Association of N-Acetyl Asparagine with QTc in Diabetes: A Metabolomics Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Metabolomics Study
2.2.1. Reference Compounds and Reagents
2.2.2. Sera Preparation for Metabolomic Analyses
2.2.3. Ultra-Performance Liquid Chromatography-Mass Spectrometry (UPLC-MS) Analyses of Serum Samples
2.3. Weighted Gene Co-Expression Network Analysis and Visualization
2.4. Electrocardiography Acquisition and QTc Analysis
2.5. Statistical Analysis
3. Results
3.1. Baseline Demographics and Clinical Characteristics
3.2. Untargeted Metabolomics Analysis Using Weighted Gene Correlation Network Analysis (WGCNA)
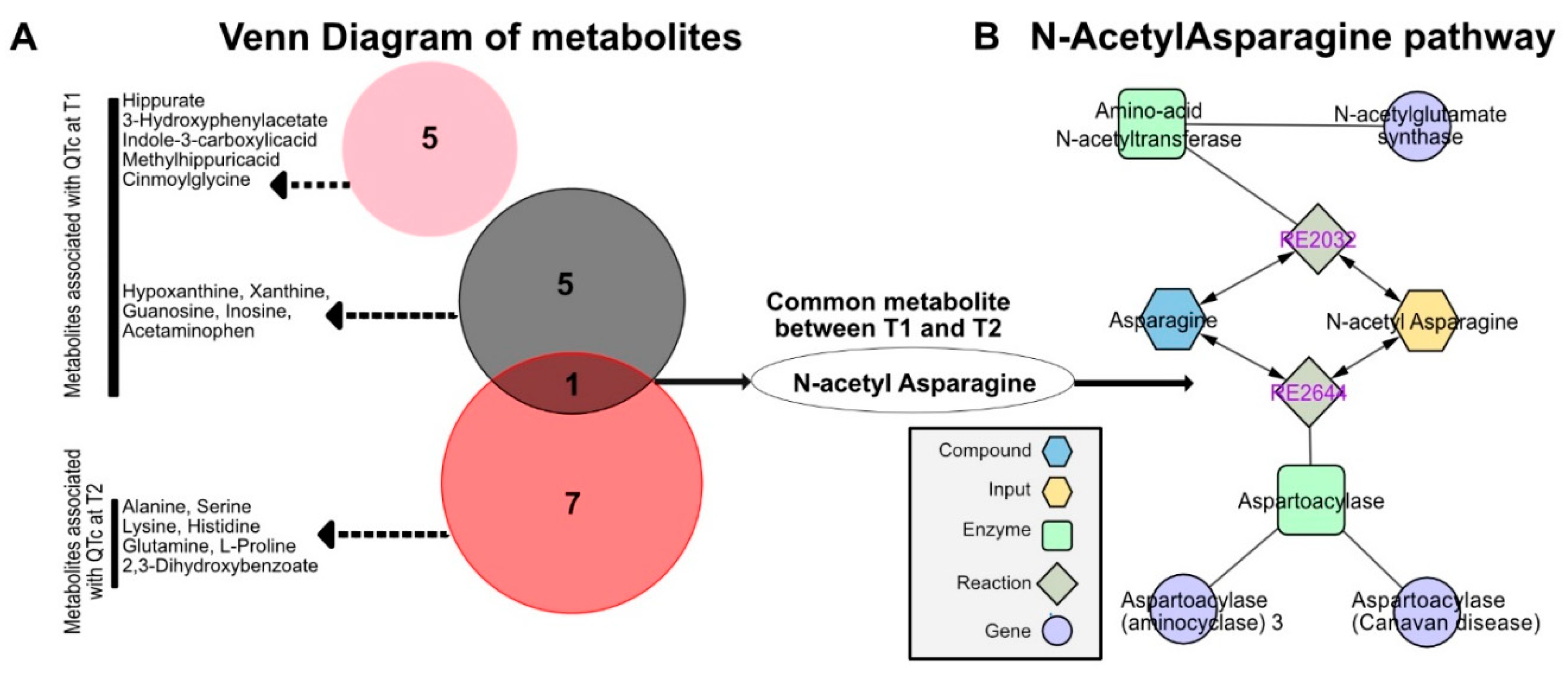
3.3. Identification of N-Acetyl Asparagine
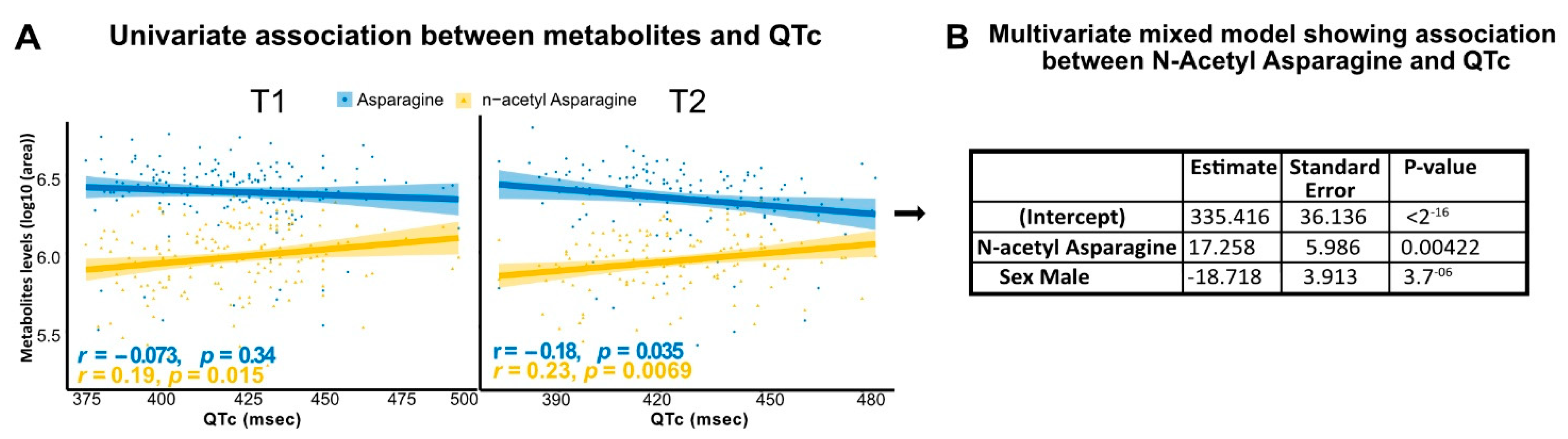
3.4. N-Acetyl Asparagine Positively Correlates with QTc
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lynge, T.H.; Svane, J.; Pedersen-Bjergaard, U.; Gislason, G.; Torp-Pedersen, C.; Banner, J.; Risgaard, B.; Winkel, B.G.; Tfelt-Hansen, J. Sudden cardiac death among persons with diabetes aged 1-49 years: A 10-year nationwide study of 14 294 deaths in Denmark. Eur. Heart J. 2020, 41, 2699–2706. [Google Scholar] [CrossRef] [PubMed]
- Rana, B.S.; Band, M.M.; Ogston, S.; Morris, A.D.; Pringle, S.D.; Struthers, A.D. Relation of QT interval dispersion to the number of different cardiac abnormalities in diabetes mellitus. Am. J. Cardiol. 2002, 90, 483–487. [Google Scholar] [CrossRef]
- Aune, D.; Schlesinger, S.; Norat, T.; Riboli, E. Diabetes mellitus and the risk of sudden cardiac death: A systematic review and meta-analysis of prospective studies. Nutr. Metab. Cardiovasc. Dis. 2018, 28, 543–556. [Google Scholar] [CrossRef] [PubMed]
- Saque, V.; Vaglio, M.; Funck-Brentano, C.; Kilani, M.; Bourron, O.; Hartemann, A.; Badilini, F.; Salem, J.E. Fast, accurate and easy-to-teach QT interval assessment: The triplicate concatenation method. Arch. Cardiovasc. Dis. 2017, 110, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Salem, J.E.; Yang, T.; Moslehi, J.J.; Waintraub, X.; Gandjbakhch, E.; Bachelot, A.; Hidden-Lucet, F.; Hulot, J.S.; Knollmann, B.C.; Lebrun-Vignes, B.; et al. Androgenic Effects on Ventricular Repolarization: A Translational Study from the International Pharmacovigilance Database to iPSC-Cardiomyocytes. Circulation 2019, 140, 1070–1080. [Google Scholar] [CrossRef]
