The Role of Matrix Metalloproteinase Single-Nucleotide Polymorphisms in the Clinicopathological Properties of Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. DNA Extraction and Polymerase Chain Reaction—Restriction Fragment Length Polymorphism (PCR-RFLP)
2.3. Statistical Analysis
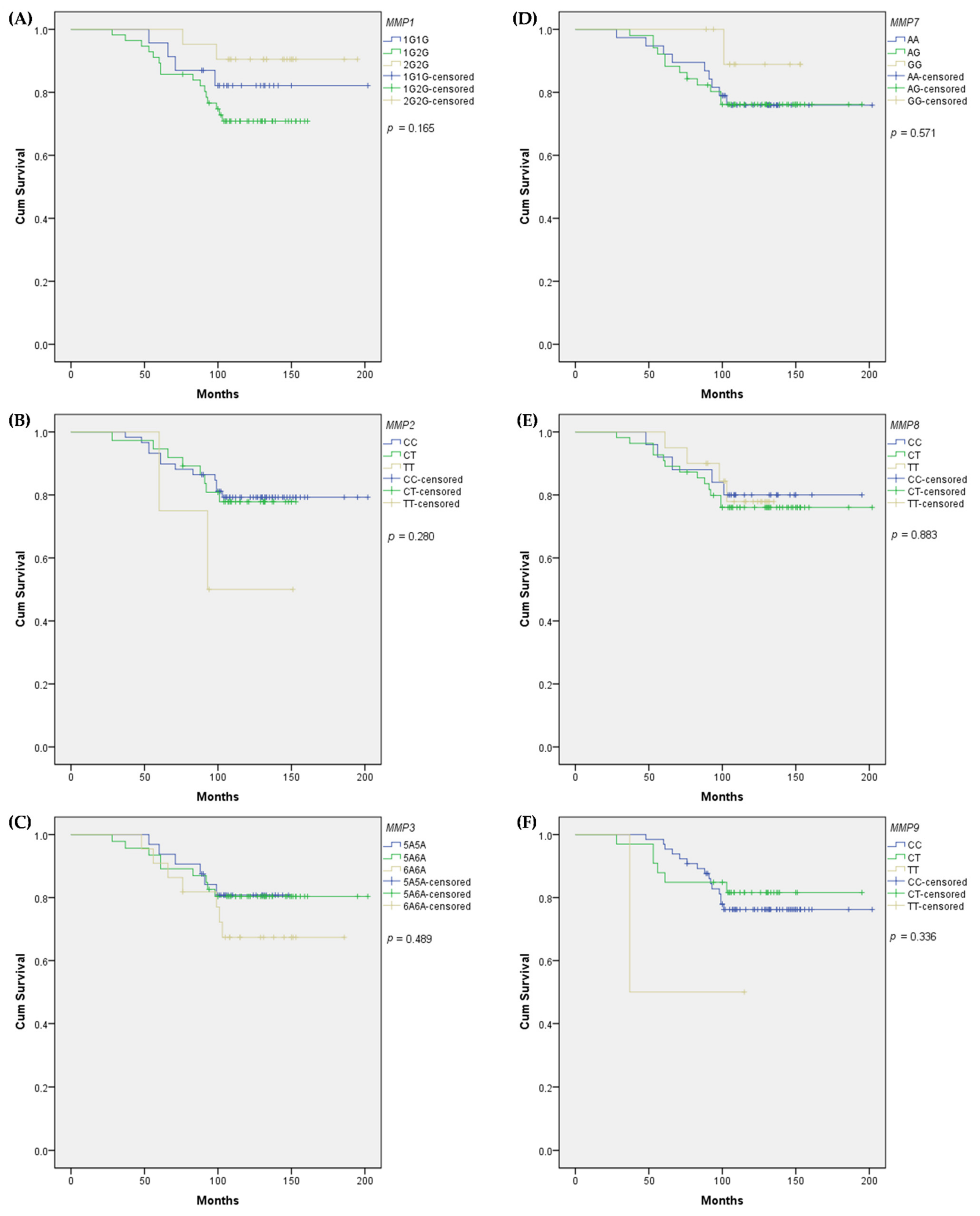
3. Results
3.1. Cohort Characteristics and Genotypes Distribution
3.2. Associations of SNPs with BC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Britt, K.L.; Cuzick, J.; Phillips, K.A. Key Steps for Effective Breast Cancer Prevention. Nat. Rev. Cancer 2020, 20, 417–436. [Google Scholar] [CrossRef]
- Zardavas, D.; Irrthum, A.; Swanton, C.; Piccart, M. Clinical Management of Breast Cancer Heterogeneity. Nat. Rev. Clin. Oncol. 2015, 12, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Turashvili, G.; Brogi, E. Tumor Heterogeneity in Breast Cancer. Front. Med. 2017, 4, 227. [Google Scholar] [CrossRef] [PubMed]
- Toriola, A.T.; Colditz, G.A. Trends in Breast Cancer Incidence and Mortality in the United States: Implications for Prevention. Breast Cancer Res. Treat. 2013, 138, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Mu, P. Targeting Breast Cancer Metastasis. Breast Cancer Basic Clin. Res. 2015, 9, 23–34. [Google Scholar] [CrossRef]
- Nicolini, A.; Ferrari, P.; Duffy, M.J. Prognostic and Predictive Biomarkers in Breast Cancer: Past, Present and Future. Semin. Cancer Biol. 2018, 52, 56–73. [Google Scholar] [CrossRef]
- Klein, T.; Bischoff, R. Physiology and Pathophysiology of Matrix Metalloproteases. Amino Acids 2011, 41, 271–290. [Google Scholar] [CrossRef]
- Laronha, H.; Caldeira, J. Structure and Function of Human Matrix Metalloproteinases. Cells 2020, 9, 1076. [Google Scholar] [CrossRef]
- Gialeli, C.; Theocharis, A.D.; Karamanos, N.K. Roles of Matrix Metalloproteinases in Cancer Progression and their Pharmacological Targeting. FEBS J. 2011, 278, 16–27. [Google Scholar] [CrossRef]
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix Metalloproteinases: Regulators of the Tumor Microenvironment. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef]
- Liu, H.; Kato, Y.; Erzinger, S.A.; Kiriakova, G.M.; Qian, Y.; Palmieri, D.; Steeg, P.; Price, J.E. The Role of MMP-1 in Breast Cancer Growth and Metastasis to the Brain in a Xenograft Model. BMC Cancer 2012, 12, 583. [Google Scholar] [CrossRef]
