Abstract
Silencing the expression of the double homeobox 4 (DUX4) gene offers great potential for the treatment of facioscapulohumeral muscular dystrophy (FSHD). Several research groups have recently reported promising results using systemic antisense therapy in a transgenic small animal model of FSHD, the ACTA1-MCM/FLExDUX4 mouse model. However, the treatment was applied in non-DUX4-induced mice or shortly after DUX4 activation, which resulted in conditions that do not correctly represent the situation in a clinic. Here, we generated progressive FSHD-like pathology in ACTA1-MCM/FLExDUX4 mice and then treated the animals with vivoPMO-PACS4, an antisense compound that efficiently downregulates DUX4. To best mimic the translation of this treatment in clinical settings, the systemic antisense oligonucleotide administration was delayed to 3 weeks after the DUX4 activation so that the pathology was established at the time of the treatment. The chronic administration of vivoPMO-PACS4 for 8 weeks downregulated the DUX4 expression by 60%. Consequently, the treated mice showed an increase by 18% in body-wide muscle mass and 32% in muscle strength, and a reduction in both myofiber central nucleation and muscle fibrosis by up to 29% and 37%, respectively. Our results in a more suitable model of FSHD pathology confirm the efficacy of vivoPMO-PACS4 administration, and highlight the significant benefit provided by the long-term treatment of the disease.
1. Introduction
Facioscapulohumeral muscular dystrophy (FSHD) is a rare hereditary autosomal dominant disease with an estimated prevalence of 5 to 13 per 100,000 [1,2]. As the name suggests, FSHD affects muscles in the face, shoulder girdle and upper arms, and often extends to the trunk and lower limbs as the disease progresses, causing about 20% of patients to become wheelchair-bound [3]. Unlike most muscular dystrophies, the muscle weakness in FSHD is clinically asymmetric [4]. The disease is also gender-specific, with a higher proportion of females than males being asymptomatic or less affected [4]. Although the life expectancy is normal, over 90% of FSHD patients experience chronic fatigue [5]. More than one third of patients have reported respiratory dysfunction, cardiac arrythmias and/or sensorineural hearing loss [6,7,8]. Sleep-related breathing disorders have been additionally identified in at least 40% of patients [9]. In two independent studies involving 80–100 FSHD patients with different disease severities, 14% of the participants required non-invasive ventilation, with the risks of pulmonary insufficiency increasing with spine deformity, severity level and wheelchair dependency [6,7]. However, it remains under debate whether the respiratory impairment in FSHD is caused by weakness of the expiratory muscles and the proximal lower extremities [10] or by weakness of both the expiratory and inspiratory muscles, and in particular the diaphragm [11].
The current treatment options for FSHD are supportive. However, therapeutic agents aiming at slowing or halting disease progression remain to be investigated or verified. Since the aberrant expression of the double homeobox 4 (DUX4) gene has been suggested as a predominant cause of FSHD pathogenesis [12,13,14], targeting DUX4 has great potential to provide a cure for the disease. Therefore, we and others have developed several approaches for suppressing DUX4 expression or its downstream activity [14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33]. Despite FSHD affecting multiple skeletal muscles, only a few studies describe the systemic effects of therapeutic strategies on FSHD-like animal models. We previously showed that a four- weekly systemic administration of vivoPMO-PACS4, an octaguanidine dendrimer-conjugated phosphorodiamidate morpholino oligomer that targets key features in the 3′UTR of DUX4, inhibits DUX4 expression and improves functionality as well as the molecular and histological features of the muscles of treated ACTA-MCM/FLExDUX4 mice [33,34]. Using the same FSHD-like transgenic mouse model, but without inducing DUX4 expression, Bouwman and colleagues have since reported that the longitudinal delivery of a constrained ethyl gapmer antisense oligonucleotide targeting the open reading frame of DUX4 is also effective at knocking down the leaky DUX4 level and alleviating muscle pathology, although with limited improvement in muscle mass and function [21]. Several pharmaceutical companies have also tested their lead antisense compounds in ACTA1-MCM/FLExDUX4 mice. For example, Arrowhead Pharmaceuticals showed that the systemic delivery of their best siRNA candidate from 2 to 10 days after tamoxifen (TMX)-mediated DUX4 induction improved the body mass, improved the animal’s performance on the rotarod test and reduced muscle fibrosis [35]. Although the results from these studies are promising, the experimental models used prevent these findings from providing real advances to clinical settings. This is because the treatments were designed to specifically address the leaky DUX4 level, or they were applied shortly after switching on DUX4 expression, which is less relevant for FSHD patients who suffer from chronic DUX4 pathogenesis.
