NPM1-Mutated Patient-Derived AML Cells Are More Vulnerable to Rac1 Inhibition
Abstract
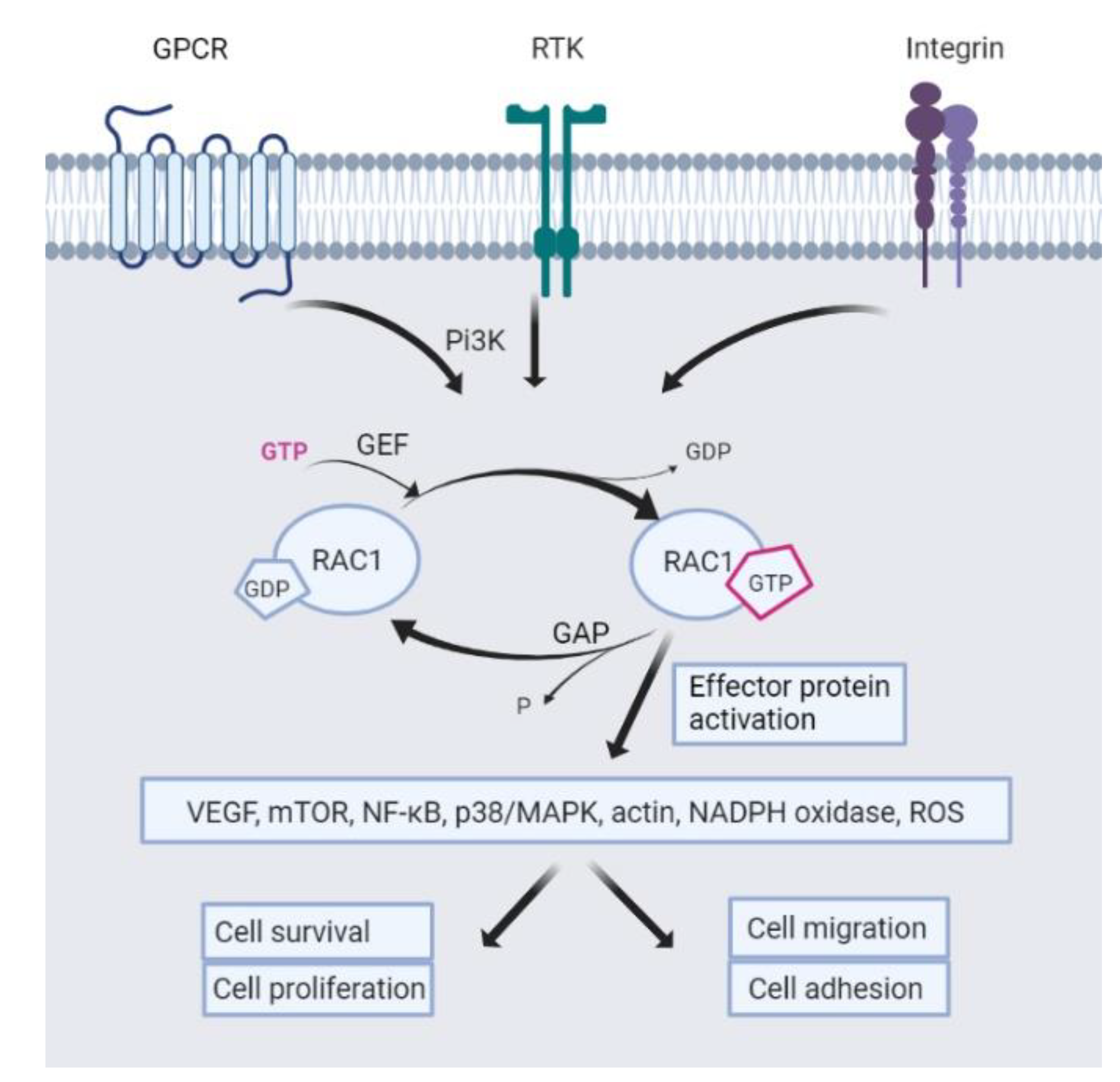
:1. Introduction
2. Materials and Methods
2.1. Primary Human AML Cells
2.2. Pharmacological Agents
2.3. Proliferation Assay
2.4. Apoptosis Assay
2.5. Cytokine Detection
2.6. Statistical Analysis and Graphical Presentation
3. Results
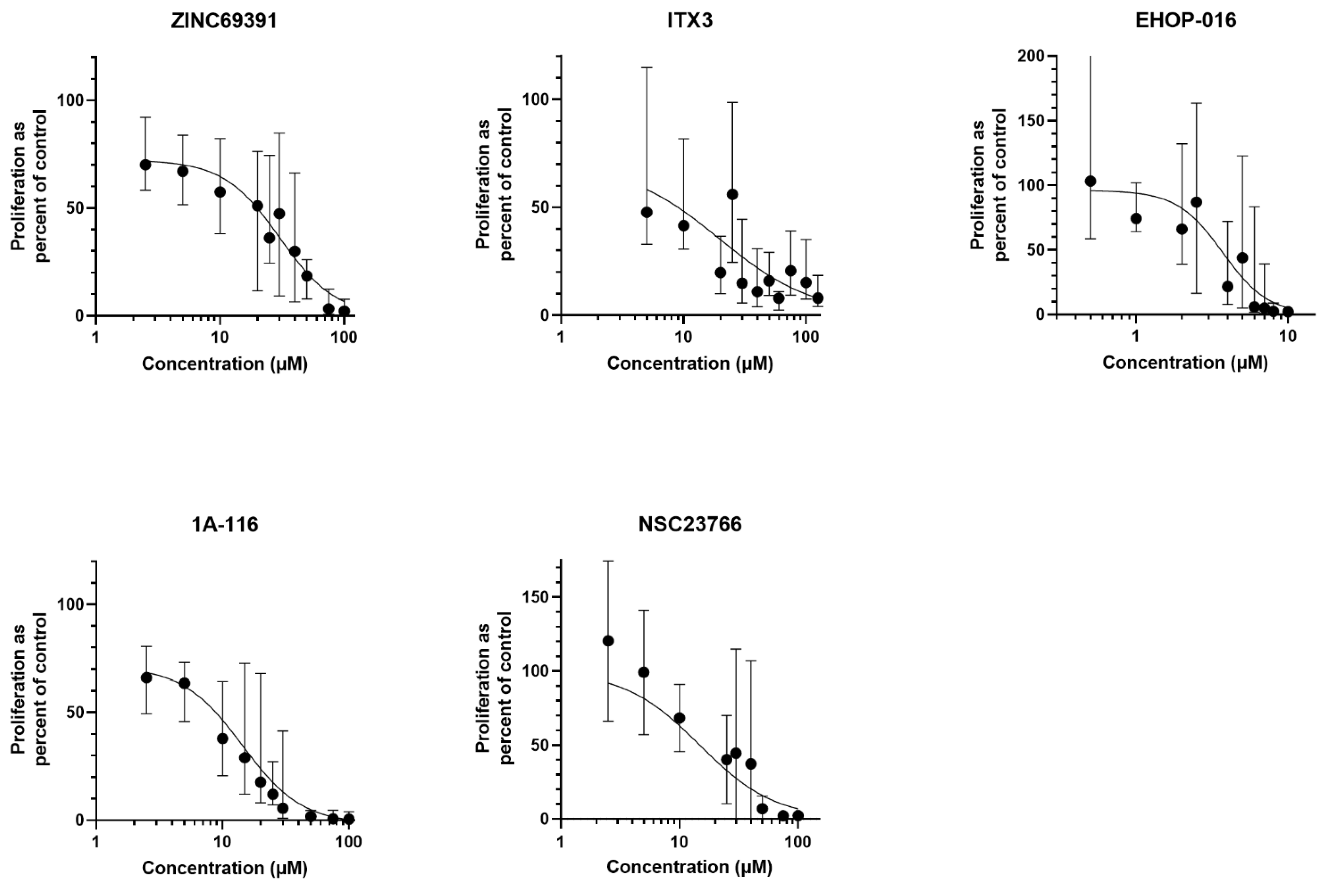
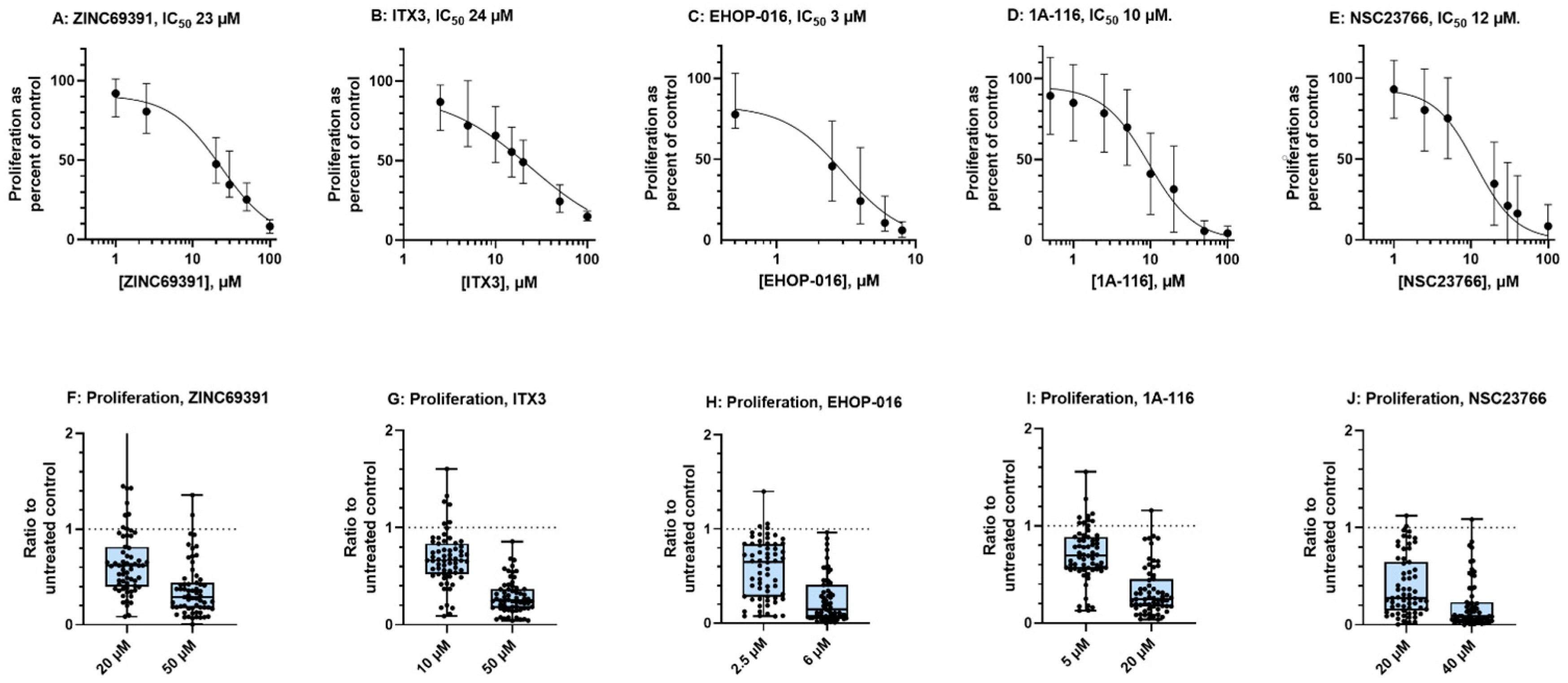
3.1. Rac1 Inhibition Demonstrates Antiproliferative Effects in Primary AML Cells
3.2. RAC1 Inhibition Demonstrates Proapoptotic Effects in Primary AML Cells
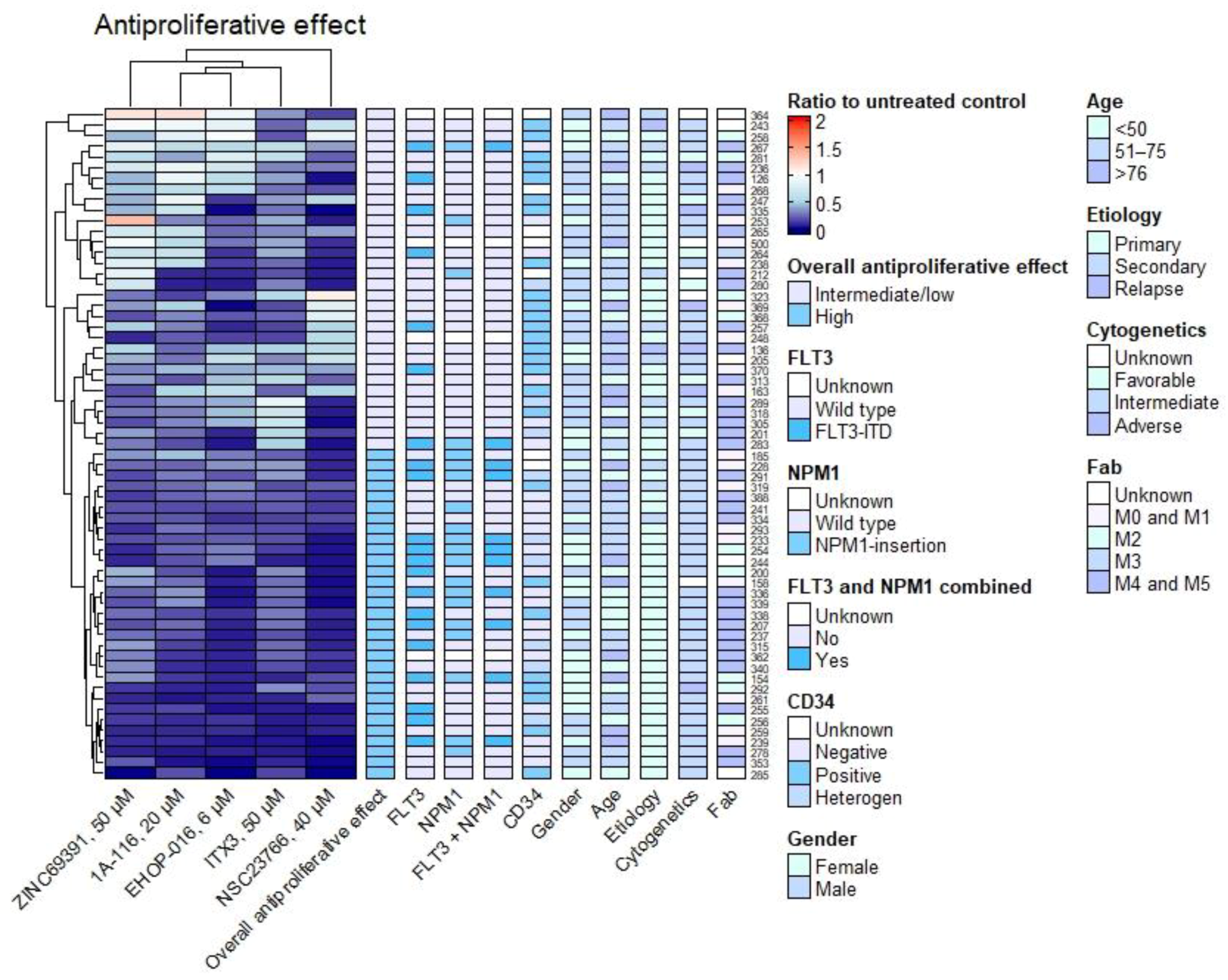
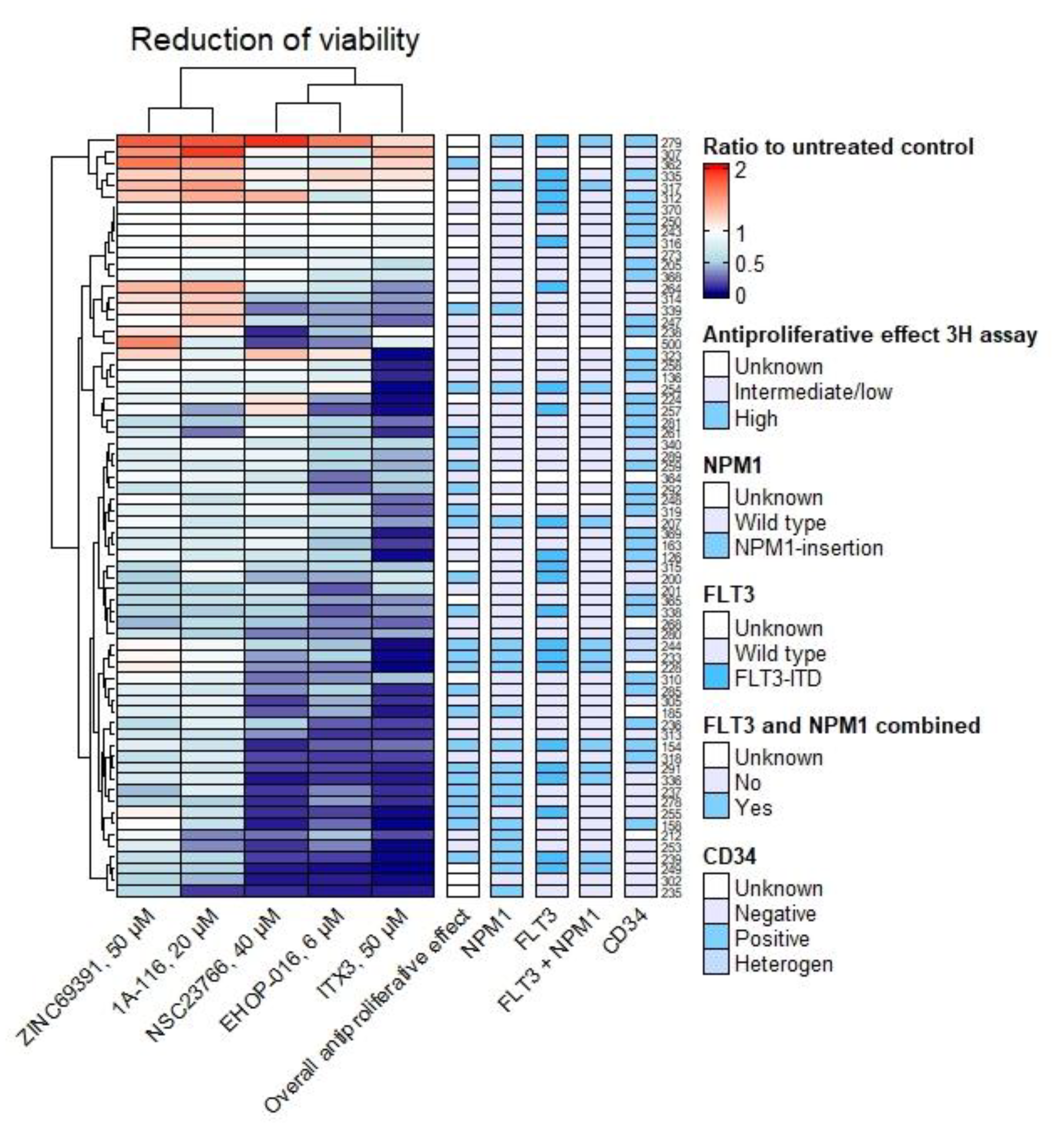
3.3. Antiproliferative and Proapoptotic Effect Varies between Patients and Is More Frequent in NPM1-Mutated Samples
3.4. Rac1 Inhibition Results in Reduced Release of Cytokines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Durand-Onaylı, V.; Haslauer, T.; Härzschel, A.; Hartmann, T. Rac GTPases in Hematological Malignancies. Int. J. Mol. Sci. 2018, 19, 4041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, J.; Oyang, L.; Rao, S.; Han, Y.; Luo, X.; Yi, P.; Lin, J.; Xia, L.; Hu, J.; Tan, S.; et al. Rac1, A Potential Target for Tumor Therapy. Front. Oncol. 2021, 11, 674426. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Li, L.; Hamaker, M.; Small, D.; Duffield, A.S. FLT3-ITD cooperates with Rac1 to modulate the sensitivity of leukemic cells to chemotherapeutic agents via regulation of DNA repair pathways. Haematologica 2019, 104, 2418–2428. [Google Scholar] [CrossRef] [PubMed]
- Reikvam, H.; Aasebø, E.; Brenner, A.K.; Bartaula-Brevik, S.; Grønningsæter, I.S.; Forthun, R.B.; Hovland, R.; Bruserud, Ø. High Constitutive Cytokine Release by Primary Human Acute Myeloid Leukemia Cells is Associated with a Specific Intercellular Communication Phenotype. J. Clin. Med. 2019, 8, 970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef] [Green Version]