- Przybylowska, K.; Kluczna, A.; Zadrozny, M.; Krawczyk, T.; Kulig, A.; Rykala, J.; Kolacinska, A.; Morawiec, Z.; Drzewoski, J.; Blasiak, J. Polymorphisms of the Promoter Regions of Matrix Metalloproteinases Genes MMP-1 and MMP-9 in Breast Cancer. Breast Cancer Res. Treat. 2006, 95, 65–72. [Google Scholar] [CrossRef]
- Jonsson, A.; Falk, P.; Angenete, E.; Hjalmarsson, C.; Ivarsson, M.L. Plasma MMP-1 Expression as a Prognostic Factor in Colon Cancer. J. Surg. Res. 2021, 266, 254–260. [Google Scholar] [CrossRef]
- Padala, C.; Tupurani, M.A.; Puranam, K.; Gantala, S.; Shyamala, N.; Kondapalli, M.S.; Gundapaneni, K.K.; Mudigonda, S.; Galimudi, R.K.; Kupsal, K.; et al. Synergistic Effect of Collagenase-1 (MMP1), Stromelysin-1 (MMP3) and Gelatinase-B (MMP9) Gene Polymorphisms in Breast Cancer. PLoS ONE 2017, 12, e0184448. [Google Scholar] [CrossRef]
- Hughes, S.; Agbaje, O.; Bowen, R.L.; Holliday, D.L.; Shaw, J.A.; Duffy, S.; Jones, J.L. Matrix Metalloproteinase Single-nucleotide Polymorphisms and Haplotypes Predict Breast Cancer Progression. Clin. Cancer Res. 2007, 13, 6673–6680. [Google Scholar] [CrossRef]
- Liao, C.H.; Tsai, C.W.; Chang, W.S.; Wang, Z.H.; Gong, C.L.; Wu, H.C.; Wang, B.R.; Hsu, S.W.; Huang, W.C.; Shen, T.C.; et al. Association of Matrix Metalloproteinase-1 Genotypes With Bladder Cancer Risk. In Vivo 2021, 35, 2535–2540. [Google Scholar] [CrossRef]
- Balkhi, S.; Mashayekhi, F.; Salehzadeh, A.; Saedi, H.S. Matrix Metalloproteinase (MMP)-1 and MMP-3 gene Variations Affect MMP-1 and -3 Serum Concentration and Associates with Breast Cancer. Mol. Biol. Rep. 2020, 47, 9637–9644. [Google Scholar] [CrossRef]
- Su, C.H.; Lane, H.Y.; Hsiao, C.L.; Liu, L.C.; Ji, H.X.; Li, H.T.; Yen, S.-T.; Su, C.-H.; Hsia, T.-C.; Chang, W.-S.; et al. Matrix Metalloproteinase-1 Genetic Polymorphism in Breast Cancer in Taiwanese. Anticancer Res. 2016, 36, 3341–3345. [Google Scholar]
- Wang, L.; Kong, B. Analysis of the Association of Matrix Metalloproteinase-1 Gene promoter (rs1799750) Polymorphism and Risk of Ovarian Cancer. Int. J. Gynecol. Cancer 2015, 25, 961–967. [Google Scholar] [CrossRef]
- Peng, Q.; Xu, Y. Association between Promoter Polymorphisms of Matrix Metalloproteinase-1 and Risk of Gastric Cancer. Onco Targets Ther. 2015, 8, 2519–2526. [Google Scholar]
- Dofara, S.G.; Chang, S.L.; Diorio, C. Gene Polymorphisms and Circulating Levels of MMP-2 and MMP-9: A Review of their Role in Breast Cancer Risk. Anticancer Res. 2020, 40, 3619–3631. [Google Scholar] [CrossRef]
- Abbas, Y.J.; Al-Tu’ma, F.J.; Al-Hemerri, A.F. Association between Matrix Metalloproteinas-2 Gene Variants and Pathogenesis of Breast Cancer in Sera of Iraqi Women. J. Contemp Med. Sci 2020, 6, 285–290. [Google Scholar] [CrossRef]