- Roden, D.M. Drug-induced prolongation of the QT interval. N. Engl. J. Med. 2004, 350, 1013–1022. [Google Scholar] [CrossRef]
- De Bruin, M.L.; Pettersson, M.; Meyboom, R.H.; Hoes, A.W.; Leufkens, H.G. Anti-HERG activity and the risk of drug-induced arrhythmias and sudden death. Eur. Heart J. 2005, 26, 590–597. [Google Scholar] [CrossRef]
- Nicholson, J.K. Global systems biology, personalized medicine and molecular epidemiology. Mol. Syst. Biol. 2006, 2, 52. [Google Scholar] [CrossRef]
- McGarrah, R.W.; Crown, S.B.; Zhang, G.F.; Shah, S.H.; Newgard, C.B. Cardiovascular Metabolomics. Circ. Res. 2018, 122, 1238–1258. [Google Scholar] [CrossRef]
- Park, J.; Noh, K.; Lee, H.W.; Lim, M.S.; Seong, S.J.; Seo, J.J.; Kim, E.J.; Kang, W.; Yoon, Y.R. Pharmacometabolomic approach to predict QT prolongation in guinea pigs. PLoS ONE 2013, 8, e60556. [Google Scholar] [CrossRef]
- Campagna, M.; Locci, E.; Piras, R.; Noto, A.; Lecca, L.I.; Pilia, I.; Cocco, P.; d’Aloja, E.; Scano, P. Metabolomic patterns associated to QTc interval in shiftworkers: An explorative analysis. Biomarkers 2016, 21, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Bourron, O.; Aubert, C.E.; Liabeuf, S.; Cluzel, P.; Lajat-Kiss, F.; Dadon, M.; Komajda, M.; Mentaverri, R.; Brazier, M.; Pierucci, A.; et al. Below-knee arterial calcification in type 2 diabetes: Association with receptor activator of nuclear factor kappaB ligand, osteoprotegerin, and neuropathy. J. Clin. Endocrinol. Metab. 2014, 99, 4250–4258. [Google Scholar] [CrossRef]
- Madhukar, R.; Jagadeesh, A.T.; Moey, M.Y.Y.; Vaglio, M.; Badilini, F.; Leban, M.; Hartemann, A.; Dureau, P.; Funck-Brentano, C.; Bourron, O.; et al. Association of thyroid-stimulating hormone with corrected QT interval variation: A prospective cohort study among patients with type 2 diabetes. Arch. Cardiovasc. Dis. 2021, 114, 656–666. [Google Scholar] [CrossRef] [PubMed]