- Cabrera, M.; Echeverria, E.; Lenicov, F.R.; Cardama, G.; Gonzalez, N.; Davio, C.; Fernández, N.; Menna, P.L. Pharmacological Rac1 inhibitors with selective apoptotic activity in human acute leukemic cell lines. Oncotarget 2017, 8, 98509–98523. [Google Scholar] [CrossRef] [Green Version]
- Cardama, G.A.; Comin, M.J.; Hornos, L.; Gonzalez, N.; DeFelipe, L.; Turjanski, A.G.; Alonso, D.F.; Gomez, D.E.; Menna, P.L. Preclinical Development of Novel Rac1-GEF Signaling Inhibitors using a Rational Design Approach in Highly Aggressive Breast Cancer Cell Lines. Anti-Cancer Agents Med. Chem. 2014, 14, 840–851. [Google Scholar] [CrossRef] [Green Version]
- Montalvo-Ortiz, B.L.; Castillo-Pichardo, L.; Hernández, E.; Humphries-Bickley, T.; De La Mota-Peynado, A.; Cubano, L.A.; Vlaar, C.P. Characterization of EHop-016, Novel Small Molecule Inhibitor of Rac GTPase. J. Biol. Chem. 2012, 287, 13228–13238. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Qiu, Q.; Qian, X.; Yi, J.; Jiao, Y.; Yu, M.; Li, X.; Mi, C.; Zhang, J.; Lu, B.; et al. Long noncoding RNA LCAT1 functions as a ceRNA to regulate RAC1 function by sponging miR-4715-5p in lung cancer. Mol. Cancer 2019, 18, 171. [Google Scholar] [CrossRef]
- Wang, J.; Rao, Q.; Wang, M.; Wei, H.; Xing, H.; Liu, H.; Wang, Y.; Tang, K.; Peng, L.; Tian, Z.; et al. Overexpression of Rac1 in leukemia patients and its role in leukemia cell migration and growth. Biochem. Biophys. Res. Commun. 2009, 386, 769–774. [Google Scholar] [CrossRef]