- Ghilardi, G.; Biondi, M.L.; Caputo, M.; Leviti, S.; DeMonti, M.; Guagnellini, E.; Scorza, R. A Single Nucleotide Polymorphism in the Matrix Metalloproteinase-3 Promoter Enhances Breast Cancer Susceptibility. Clin. Cancer Res. 2002, 8, 3820–3823. [Google Scholar]
- AbdRaboh, N.R.; Bayoumi, F.A. Gene Polymorphism of Matrix Metalloproteinases 3 and 9 in Breast Cancer. Gene Rep. 2016, 5, 151–156. [Google Scholar] [CrossRef]
- Beeghly-Fadiel, A.; Shu, X.O.; Long, J.; Li, C.; Cai, Q.; Cai, H.; Gao, Y.-T.; Zheng, W. Genetic Polymorphisms in the MMP-7 Gene and Breast Cancer Survival. Int. J. Cancer 2009, 124, 208–214. [Google Scholar] [CrossRef]
- Zhou, P.; Du, L.F.; Lv, G.Q.; Yu, X.M.; Gu, Y.L.; Li, J.P.; Zhang, C. Current Evidence on the Relationship Between Four Polymorphisms in the Matrix Metalloproteinases (MMP) Gene and Breast Cancer Risk: A Meta-analysis. Breast Cancer Res. Treat. 2011, 127, 813–818. [Google Scholar] [CrossRef]
- Feng, J.; Chen, Y.; Hua, W.; Sun, X.; Chen, Y.; Liu, Y.; Fan, J.; Zhao, Y.; Xu, X.; Yang, X. The MMP-8 rs11225395 Promoter Polymorphism Increases Cancer Risk of Non-Asian Populations: Evidence from a Meta-Analysis. Biomolecules 2019, 9, 570. [Google Scholar] [CrossRef]
- Decock, J.; Long, J.R.; Laxton, R.C.; Shu, X.O.; Hodgkinson, C.; Hendrickx, W.; Pearce, E.G.; Gao, Y.-T.; Pereira, A.C.; Paridaens, R.; et al. Association of Matrix Metalloproteinase-8 Gene Variation with Breast Cancer Prognosis. Cancer Res. 2007, 67, 10214–10221. [Google Scholar] [CrossRef]
- Wang, K.; Zhou, Y.; Li, G.; Wen, X.; Kou, Y.; Yu, J.; He, H.; Zhao, Q.; Xue, F.; Wang, J.; et al. MMP8 and MMP9 Gene Polymorphisms Were Associated with Breast Cancer Risk in a Chinese Han Population. Sci Rep. 2018, 8, 13422. [Google Scholar] [CrossRef]
- Lei, H.; Hemminki, K.; Altieri, A.; Johansson, R.; Enquist, K.; Hallmans, G.; Lenner, P.; Försti, A. Promoter Polymorphisms in Matrix Metalloproteinases and Their Inhibitors: Few Associations with Breast Cancer Susceptibility and Progression. Breast Cancer Res. Treat. 2007, 103, 61–69. [Google Scholar] [CrossRef]
- Tsironi, E.E.; Pefkianaki, M.; Tsezou, A.; Kotoula, M.G.; Dardiotis, E.; Almpanidou, P.; Papathanasiou, A.A.; Rodopoulou, P.; Chatzoulis, D.Z.; Hadjigeorgiou, G.M. Evaluation of MMP1 and MMP3 Gene Polymorphisms in Exfoliation Syndrome and Exfoliation Glaucoma. Mol. Vis. 2009, 15, 2890–2895. [Google Scholar] [PubMed]