- Bourron, O.; Phan, F.; Diallo, M.H.; Hajage, D.; Aubert, C.E.; Carlier, A.; Salem, J.E.; Funck-Brentano, C.; Kemel, S.; Cluzel, P.; et al. Circulating Receptor Activator of Nuclear Factor kB Ligand and triglycerides are associated with progression of lower limb arterial calcification in type 2 diabetes: A prospective, observational cohort study. Cardiovasc. Diabetol. 2020, 19, 140. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.S.; Prifti, E.; Ichou, F.; Leban, M.; Funck-Brentano, C.; Touraine, P.; Salem, J.E.; Bachelot, A. Effect of congenital adrenal hyperplasia treated by glucocorticoids on plasma metabolome: A machine-learning-based analysis. Sci. Rep. 2020, 10, 8859. [Google Scholar] [CrossRef]
- Kessner, D.; Chambers, M.; Burke, R.; Agus, D.; Mallick, P. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics 2008, 24, 2534–2536. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Giacomoni, F.; Le Corguille, G.; Monsoor, M.; Landi, M.; Pericard, P.; Petera, M.; Duperier, C.; Tremblay-Franco, M.; Martin, J.F.; Jacob, D.; et al. Workflow4Metabolomics: A collaborative research infrastructure for computational metabolomics. Bioinformatics 2015, 31, 1493–1495. [Google Scholar] [CrossRef]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef]
- Boudah, S.; Olivier, M.F.; Aros-Calt, S.; Oliveira, L.; Fenaille, F.; Tabet, J.C.; Junot, C. Annotation of the human serum metabolome by coupling three liquid chromatography methods to high-resolution mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2014, 966, 34–47. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Karnovsky, A.; Weymouth, T.; Hull, T.; Tarcea, V.G.; Scardoni, G.; Laudanna, C.; Sartor, M.A.; Stringer, K.A.; Jagadish, H.V.; Burant, C.; et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics 2012, 28, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Woosley, R.L.; Heise, C.W.; Gallo, T.; Tate, J.; Woosley, D.; Romero, K.A. QTdrugs List [Internet]. Available online: https://crediblemeds.org/ (accessed on 3 May 2020).
