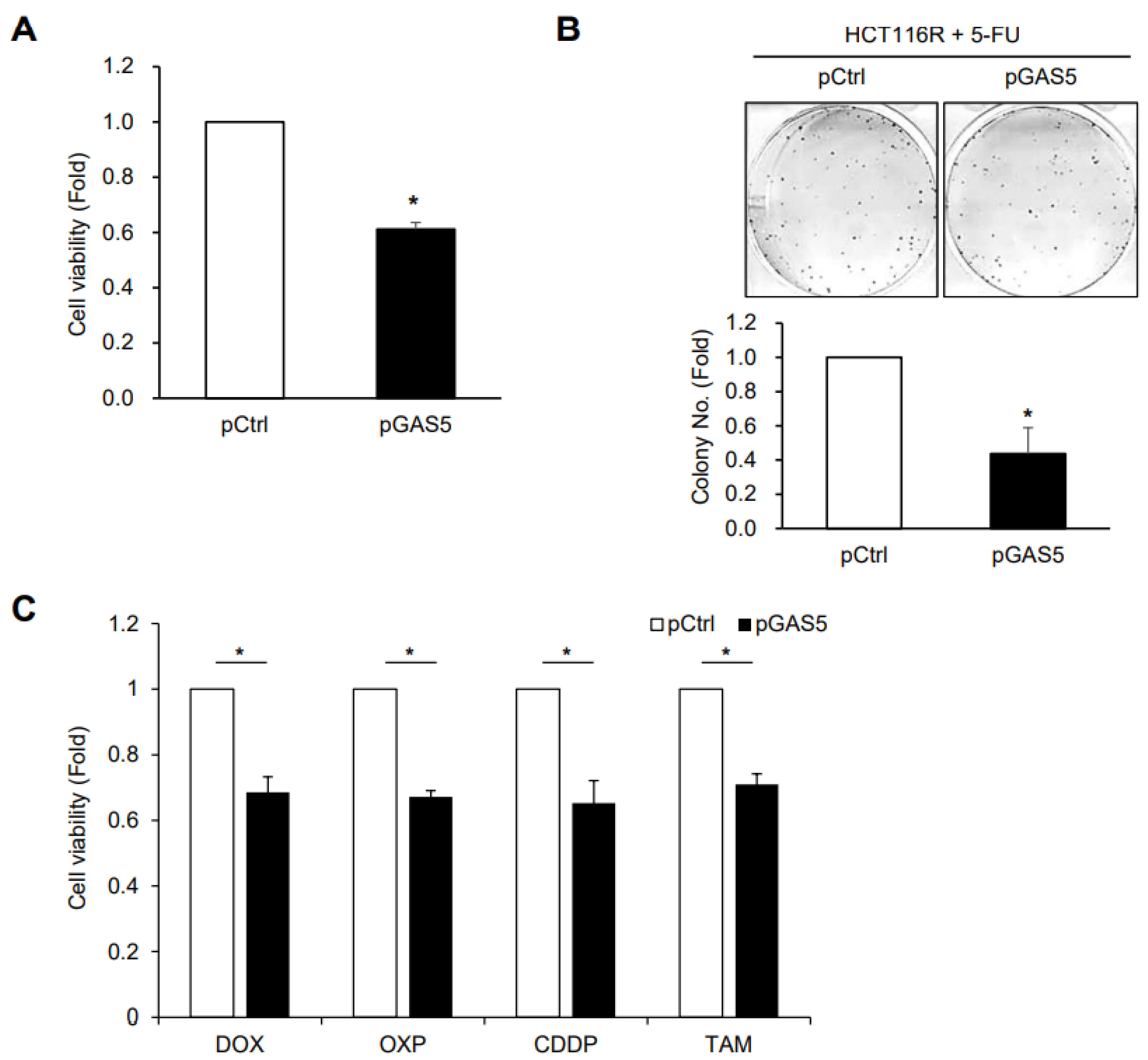
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zahreddine, H.; Borden, K.L. Mechanisms and insights into drug resistance in cancer. Front. Pharmacol. 2013, 4, 28. [Google Scholar] [CrossRef] [PubMed]
- Cree, I.A.; Charlton, P. Molecular chess? Hallmarks of anti-cancer drug resistance. BMC Cancer 2017, 17, 10. [Google Scholar] [CrossRef] [PubMed]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug resistance in cancer: An overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef] [PubMed]
- Vasan, N.; Baselga, J.; Hyman, D.M. A view on drug resistance in cancer. Nature 2019, 575, 299–309. [Google Scholar] [CrossRef]
- Longley, D.B.; Johnston, P.G. Molecular mechanisms of drug resistance. J. Pathol. 2005, 205, 275–292. [Google Scholar] [CrossRef] [PubMed]
- Holohan, C.; Van Schaeybroeck, S.; Longley, D.B.; Johnston, P.G. Cancer drug resistance: An evolving paradigm. Nat. Rev. Cancer 2013, 13, 714–726. [Google Scholar] [CrossRef]
- Thomas, D.M.; Zalcberg, J.R. 5-fluorouracil: A pharmacological paradigm in the use of cytotoxics. Clin. Exp. Pharmacol. Physiol. 1998, 25, 887–895. [Google Scholar] [CrossRef]
- Zhang, N.; Yin, Y.; Xu, S.J.; Chen, W.S. 5-Fluorouracil: Mechanisms of resistance and reversal strategies. Molecules 2008, 13, 1551–1569. [Google Scholar] [CrossRef]
- Ward, R.A.; Fawell, S.; Floc’h, N.; Flemington, V.; McKerrecher, D.; Smith, P.D. Challenges and Opportunities in Cancer Drug Resistance. Chem. Rev. 2021, 121, 3297–3351. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Abak, A.; Tondro Anamag, F.; Shoorei, H.; Fattahi, F.; Javadinia, S.A.; Basiri, A.; Taheri, M. 5-Fluorouracil: A Narrative Review on the Role of Regulatory Mechanisms in Driving Resistance to This Chemotherapeutic Agent. Front. Oncol. 2021, 11, 658636. [Google Scholar] [CrossRef]
- Liu, K.; Gao, L.; Ma, X.; Huang, J.J.; Chen, J.; Zeng, L.; Ashby, C.R., Jr.; Zou, C.; Chen, Z.S. Long non-coding RNAs regulate drug resistance in cancer. Mol. Cancer 2020, 19, 54. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef]
- Zhou, X.; Ao, X.; Jia, Z.; Li, Y.; Kuang, S.; Du, C.; Zhang, J.; Wang, J.; Liu, Y. Non-coding RNA in cancer drug resistance: Underlying mechanisms and clinical applications. Front. Oncol. 2022, 12, 951864. [Google Scholar] [CrossRef]
- Chen, B.; Dragomir, M.P.; Yang, C.; Li, Q.; Horst, D.; Calin, G.A. Targeting non-coding RNAs to overcome cancer therapy resistance. Signal Transduct. Target Ther. 2022, 7, 121. [Google Scholar] [CrossRef]