- Gremlich, S.; Fratta, S.; Rebellato, E.; Uras, R.; Reymondin, D.; Damnon, F.; Germond, M.; Gerber, S. Interleukin-1 Receptor Antagonist Gene (IL-1RN) Polymorphism is a Predictive Factor of Clinical Pregnancy after IVF. Hum. Reprod. 2008, 23, 1200–1206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jin, X.; Fang, S.; Li, Y.; Wang, R.; Guo, W.; Wang, N.; Wang, Y.; Wen, D.; Wei, L.; et al. The Functional SNP in the Matrix Metalloproteinase-3 Promoter Modifies Susceptibility and Lymphatic Metastasis in Esophageal Squamous Cell Carcinoma but not in Gastric Cardiac Adenocarcinoma. Carcinogenesis 2004, 25, 2519–2524. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Ortiz, J.M.; Gutiérrez-Angulo, M.; Partida-Pérez, M.; Peregrina-Sandoval, J.; Ramírez-Ramírez, R.; Muñiz-Mendoza, R.; Suárez-Villanueva, S.; Centeno-Flores, M.; Maciel-Gutiérrez, V.; Cabrales-Vazquez, J.; et al. Association of MMP7-181A/G and MMP13-77A/G Polymorphisms with Colorectal Cancer in a Mexican Population. Genet. Mol. Res. 2014, 13, 3537–3544. [Google Scholar] [CrossRef]
- Shen, T.C.; Hsia, T.C.; Chao, C.Y.; Chen, W.C.; Chen, C.Y.; Chen, W.C.; Lin, Y.T.; Hsiao, C.L.; Chang, W.S.; Tsai, C.W.; et al. The Contribution of MMP-8 Promoter Polymorphisms in Lung Cancer. Anticancer Res. 2017, 37, 3563–3567. [Google Scholar]
- Wang, L.F.; Chien, C.Y.; Tai, C.F.; Kuo, W.R.; Hsi, E.; Juo, S.H.H. Matrix Metalloproteinase-9 Gene Polymorphisms in Nasal Polyposis. BMC Med. Genet. 2010, 11, 85. [Google Scholar] [CrossRef]
- Radisky, E.S.; Radisky, D.C. Matrix Metalloproteinases as Breast Cancer Drivers and Therapeutic Targets. Front. Biosci. Landmark 2015, 20, 1144–1163. [Google Scholar] [CrossRef]
- Liu, D.; Guo, H.; Li, Y.; Xu, X.; Yang, K.; Bai, Y. Association Between Polymorphisms in the Promoter Regions of Matrix Metalloproteinases (MMPs) and Risk of Cancer Metastasis: A Meta-analysis. PLoS ONE 2012, 7, e31251. [Google Scholar] [CrossRef]
- Zhou, J.; Brinckerhoff, C.; Lubert, S.; Yang, K.; Saini, J.; Hooke, J.; Mural, R.; Shriver, C.; Somiari, S. Analysis of Matrix Metalloproteinase-1 Gene Polymorphisms and Expression in Benign and Malignant Breast Tumors. Cancer Invest. 2011, 29, 599–607. [Google Scholar] [CrossRef][Green Version]
- Habel, A.F.; Ghali, R.M.; Bouaziz, H.; Daldoul, A.; Hadj-Ahmed, M.; Mokrani, A.; Zaied, S.; Hechiche, M.; Rahal, K.; Yacoubi-Loueslati, B.; et al. Common Matrix Metalloproteinase-2 gene Variants and Altered Susceptibility to Breast Cancer and Associated Features in Tunisian Women. Tumor Biol. 2019, 41, 1010428319845749. [Google Scholar] [CrossRef]