- Cardama, G.; Alonso, D.; Gonzalez, N.; Maggio, J.; Gomez, D.; Rolfo, C.; Menna, P. Relevance of small GTPase Rac1 pathway in drug and radio-resistance mechanisms: Opportunities in cancer therapeutics. Crit. Rev. Oncol. 2018, 124, 29–36. [Google Scholar] [CrossRef]
- Bouquier, N.; Vignal, E.; Charrasse, S.; Weill, M.; Schmidt, S.; Léonetti, J.-P.; Blangy, A.; Fort, P. A Cell Active Chemical GEF Inhibitor Selectively Targets the Trio/RhoG/Rac1 Signaling Pathway. Chem. Biol. 2009, 16, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Martin, H.; Mali, R.S.; Ma, P.; Chatterjee, A.; Ramdas, B.; Sims, E.; Munugalavadla, V.; Ghosh, J.; Mattingly, R.R.; Visconte, V.; et al. Pak and Rac GTPases promote oncogenic KIT-induced neoplasms. J. Clin. Investig. 2013, 123, 4449–4463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dipankar, P.; Kumar, P.; Sarangi, P.P. In silico identification and characterization of small-molecule inhibitors specific to RhoG/Rac1 signaling pathway. J. Biomol. Struct. Dyn. 2021. [Google Scholar] [CrossRef] [PubMed]
- González, N.; Cardama, G.A.; Chinestrad, P.; Robles-Valero, J.; Rodríguez-Fdez, S.; Lorenzo-Martín, L.F.; Bustelo, X.R.; Menna, P.L.; Gomez, D.E. Computational and in vitro Pharmacodynamics Characterization of 1A-116 Rac1 Inhibitor: Relevance of Trp56 in Its Biological Activity. Front. Cell Dev. Biol. 2020, 8, 240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stapnes, C.; Døskeland, A.P.; Hatfield, K.; Ersvær, E.; Ryningen, A.; Lorens, J.B.; Gjertsen, B.T.; Bruserud, Ø. The proteasome inhibitors bortezomib and PR-171 have antiproliferative and proapoptotic effects on primary human acute myeloid leukaemia cells. Br. J. Haematol. 2007, 136, 814–828. [Google Scholar] [CrossRef]