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. LNCcation: lncRNA localization and function. J. Cell. Biol. 2021, 220, e202009045. [Google Scholar] [CrossRef]
- Lagarde, J.; Uszczynska-Ratajczak, B.; Carbonell, S.; Perez-Lluch, S.; Abad, A.; Davis, C.; Gingeras, T.R.; Frankish, A.; Harrow, J.; Guigo, R.; et al. High-throughput annotation of full-length long noncoding RNAs with capture long-read sequencing. Nat. Genet. 2017, 49, 1731–1740. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell. Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Bao, Z.; Yang, Z.; Huang, Z.; Zhou, Y.; Cui, Q.; Dong, D. LncRNADisease 2.0: An updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019, 47, D1034–D1037. [Google Scholar] [CrossRef]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigo, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet 2018, 19, 535–548. [Google Scholar] [CrossRef]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Furio-Tari, P.; Tarazona, S.; Gabaldon, T.; Enright, A.J.; Conesa, A. spongeScan: A web for detecting microRNA binding elements in lncRNA sequences. Nucleic Acids Res. 2016, 44, W176–W180. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.; King, R.M.; Philipson, L. Genes specifically expressed at growth arrest of mammalian cells. Cell 1988, 54, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Salehen, N.; Norazit, A.; Rahman, A.A.; Murad, N.A.A.; Rahman, N.; Ibrahim, K. Tumor Suppressive Effects of GAS5 in Cancer Cells. Noncoding RNA 2022, 8, 39. [Google Scholar] [CrossRef]
- Zhou, Z.; Chen, J.; Huang, Y.; Liu, D.; Chen, S.; Qin, S. Long Noncoding RNA GAS5: A New Factor Involved in Bone Diseases. Front. Cell. Dev. Biol. 2021, 9, 807419. [Google Scholar] [CrossRef]
- Lin, G.; Wu, T.; Gao, X.; He, Z.; Nong, W. Research Progress of Long Non-Coding RNA GAS5 in Malignant Tumors. Front. Oncol. 2022, 12, 846497. [Google Scholar] [CrossRef]
- Wang, C.; Ke, S.; Li, M.; Lin, C.; Liu, X.; Pan, Q. Downregulation of LncRNA GAS5 promotes liver cancer proliferation and drug resistance by decreasing PTEN expression. Mol. Genet. Genom. 2020, 295, 251–260. [Google Scholar] [CrossRef]
- Gu, J.; Wang, Y.; Wang, X.; Zhou, D.; Wang, X.; Zhou, M.; He, Z. Effect of the LncRNA GAS5-MiR-23a-ATG3 Axis in Regulating Autophagy in Patients with Breast Cancer. Cell. Physiol. Biochem. 2018, 48, 194–207. [Google Scholar] [CrossRef]