- Krippl, P.; Langsenlehner, U.; Renner, W.; Yazdani-Biuki, B.; Köppel, H.; Leithner, A.; Wascher, T.C.; Paulweber, B.; Samonigg, H. The 5A/6A Polymorphism of the Matrix Metalloproteinase 3 Gene Promoter and Breast Cancer. Clin. Cancer Res. 2004, 10, 3518–3520. [Google Scholar] [CrossRef]
- Yari, K.; Rahimi, Z.; Payandeh, M.; Rahimi, Z. MMP-7 A-181G Polymorphism in Breast Cancer Patients from Western Iran. Breast Care 2015, 10, 398–402. [Google Scholar] [CrossRef]
| Polymorphism | Primer Sequence * | Length of Fragments, Bp | Restriction Enzymes | Ref. | |
|---|---|---|---|---|---|
| MMP1 rs1799750 | F: 5′-TGACTTTTAAAACATAGTCTATGTTCA-3′ R: 5′-TCTTGGATTGATTTGAGATAAGTCATAGC-3′ | 2G allele 270 | 1G allele 241 + 28 | FastDigest AluI | [31] |
| MMP2 rs243865 | F: 5′-ATATTCCCCACCCAGCAGTC-3′ R: 5′-TTGGGAACGCCTGACTTCAG-3′ | C allele 122 | T allele 103 + 19 | AccI | [32] |
| MMP3 rs3025058 | F: 5′-GGTTCTCCATTCCTTTGATGGGGGGAAAGA-3′ R: 5′-CTTCCTGGAATTCACATCACTGCCACCACT-3′ | 5A allele 96 + 33 | 6A allele 130 | FastDigest PsyI | [31,33] |
| MMP7 rs11568818 | F: 5′-TGGTACCATAATGTCCTGAATG-3′ R: 5′-TCGTTATTGGCAGGCCGCACACAATGAATT-3′ | A allele 150 | G allele 130 + 20 | EcoRI | [34] |
| MMP8 rs11225395 | F: 5′-CCATCTTCACATAGCCTTGG-3′ R: 5′-CCTTGTCTTCTGCCTGTGAA-3′ | C allele 172 + 113 | T allele 285 | BfmI | [35] |
| MMP9 rs3918242 | F: 5′-GCCTGGCACATAGTAGGCCC-3′ R: 5′-CTTCCTAGCCAGCCGGCATC-3′ | C allele 436 | T allele 242 + 194 | SphI | [36] |
| Characteristic | Subgroups and Frequencies |
|---|---|
| Patient age at diagnosis | 30–40 years—35%, 40–50 years—65% |
| Tumor size (T) | 0–2 cm—66%, >2–5 cm—34% |
| Lymph node involvement (N) | negative—54%, positive—46% |
| Estrogen receptor (ER) | negative—43%, positive—57% |
| Progesterone receptor (PR) | negative—52%, positive—48% |
| Human epidermal growth factor receptor 2 (HER2) | negative—78%, positive—22% |
| Tumor histological type | ductal—88%, lobular—4%, medullary—3%, other—5% |
| Tumor histological grade (G) | G1 or G2—71%, G3—29% |
| Progression | absent—69%, present—31% |
| Metastasis | absent—74%, present—26% |
| Death | absent—78%, present—22% |
| SNP | Genotype Frequency | ||
|---|---|---|---|
| MMP1 rs1799750 | 2G2G 0.21 | 1G2G 0.56 | 1G1G 0.23 |
| MMP2 rs243865 | CC 0.59 | CT 0.37 | TT 0.04 |
| MMP3 rs3025058 | 5A5A 0.32 | 5A6A 0.46 | 6A6A 0.22 |
| MMP7 rs11568818 | AA 0.38 | AG 0.51 | GG 0.11 |
| MMP8 rs11225395 | CC 0.25 | CT 0.55 | TT 0.20 |