- Bruserud, O.; Ryningen, A.; Olsnes, A.M.; Stordrange, L.; Oyan, A.M.; Kalland, K.H.; Gjertsen, B.T. Subclassification of patients with acute myelogenous leukemia based on chemokine responsiveness and constitutive chemokine release by their leukemic cells. Haematologica 2007, 92, 332–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brenner, A.K.; Tvedt, T.H.A.; Nepstad, I.; Rye, K.P.; Hagen, K.M.; Reikvam, H.; Bruserud, Ø. Patients with acute myeloid leukemia can be subclassified based on the constitutive cytokine release of the leukemic cells; the possible clinical relevance and the importance of cellular iron metabolism. Expert Opin. Ther. Targets 2017, 21, 357–369. [Google Scholar] [CrossRef]
- Song, J.H.; Kim, S.H.; Cho, D.; Lee, I.-K.; Kim, H.-J.; Kim, T.S. Enhanced invasiveness of drug-resistant acute myeloid leukemia cells through increased expression of matrix metalloproteinase-2. Int. J. Cancer 2009, 125, 1074–1081. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Su, Y.; Wu, K.; Pan, F.; Wang, A. DOCK2 contributes to endotoxemia-induced acute lung injury in mice by activating proinflammatory macrophages. Biochem. Pharmacol. 2021, 184, 114399. [Google Scholar] [CrossRef]
- Ladikou, E.E.; Chevassut, T.; Pepper, C.J.; Pepper, A.G. Dissecting the role of the CXCL12/CXCR4 axis in acute myeloid leukaemia. Br. J. Haematol. 2020, 189, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, M.P.; Vallée, A.; Robert, G.; Bonneau, J.; Leroy, C.; Varin-Blank, N.; Rio, A.-G.; Troadec, M.-B.; Galibert, M.-D.; Gandemer, V. CD9, a key actor in the dissemination of lymphoblastic leukemia, modulating CXCR4-mediated migration via RAC1 signaling. Blood 2015, 126, 1802–1812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zoughlami, Y.; van Stalborgh, A.M.; van Hennik, P.B.; Hordijk, P.L. Nucleophosmin1 is a Negative Regulator of the Small GTPase Rac1. PLoS ONE 2013, 8, e68477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bustelo, X.R.; Sauzeau, V.; Berenjeno, I.M. GTP-binding proteins of the Rho/Rac family: Regulation, effectors and functions in vivo. Bioessays 2007, 29, 356–370. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Qin, T.; Bi, Z.; Hong, H.; Ding, L.; Chen, J.; Wu, W.; Lin, X.; Fu, W.; Zheng, F.; et al. Rac1 activates non-oxidative pentose phosphate pathway to induce chemoresistance of breast cancer. Nat. Commun. 2020, 11, 1456. [Google Scholar] [CrossRef] [Green Version]
- Garitano-Trojaola, A.; Sancho, A.; Götz, R.; Eiring, P.; Walz, S.; Jetani, H.; Gil-Pulido, J.; da Via, M.C.; Teufel, E.; Rhodes, N.; et al. Actin cytoskeleton deregulation confers midostaurin resistance in FLT3-mutant acute myeloid leukemia. Commun. Biol. 2021, 4, 799. [Google Scholar] [CrossRef]
- Shutes, A.; Onesto, C.; Picard, V.; Leblond, B.; Schweighoffer, F.; Der, C.J. Specificity and Mechanism of Action of EHT 1864, a Novel Small Molecule Inhibitor of Rac Family Small GTPases. J. Biol. Chem. 2007, 282, 35666–35678. [Google Scholar] [CrossRef] [Green Version]
- Parapini, S.; Paone, S.; Erba, E.; Cavicchini, L.; Pourshaban, M.; Celani, F.; Contini, A.; D’Alessandro, S.; Olivieri, A. In Vitro Antimalarial Activity of Inhibitors of the Human GTPase Rac1. Antimicrob. Agents Chemother. 2022, 66, e01498-21. [Google Scholar] [CrossRef]
- Humphries, B.A.; Wang, Z.; Yang, C. MicroRNA Regulation of the Small Rho GTPase Regulators—Complexities and Opportunities in Targeting Cancer Metastasis. Cancers 2020, 12, 1092. [Google Scholar] [CrossRef]
- Navarro-Lérida, I.; Pellinen, T.; Sanchez, S.A.; Guadamillas, M.C.; Wang, Y.; Mirtti, T.; Calvo, E.; Del Pozo, M.A. Rac1 Nucleocytoplasmic Shuttling Drives Nuclear Shape Changes and Tumor Invasion. Dev. Cell 2015, 32, 318–334. [Google Scholar] [CrossRef] [Green Version]