- Gao, Z.Q.; Wang, J.F.; Chen, D.H.; Ma, X.S.; Wu, Y.; Tang, Z.; Dang, X.W. Long non-coding RNA GAS5 suppresses pancreatic cancer metastasis through modulating miR-32-5p/PTEN axis. Cell. Biosci. 2017, 7, 66. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Z.; Wang, H. Downregulation of lncRNA GAS5 prevents mitochondrial apoptosis and hypoxic-ischemic brain damage in neonatal rats through the microRNA-128-3p/Bax/Akt/GSK-3beta axis. Neuroreport 2021, 32, 1395–1402. [Google Scholar] [CrossRef]
- Peng, T.; Ji, D.; Jiang, Y. Long non-coding RNA GAS5 suppresses rheumatoid arthritis progression via miR-128-3p/HDAC4 axis. Mol. Cell. Biochem. 2021, 476, 2491–2501. [Google Scholar] [CrossRef]
- Zhao, H.; Yu, H.; Zheng, J.; Ning, N.; Tang, F.; Yang, Y.; Wang, Y. Lowly-expressed lncRNA GAS5 facilitates progression of ovarian cancer through targeting miR-196-5p and thereby regulating HOXA5. Gynecol. Oncol. 2018, 151, 345–355. [Google Scholar] [CrossRef]
- Zhang, X.F.; Ye, Y.; Zhao, S.J. LncRNA Gas5 acts as a ceRNA to regulate PTEN expression by sponging miR-222-3p in papillary thyroid carcinoma. Oncotarget 2018, 9, 3519–3530. [Google Scholar] [CrossRef]
- Zheng, S.; Li, M.; Miao, K.; Xu, H. lncRNA GAS5-promoted apoptosis in triple-negative breast cancer by targeting miR-378a-5p/SUFU signaling. J. Cell. Biochem. 2020, 121, 2225–2235. [Google Scholar] [CrossRef]
- Shin, Y.-K.; Yoo, B.C.; Chang, H.J.; Jeon, E.; Hong, S.-H.; Jung, M.-S.; Lim, S.-J.; Park, J.-G. Down-regulation of Mitochondrial F1F0-ATP Synthase in Human Colon Cancer Cells with Induced 5-Fluorouracil Resistance. Cancer Res. 2005, 65, 3162–3170. [Google Scholar] [CrossRef]
- Kim, C.; Hong, Y.; Lee, H.; Kang, H.; Lee, E.K. MicroRNA-195 desensitizes HCT116 human colon cancer cells to 5-fluorouracil. Cancer Lett. 2018, 412, 264–271. [Google Scholar] [CrossRef]
- Yoon, J.H.; Gorospe, M. Identification of mRNA-Interacting Factors by MS2-TRAP (MS2-Tagged RNA Affinity Purification). Methods Mol. Biol. 2016, 1421, 15–22. [Google Scholar] [CrossRef]
- Hong, Y.; Tak, H.; Kim, C.; Kang, H.; Ji, E.; Ahn, S.; Jung, M.; Kim, H.L.; Lee, J.H.; Kim, W.; et al. RNA binding protein HuD contributes to beta-cell dysfunction by impairing mitochondria dynamics. Cell Death Differ. 2020, 27, 1633–1643. [Google Scholar] [CrossRef]
- Lee, H.; Kim, C.; Ku, J.L.; Kim, W.; Yoon, S.K.; Kuh, H.J.; Lee, J.H.; Nam, S.W.; Lee, E.K. A long non-coding RNA snaR contributes to 5-fluorouracil resistance in human colon cancer cells. Mol. Cells 2014, 37, 540–546. [Google Scholar] [CrossRef]