| MMP9 rs3918242 | CC 0.65 | CT 0.33 | TT 0.02 |
| Characteristic | SNP | Genotype | OR | 95% CI | p |
|---|---|---|---|---|---|
| G3 histological grade | MMP1 rs1799750 | 2G2G | 1.000 | (ref.) | - |
| 1G2G | 0.600 | 0.349–1.031 | 0.064 | ||
| 1G1G | 0.095 | 0.022–0.406 | 0.001 | ||
| 1G1G | 1.000 | (ref.) | - | ||
| 1G2G + 2G2G | 5.670 | 1.235–26.030 | 0.026 | ||
| 1G2G + 2G2G | 1.000 | (ref.) | - | ||
| 1G1G | 0.176 | 0.038–0.810 | 0.026 | ||
| G3 histological grade | MMP7 rs11568818 | AA | 1.000 | (ref.) | - |
| AG | 1.419 | 0.526–3.831 | 0.490 | ||
| GG | 6.562 | 1.532–28.120 | 0.011 | ||
| GG | 1.000 | (ref.) | - | ||
| AA + AG | 0.328 | 0.203–0.532 | 0.000 | ||
| AA + AG | 1.000 | (ref.) | - | ||
| GG | 5.330 | 1.424–19.941 | 0.013 | ||
| Negative PR | MMP8 rs11225395 | TT | 1.000 | (ref.) | - |
| CC + CT | 1.353 | 0.868–2.108 | 0.181 | ||
| CC + CT | 1.000 | (ref.) | - | ||
| TT | 0.429 | 0.165–1.115 | 0.082 | ||
| Positive HER2 | MMP9 rs3918242 | CC | 1.000 | (ref.) | - |
| CT | 0.422 | 0.129–1.386 | 0.155 | ||
| TT | 4.947 × 109 | 0.000-inf | NA | ||
| TT | 1.000 | (ref.) | - | ||
| CC + CT | 0.256 | 0.157–0.419 | 0.000 | ||
| CC + CT | 1.000 | (ref.) | - | ||
| TT | 6.300 × 109 | 0.000-inf | NA |
| Dependent | Covariates | Model 1 | Model 2 | Model 3 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | p | OR | 95% CI | p | OR | 95% CI | p | |||
| G3 histological grade | MMP1 | 1G2G vs. 2G2G | 0.617 | 0.320–1.191 | 0.150 | 1.636 | 0.488–5.486 | 0.425 | 1.171 | 0.323–4.245 | 0.811 |
| 1G1G vs. 2G2G | 0.098 | 0.022–0.430 | 0.002 | 0.381 | 0.060–2.422 | 0.307 | 0.295 | 0.042–2.098 | 0.223 | ||
| Age (30–40 vs. 41–50 years) | 0.932 | 0.373–2.332 | 0.881 | 0.784 | 0.273–2.252 | 0.651 | 0.724 | 0.233–2.249 | 0.577 | ||
| ER (− vs. +) | 1.029 | 0.329–3.223 | 0.961 | 1.517 | 0.458–5.022 | 0.495 | |||||
| PR (− vs. +) | 5.725 | 1.585–20.685 | 0.008 | 4.937 | 1.324–18.408 | 0.017 | |||||
| HER2 (+ vs. −) | 0.411 | 0.111–1.521 | 0.183 | 0.312 | 0.077–1.261 | 0.102 | |||||
| T (>2–5 cm vs. 0–2 cm) | 2.785 | 0.923–8.406 | 0.069 | ||||||||
| N (N1 vs. N0) | 2.208 | 0.759–6.421 | 0.146 | ||||||||
| G3 histological grade | MMP1 | 1G2G + 2G2G vs. 1G1G | 5.679 | 1.236–26.092 | 0.026 | 3.786 | 0.762–18.801 | 0.104 | 3.811 | 0.692–20.976 | 0.124 |
| Age (30–40 vs. 41–50 years) | 1.207 | 0.478–3.048 | 0.690 | 0.856 | 0.306–2.396 | 0.767 | 0.746 | 0.247–2.254 | 0.603 | ||
| ER (− vs. +) | 1.064 | 0.343–3.303 | 0.915 | 1.545 | 0.472–5.053 | 0.472 | |||||