- Müller, P.M.; Rademacher, J.; Bagshaw, R.D.; Wortmann, C.; Barth, C.; van Unen, J.; Alp, K.M.; Giudice, G.; Eccles, R.L.; E Heinrich, L.; et al. Systems analysis of RhoGEF and RhoGAP regulatory proteins reveals spatially organized RAC1 signalling from integrin adhesions. Nat. Cell Biol. 2020, 22, 498–511. [Google Scholar] [CrossRef] [PubMed]
- Falini, B.; Brunetti, L.; Sportoletti, P.; Martelli, M.P. NPM1-mutated acute myeloid leukemia: From bench to bedside. Blood 2020, 136, 1707–1721. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [Green Version]



| PATIENT CHARACTERISTICS | OBSERVATIONS | |
|---|---|---|
| Demographic Data and Disease History | ||
| Gender (numbers) | Female/male | 33/46 |
| Age, years (median, range) | 59 (17–90) | |
| History (number, percent) | De novo | 61 (77) |
| Secondary | 16 (20) | |
| Relapse | 2 (3) | |
| Hematology (mean, range) | ||
| Hemoglobin, g/dL | 9.6 (3.8–15.4) | |
| Platelets, ×109/L | 76 (11–258) | |
| White blood cells, ×109/L | 71.8 (5–660) | |
| AML cell differentiation (number, percent) | ||
| FAB | M0-1 | 18 (23) |
| M2 | 10 (13) | |
| M3 | 1 (1) | |
| M4-5 | 45 (57) | |
| Unclassified | 5 (6) | |
| Genetic abnormalities (number, percent) | ||
| Cytogenetics | Favorable | 11 (14) |
| Intermediate * | 56 (71) | |
| Adverse | 7 (9) | |
| Unknown | 5 (6) | |
| FLT3 | Wild type | 48 (61) |
| ITD/TKD | 25/1 (33) | |
| Unknown | 5 (6) | |
| NPM1 | Wild type | 49 (62) |
| Insertion | 25 (32) | |
| Unknown | 5 (6) | |
| Inhibitor | ZINC69391 | ITX3 | EHOP-016 | 1A-116 | NSC23766 |
|---|---|---|---|---|---|
| Chemical formula | C14H14F3N5 | C22H17N3OS | C25H30N6O | C16H16F3N3 | C24H35N7.3HCl |
| Molecular weight (g/mol) | 309.29 | 371.45 | 430.55 | 307.31 | 530.9 |