- Adlakha, Y.K.; Saini, N. MicroRNA-128 downregulates Bax and induces apoptosis in human embryonic kidney cells. Cell. Mol. Life Sci. 2011, 68, 1415–1428. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Shao, G.; Lv, X.; Liu, Y.; Fan, Y.; Wu, A.; Hu, H. Downregulation of miRNA-128 sensitises breast cancer cell to chemodrugs by targeting Bax. Cell. Biol. Int. 2013, 37, 653–658. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Liu, L.; Li, G.; Cai, M.; Tan, C.; Han, X.; Han, L. LncRNA GAS5 confers the radio sensitivity of cervical cancer cells via regulating miR-106b/IER3 axis. Int. J. Biol. Macromol. 2019, 126, 994–1001. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Pan, T.; Jiang, D.; Jin, L.; Geng, Y.; Feng, X.; Shen, A.; Zhang, L. The lncRNA-GAS5/miR-221-3p/DKK2 Axis Modulates ABCB1-Mediated Adriamycin Resistance of Breast Cancer via the Wnt/beta-Catenin Signaling Pathway. Mol. Ther. Nucleic Acids 2020, 19, 1434–1448. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Liu, Z.; Liao, S.; Li, E.; Wu, X.; Zeng, W. Elevation of long non-coding RNA GAS5 and knockdown of microRNA-21 up-regulate RECK expression to enhance esophageal squamous cell carcinoma cell radio-sensitivity after radiotherapy. Genomics 2020, 112, 2173–2185. [Google Scholar] [CrossRef]
- Yang, X.; Meng, L.; Zhong, Y.; Hu, F.; Wang, L.; Wang, M. The long intergenic noncoding RNA GAS5 reduces cisplatin-resistance in non-small cell lung cancer through the miR-217/LHPP axis. Aging 2021, 13, 2864–2884. [Google Scholar] [CrossRef]
- Mourtada-Maarabouni, M.; Hasan, A.M.; Farzaneh, F.; Williams, G.T. Inhibition of human T-cell proliferation by mammalian target of rapamycin (mTOR) antagonists requires noncoding RNA growth-arrest-specific transcript 5 (GAS5). Mol. Pharmacol. 2010, 78, 19–28. [Google Scholar] [CrossRef]
- Tani, H.; Torimura, M.; Akimitsu, N. The RNA degradation pathway regulates the function of GAS5 a non-coding RNA in mammalian cells. PLoS ONE 2013, 8, e55684. [Google Scholar] [CrossRef]
- Li, J.; Li, L.; Yuan, H.; Huang, X.W.; Xiang, T.; Dai, S. Up-regulated lncRNA GAS5 promotes chemosensitivity and apoptosis of triple-negative breast cancer cells. Cell Cycle 2019, 18, 1965–1975. [Google Scholar] [CrossRef]
- Tian, D.; Tian, M.; Han, G.; Li, J.L. Increased glucocorticoid receptor activity and proliferation in metastatic colon cancer. Sci. Rep. 2019, 9, 11257. [Google Scholar] [CrossRef]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010, 3, ra8. [Google Scholar] [CrossRef]