| PR (− vs. +) | 5.483 | 1.541–19.514 | 0.009 | 4.886 | 1.318–18.113 | 0.018 | |||||
| HER2 (+ vs. −) | 0.429 | 0.117–1.569 | 0.201 | 0.314 | 0.078–1.266 | 0.103 | |||||
| T (>2–5 cm vs. 0–2 cm) | 2.859 | 0.965–8.471 | 0.058 | ||||||||
| N (N1 vs. N0) | 2.216 | 0.762–6.446 | 0.144 | ||||||||
| G3 histological grade | MMP1 | 1G1G vs. 1G2G + 2G2G | 0.176 | 0.038–0.809 | 0.026 | 0.264 | 0.053–1.312 | 0.104 | 0.262 | 0.048–1.444 | 0.124 |
| Age (30–40 vs. 41–50 years) | 1.207 | 0.478–3.048 | 0.690 | 0.856 | 0.306–2.396 | 0.767 | 0.746 | 0.247–2.254 | 0.603 | ||
| ER (− vs. +) | 1.064 | 0.343–3.303 | 0.915 | 1.545 | 0.472–5.053 | 0.472 | |||||
| PR (− vs. +) | 5.483 | 1.541–19.514 | 0.009 | 4.886 | 1.318–18.113 | 0.018 | |||||
| HER2 (+ vs. −) | 0.429 | 0.117–1.569 | 0.201 | 0.314 | 0.078–1.266 | 0.103 | |||||
| T (>2–5 cm vs. 0–2 cm) | 2.859 | 0.965–8.471 | 0.058 | ||||||||
| N (N1 vs. N0) | 2.216 | 0.762–6.446 | 0.144 | ||||||||
| G3 histological grade | MMP7 | AG vs. AA | 1.429 | 0.529–3.865 | 0.481 | 1.140 | 0.381–3.406 | 0.815 | 0.821 | 0.250–2.693 | 0.745 |
| GG vs. AA | 6.593 | 1.536–28.304 | 0.011 | 7.099 | 1.382–36.456 | 0.019 | 8.425 | 1.286–55.200 | 0.026 | ||
| Age (30–40 vs. 41–50 years) | 1.213 | 0.476–3.087 | 0.686 | 0.859 | 0.303–2.438 | 0.775 | 0.829 | 0.270–2.550 | 0.744 | ||
| ER (− vs. +) | 0.905 | 0.282–2.900 | 0.866 | 1.343 | 0.397–4.546 | 0.635 | |||||
| PR (− vs. +) | 8.069 | 2.084–31.249 | 0.003 | 6.587 | 1.697–25.563 | 0.006 | |||||
| HER2 (+ vs. −) | 0.367 | 0.099–1.359 | 0.133 | 0.231 | 0.052–1.026 | 0.054 | |||||
| T (>2–5 cm vs. 0–2 cm) | 3.019 | 0.945–9.649 | 0.062 | ||||||||
| N (N1 vs. N0) | 2.595 | 0.810–8.317 | 0.109 | ||||||||
| G3 histological grade | MMP7 | AA + AG vs. GG | 0.297 | 0.165–0.535 | 0.000 | 0.124 | 0.048–0.324 | 0.000 | 0.059 | 0.017–0.205 | 0.000 |
| Age (30–40 vs. 41–50 years) | 1.319 | 0.543–3.207 | 0.541 | 0.841 | 0.303–2.335 | 0.740 | 0.759 | 0.251–2.289 | 0.624 | ||
| ER (− vs. +) | 0.868 | 0.276–2.736 | 0.809 | 1.283 | 0.383–4.300 | 0.686 | |||||
| PR (− vs. +) | 7.907 | 2.103–29.731 | 0.002 | 6.117 | 1.598–23.413 | 0.008 | |||||
| HER2 (+ vs. −) | 0.362 | 0.097–1.343 | 0.129 | 0.242 | 0.055–1.058 | 0.059 | |||||
| T (>2–5 cm vs. 0–2 cm) | 2.833 | 0.892–8.998 | 0.077 | ||||||||
| N (N1 vs. N0) | 2.395 | 0.764–7.504 | 0.134 | ||||||||
| G3 histological grade | MMP7 | GG vs. AA + AG | 5.330 | 1.423–19.968 | 0.013 | 6.568 | 1.471–29.320 | 0.014 | 9.307 | 1.568–55.253 | 0.014 |