| CAS number | 303094-67-9 | 347323-96-0 | 1380432-32-5 | 1430208-73-3 | 1177865-17-6 |
| IC50 against cell lines | IC50 41.7–54.1 µM, AML cell lines U937, HL60 and KG1a [6]. IC50 31–61 µM, breast cancer cell lines MCF7, F3II and MDA-MB-231 [7]. | Unknown | IC50 1.1–3 µM, breast cancer cell lines MDA-MB-435 and MDA-MB-231 [8]. IC50 4.3–9.1 µM, lung cancer cell lines Calu1, A549 and HOP62 [9]. | IC50 26–63 µM, AML cell lines U937, HL60 and KG1a [6]. IC50 4–21 µM, breast cancer cell lines F3II and MDA-MB-231 [7]. | IC50 25–100 µM, AML cell lines HL60 and KG1a [10]. IC50 140uM, breast cancer cell line F3II [7]. |
| Specificity | Inhibit Tiam1 and Dock180 (=Dock1) [7,11]. | Inhibit TrioN. No effect on Tiam1 and Vav2 [12]. | Inhibit Rac2, Rac3, Vav1, Vav2, RhoG, and Cdc42 at higher concentrations of 5–10 µM. No effect on Tiam-1 and TrioN [8,13,14]. | Inhibit P-Rex1, Vav1, Vav2, Vav3, Tiam1 and Dbl [7,15]. | Inhibit TrioN, Tiam1, Rac2, RhoG, TrioN and Tiam-1. No effect on Vav [1,8,14]. |
| Inhibitor | ZINC69391 | ITX3 | EHOP-016 | 1A-116 | NSC23766 |
|---|---|---|---|---|---|
| Highest concentration (µM) | 50 | 50 | 6 | 20 | 40 |
| Median | 0.29 | 0.25 | 0.15 | 0.25 | 0.09 |
| 95% CI of median | 0.20–0.28 | 0.22–0.32 | 0.08–0.27 | 0.20–0.31 | 0.07–0.15 |
| Min-Max | 0.004–1.40 | 0.04–0.86 | 0.002–0.97 | 0.04–1.16 | 0.001–1.09 |
| Mean | 0.36 | 0.29 | 0.25 | 0.33 | 0.21 |
| Lowest concentration (µM) | 20 | 10 | 2.5 | 5 | 20 |
| Median | 0.62 | 0.66 | 0.65 | 0.69 | 0.28 |
| 95% CI of median | 0.49–0.65 | 0.61–0.72 | 0.43–0.73 | 0.60–0.79 | 0.22–0.41 |
| Min-Max | 0.09–2.50 | 0.09–1.61 | 0.07–1.40 | 0.13–1.56 | 0.004–1.12 |
| Mean | 0.66 | 0.68 | 0.58 | 0.71 | 0.40 |
| Cytokine | Number of Patients with Detectable Levels | Median Concentration and Range (pg/mL) |