- Cheng, S.; Shah, S.H.; Corwin, E.J.; Fiehn, O.; Fitzgerald, R.L.; Gerszten, R.E.; Illig, T.; Rhee, E.P.; Srinivas, P.R.; Wang, T.J.; et al. Potential Impact and Study Considerations of Metabolomics in Cardiovascular Health and Disease: A Scientific Statement from the American Heart Association. Circ. Cardiovasc. Genet. 2017, 10, e000032. [Google Scholar] [CrossRef] [PubMed]
- Vieira-Silva, S.; Falony, G.; Belda, E.; Nielsen, T.; Aron-Wisnewsky, J.; Chakaroun, R.; Forslund, S.K.; Assmann, K.; Valles-Colomer, M.; Nguyen, T.T.D.; et al. Statin therapy is associated with lower prevalence of gut microbiota dysbiosis. Nature 2020, 581, 310–315. [Google Scholar] [CrossRef]
- Wang, D.; Wang, X.; Wu, J.; Su, R.; Kong, J.; Yu, X. Metabolic risk factors associated with sudden cardiac death (SCD) during acute myocardial ischemia. Forensic Sci. Res. 2017, 2, 126–131. [Google Scholar] [CrossRef]
- Mehta, A.; Liu, C.; Nayak, A.; Tahhan, A.S.; Ko, Y.A.; Dhindsa, D.S.; Kim, J.H.; Hayek, S.S.; Sperling, L.S.; Mehta, P.K.; et al. Untargeted high-resolution plasma metabolomic profiling predicts outcomes in patients with coronary artery disease. PLoS ONE 2020, 15, e0237579. [Google Scholar] [CrossRef]
- Holmes, E.; Wilson, I.D.; Nicholson, J.K. Metabolic phenotyping in health and disease. Cell 2008, 134, 714–717. [Google Scholar] [CrossRef]
- Owen, O.E.; Kalhan, S.C.; Hanson, R.W. The key role of anaplerosis and cataplerosis for citric acid cycle function. J. Biol. Chem. 2002, 277, 30409–30412. [Google Scholar] [CrossRef]
- Julia, P.; Young, H.H.; Buckberg, G.D.; Kofsky, E.R.; Bugyi, H.I. Studies of myocardial protection in the immature heart. II. Evidence for importance of amino acid metabolism in tolerance to ischemia. J. Thorac. Cardiovasc. Surg. 1990, 100, 888–895. [Google Scholar] [CrossRef]
- Zhang, Y.; Han, H.; Wang, J.; Wang, H.; Yang, B.; Wang, Z. Impairment of human ether-a-go-go-related gene (HERG) K+ channel function by hypoglycemia and hyperglycemia. Similar phenotypes but different mechanisms. J. Biol. Chem. 2003, 278, 10417–10426. [Google Scholar] [CrossRef] [PubMed]
- Vinik, A.I.; Ziegler, D. Diabetic cardiovascular autonomic neuropathy. Circulation 2007, 115, 387–397. [Google Scholar] [CrossRef] [PubMed]
- van Dongen, L.H.; Harms, P.P.; Hoogendoorn, M.; Zimmerman, D.S.; Lodder, E.M.; M’t Hart, L.M.; Herings, R.; van Weert, H.; Nijpels, G.; Swart, K.M.A.; et al. Discovery of predictors of sudden cardiac arrest in diabetes: Rationale and outline of the RESCUED (REcognition of Sudden Cardiac arrest vUlnErability in Diabetes) project. Open Heart 2021, 8, e001554. [Google Scholar] [CrossRef] [PubMed]