| Age (30–40 vs. 41–50 years) | 1.198 | 0.472–5.014 | 0.704 | 0.869 | 0.309–2.448 | 0.790 | 0.807 | 0.264–2.465 | 0.706 | ||
| ER (− vs. +) | 0.891 | 0.208–2.831 | 0.845 | 1.379 | 0.410–4.640 | 0.604 | |||||
| PR (− vs. +) | 8.187 | 2.135–31.402 | 0.002 | 6.418 | 1.662–24.785 | 0.007 | |||||
| HER2 (+ vs. −) | 0.364 | 0.099–1.346 | 0.130 | 0.239 | 0.055–1.039 | 0.056 | |||||
| T (>2–5 cm vs. 0–2 cm) | 2.977 | 0.935–9.480 | 0.065 | ||||||||
| N (N1 vs. N0) | 2.506 | 0.798–7.875 | 0.116 | ||||||||
| Positive HER2 | MMP9 | CC + CT vs. TT | 0.200 | 0.104–0.382 | 0.000 | 0.143 | 0.061–0.336 | 0.000 | 0.104 | 0.036–0.301 | 0.000 |
| Age (30–40 vs. 41–50 years) | 1.898 | 0.718–5.014 | 0.196 | 1.714 | 0.624–4.707 | 0.296 | 1.618 | 0.563–4.651 | 0.372 | ||
| ER (− vs. +) | 1.688 | 0.515–5.535 | 0.387 | 2.090 | 0.619–7.062 | 0.235 | |||||
| PR (− vs. +) | 1.254 | 0.376–4.184 | 0.712 | 1.461 | 0.411–5.187 | 0.558 | |||||
| G (G3 vs. G1/G2) | 0.276 | 0.071–1.076 | 0.064 | ||||||||
| T (>2–5 cm vs. 0–2 cm) | 1.563 | 0.513–4.767 | 0.432 | ||||||||
| N (N1 vs. N0) | 1.940 | 0.650–5.790 | 0.235 | ||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bartnykaitė, A.; Savukaitytė, A.; Bekampytė, J.; Ugenskienė, R.; Laukaitienė, D.; Korobeinikova, E.; Gudaitienė, J.; Juozaitytė, E. The Role of Matrix Metalloproteinase Single-Nucleotide Polymorphisms in the Clinicopathological Properties of Breast Cancer. Biomedicines 2022, 10, 1891. https://doi.org/10.3390/biomedicines10081891
Bartnykaitė A, Savukaitytė A, Bekampytė J, Ugenskienė R, Laukaitienė D, Korobeinikova E, Gudaitienė J, Juozaitytė E. The Role of Matrix Metalloproteinase Single-Nucleotide Polymorphisms in the Clinicopathological Properties of Breast Cancer. Biomedicines. 2022; 10(8):1891. https://doi.org/10.3390/biomedicines10081891
Chicago/Turabian StyleBartnykaitė, Agnė, Aistė Savukaitytė, Justina Bekampytė, Rasa Ugenskienė, Danguolė Laukaitienė, Erika Korobeinikova, Jurgita Gudaitienė, and Elona Juozaitytė. 2022. "The Role of Matrix Metalloproteinase Single-Nucleotide Polymorphisms in the Clinicopathological Properties of Breast Cancer" Biomedicines 10, no. 8: 1891. https://doi.org/10.3390/biomedicines10081891
APA StyleBartnykaitė, A., Savukaitytė, A., Bekampytė, J., Ugenskienė, R., Laukaitienė, D., Korobeinikova, E., Gudaitienė, J., & Juozaitytė, E. (2022). The Role of Matrix Metalloproteinase Single-Nucleotide Polymorphisms in the Clinicopathological Properties of Breast Cancer. Biomedicines, 10(8), 1891. https://doi.org/10.3390/biomedicines10081891