|---|---|---|
| CCL2 | 13 of 14 | 4749 (11–19,733) |
| CCL3 | 14 of 14 | 5662 (32–82,404) |
| CCL4 | 14 of 14 | 2076 (93–33,507) |
| CCL5 | 14 of 14 | 273 (5–1338) |
| CXCL1 | 10 of 14 | 2113 (64–39,153) |
| CXCL5 | 14 of 14 | 1377 (93–89,755) |
| CXCL10 | 13 of 14 | 34 (4–1333) |
| CXCL12 | 14 of 14 | 366 (291–471) |
| IL-1 RA | 10 of 14 | 7821 (56–28,546) |
| IL-8 | 16 of 16 | 5207 (146–268,468) |
| IL-1β | 11 of 14 | 47 (2–1721) |
| IL-6 | 10 of 14 | 63 (2–736) |
| G-CSF | 6 of 14 | 82 (4–32,133) |
| HGF | 14 of 14 | 84 (8–704) |
| TNF-α | 12 of 14 | 70 (3–1233) |
| GM-CSF | 7 of 14 | 1820 (1310–4036) |
| Cystatin C | 14 of 14 | 7239 (1075–10,621) |
| Serpin-E1 | 14 of 14 | 1477 (73–9619) |
| MMP-1 | 12 of 14 | 294 (12–16,253) |
| MMP-2 | 14 of 14 | 3013 (213–8105) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hemsing, A.L.; Rye, K.P.; Hatfield, K.J.; Reikvam, H. NPM1-Mutated Patient-Derived AML Cells Are More Vulnerable to Rac1 Inhibition. Biomedicines 2022, 10, 1881. https://doi.org/10.3390/biomedicines10081881
Hemsing AL, Rye KP, Hatfield KJ, Reikvam H. NPM1-Mutated Patient-Derived AML Cells Are More Vulnerable to Rac1 Inhibition. Biomedicines. 2022; 10(8):1881. https://doi.org/10.3390/biomedicines10081881
Chicago/Turabian StyleHemsing, Anette Lodvir, Kristin Paulsen Rye, Kimberley Joanne Hatfield, and Håkon Reikvam. 2022. "NPM1-Mutated Patient-Derived AML Cells Are More Vulnerable to Rac1 Inhibition" Biomedicines 10, no. 8: 1881. https://doi.org/10.3390/biomedicines10081881
APA StyleHemsing, A. L., Rye, K. P., Hatfield, K. J., & Reikvam, H. (2022). NPM1-Mutated Patient-Derived AML Cells Are More Vulnerable to Rac1 Inhibition. Biomedicines, 10(8), 1881. https://doi.org/10.3390/biomedicines10081881