| Measurement at T1 (N = 170) | Measurement at T2 (N = 139) | |
|---|---|---|
| General Characteristics | ||
| Age, years (mean ± SD) | 63.9 ± 8.4 | 66.8 ± 8.4 |
| Male, n (%) | 132 (77.7) | 112 (80.6) |
| Weight, kg (mean ± SD) | 83.4 ± 15.3 | 83.2 ± 15.8 |
| Height, m (median [IQR]) | 1.71 (1.65–1.76) | 1.71 (1.65–1.76) |
| BMI, kg/m2 (mean ± SD) | 28.9 ± 4.7 | 28.8 ± 4.9 |
| History of CAD a, n (%) | 110 (64.7) | 90 (64.7) |
| Hypertension, n (%) | 137 (80.6) | 119 (85.7) |
| Metabolic Biochemistry Profile | ||
| HbA1c, % (median [IQR]) | 7.5 (7.0–8.3) | 7.6 (6.9–8.3) |
| Blood glucose, mmol/L (median (IQR)) | 7.8 (6.4–9.3) | 8.1 (6.6–10.4) |
| Triglycerides, mmol/L (median (IQR)) | 1.2 (0.8–2.0) | 1.4 (0.9–2.0) |
| Total cholesterol, mmol/L(median (IQR)) | 3.7 (3.2–4.4) | 3.8 (3.4–4.4) |
| HDL cholesterol, mmol/L(median (IQR)) | 1.1 (0.9–1.3) | 1.1 (0.9–1.3) |
| LDL cholesterol, mmol/L (median (IQR)) | 1.9 (1.5–2.4) | 1.9 (1.6–2.4) |
| QT Prolonging Drugsb | ||
| Present, n (%) | 8 (4.7) | 8 (5.8) |
| Basic Metabolic Profile | ||
| Calcium c, mmol/L (mean ± SD) | 2.3 ± 0.1 | 2.3 ± 0.1 |
| Potassium, mmol/L (mean ± SD) | 4.7 ± 0.4 | 4.6 ± 0.4 |
| Creatinine, mmol/L (median [IQR]) | 84 (74–101) | 87 (76–105) |
| Albumin, g/L (median [IQR]) | 42.5 (39.8–44.4) | 43 (40.5–45) |
| Electrocardiogram Variables | ||
| Heart rate, beats/min (mean ± SD) | 69.7 ± 11.1 | 69.1 ± 11.8 |
| QTc, ms (mean ± SD) | 422 ± 24.9 | 424.9 ± 24.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gravina, G.; Moey, M.Y.Y.; Prifti, E.; Ichou, F.; Bourron, O.; Balse, E.; Badillini, F.; Funck-Brentano, C.; Salem, J.-E. Association of N-Acetyl Asparagine with QTc in Diabetes: A Metabolomics Study. Biomedicines 2022, 10, 1955. https://doi.org/10.3390/biomedicines10081955
Gravina G, Moey MYY, Prifti E, Ichou F, Bourron O, Balse E, Badillini F, Funck-Brentano C, Salem J-E. Association of N-Acetyl Asparagine with QTc in Diabetes: A Metabolomics Study. Biomedicines. 2022; 10(8):1955. https://doi.org/10.3390/biomedicines10081955
Chicago/Turabian StyleGravina, Giacomo, Melissa Y. Y. Moey, Edi Prifti, Farid Ichou, Olivier Bourron, Elise Balse, Fabio Badillini, Christian Funck-Brentano, and Joe-Elie Salem. 2022. "Association of N-Acetyl Asparagine with QTc in Diabetes: A Metabolomics Study" Biomedicines 10, no. 8: 1955. https://doi.org/10.3390/biomedicines10081955
APA StyleGravina, G., Moey, M. Y. Y., Prifti, E., Ichou, F., Bourron, O., Balse, E., Badillini, F., Funck-Brentano, C., & Salem, J.-E. (2022). Association of N-Acetyl Asparagine with QTc in Diabetes: A Metabolomics Study. Biomedicines, 10(8), 1955. https://doi.org/10.3390/biomedicines10081955