- Mayama, T.; Marr, A.K.; Kino, T. Differential Expression of Glucocorticoid Receptor Noncoding RNA Repressor Gas5 in Autoimmune and Inflammatory Diseases. Horm. Metab. Res. 2016, 48, 550–557. [Google Scholar] [CrossRef]
- Lambrou, G.I.; Hatziagapiou, K.; Zaravinos, A. The Non-Coding RNA GAS5 and Its Role in Tumor Therapy-Induced Resistance. Int. J. Mol. Sci. 2020, 21, 7633. [Google Scholar] [CrossRef]
- Xu, X.; Jiang, W.; Han, P.; Zhang, J.; Tong, L.; Sun, X. MicroRNA-128-3p Mediates Lenvatinib Resistance of Hepatocellular Carcinoma Cells by Downregulating c-Met. J. Hepatocell. Carcinoma 2022, 9, 113–126. [Google Scholar] [CrossRef]
- Zhao, C.; Guo, R.; Guan, F.; Ma, S.; Li, M.; Wu, J.; Liu, X.; Li, H.; Yang, B. MicroRNA-128-3p Enhances the Chemosensitivity of Temozolomide in Glioblastoma by Targeting c-Met and EMT. Sci. Rep. 2020, 10, 9471. [Google Scholar] [CrossRef]
- Koh, H.; Park, H.; Chandimali, N.; Huynh, D.L.; Zhang, J.J.; Ghosh, M.; Gera, M.; Kim, N.; Bak, Y.; Yoon, D.Y.; et al. MicroRNA-128 suppresses paclitaxel-resistant lung cancer by inhibiting MUC1-C and BMI-1 in cancer stem cells. Oncotarget 2017, 8, 110540–110551. [Google Scholar] [CrossRef]
- Li, B.; Chen, H.; Wu, N.; Zhang, W.J.; Shang, L.X. Deregulation of miR-128 in ovarian cancer promotes cisplatin resistance. Int. J. Gynecol. Cancer 2014, 24, 1381–1388. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.; Kang, H.; Kim, C.; Ku, J.-L.; Nam, S.; Lee, E.K. Long Non-Coding RNA GAS5 Promotes BAX Expression by Competing with microRNA-128-3p in Response to 5-Fluorouracil. Biomedicines 2023, 11, 58. https://doi.org/10.3390/biomedicines11010058
Lee H, Kang H, Kim C, Ku J-L, Nam S, Lee EK. Long Non-Coding RNA GAS5 Promotes BAX Expression by Competing with microRNA-128-3p in Response to 5-Fluorouracil. Biomedicines. 2023; 11(1):58. https://doi.org/10.3390/biomedicines11010058
Chicago/Turabian StyleLee, Heejin, Hoin Kang, Chongtae Kim, Ja-Lok Ku, Sukwoo Nam, and Eun Kyung Lee. 2023. "Long Non-Coding RNA GAS5 Promotes BAX Expression by Competing with microRNA-128-3p in Response to 5-Fluorouracil" Biomedicines 11, no. 1: 58. https://doi.org/10.3390/biomedicines11010058
APA StyleLee, H., Kang, H., Kim, C., Ku, J.-L., Nam, S., & Lee, E. K. (2023). Long Non-Coding RNA GAS5 Promotes BAX Expression by Competing with microRNA-128-3p in Response to 5-Fluorouracil. Biomedicines, 11(1), 58. https://doi.org/10.3390/biomedicines11010058