Hence, in this study we investigated the long-term inhibitory effect of vivoPMO-PACS4 on DUX4 in the ACTA1-MCM/FLExDUX4 mouse model by starting the treatment after the DUX4-mediated pathology was already established. We injected the mice with TMX to induce DUX4 expression and repeated the TMX delivery biweekly to maintain an FSHD-like pathology. A systemic antisense treatment with vivoPMO-PACS4 was started 3 weeks after the DUX4 activation. The chronic administration of the antisense chemistry for over 8 weeks efficiently suppressed the DUX4 expression at both the mRNA and protein levels. Consequently, we observed a significant increase in the mass of five examined muscle types, an enhancement of muscle function and an improvement in the histopathological features of the tibialis anterior and diaphragm muscles of the treated mice. Our data therefore confirm the benefit provided by systemic treatment with vivoPMO-PACS4, further highlighting the significant impact of long-term treatment with the conjugated PMO chemistry and the likely relevance to FSHD patients. These results support the further development of the antisense strategy as a treatment for FSHD.
2. Materials and Methods
2.1. Antisense Chemistries
Phosphorodiamidate morpholino oligomers conjugated to cell-penetrating moiety octaguanidine dendrimers (vivoPMOs) were purchased from GeneTools (Philomath, OR, USA). vivoPMO-PACS4 (AGGATCCACAGGGAGGAGGCATTTTAAT) targets both the polyadenylation signal and the cleavage site of DUX4 mRNA [33]. vivoPMO-SCR (CCTCTTACCTCAGTTACAATTTATA) is the standard control of GeneTools; it targets the HBB mutation that causes β thalassemia, and was used here as a negative control. vivoPMOs were dissolved in sterile ddH2O and were further diluted to the desired concentrations in sterile 0.9% saline (Sigma, Dorset, UK) immediately prior to their use in mice.
2.2. Animals
This study was conducted in accordance with the UK Animals (Scientific Procedures) Act 1986. Ethical and operational permissions were granted by the UK Home Office (Project Licence P36A9994E) and the Animal Welfare Committee of the Royal Holloway University of London. Mice were bred in a minimal disease facility at the Royal Holloway University of London and kept under a standard 12 h light/dark cycle with free access to food and water. Animal welfare was maintained according to the UK Home Office’s Code of Practice for the Housing and Care of Animals Bred, Supplied or Used for Scientific Purposes. FLExDUX4 (JAX 028710) and ACTA1-MCM (JAX 025750) mice were purchased from The Jackson Laboratory (Maine, USA). The FLExDUX4 colony was maintained as homozygous for Gt(ROSA)26Sortm1.1(DUX4*)Plj while the ACTA1-MCM colony was maintained as hemizygous for Tg(ACTA1-cre/Esr1*)2Kesr on a C57BL/6J background. A tamoxifen (TMX)-inducible bi-transgenic model named ACTA1-MCM/FLExDUX4 was generated by crossing ACTA1-MCM males with FLExDUX4 females. Genotyping was performed using DNA isolated from the murine ear notches, based on The Jackson Laboratory’s established protocols. Due to the sex specific-DUX4 pathology of the ACTA1-MCM/FLExDUX4 model [36], only males were used in the study and littermates were randomized between experimental groups.
2.3. Study Design
This study involved two groups of ACTA1-MCM/FLExDUX4 mice and one group of ACTA1-MCM mice, with 6 mice per group. All animals received the first dose of TMX, prepared as described previously [37], at 1.5 mg/kg via an intraperitoneal (IP) injection at 10 weeks old (considered as week 1 of the study). TMX was re-administered every 2 weeks for a total of 6 times. The ACTA1-MCM/FLExDUX4 mice were further IP injected with 10 mg/kg of either vivoPMO-PACS4 (PACS4) or vivoPMO-SCR (SCR) while the ACTA1-MCM mice received volume-matched saline, which was considered as the positive control (CTRL). Either vivoPMOs or saline was delivered one week after the second TMX dose and repeated once/twice per week thereafter for a total of 12 injections. During the study, the animals underwent several behavioral and functional tests. Treadmill exhaustion tests were conducted one week prior to the first TMX injection and on weeks 3, 7 and 11. Grip strength tests were conducted on weeks 8 and 11. Locomotor behavioral tests were conducted every 2 days during week 11 (prior to any injection or functional test). In situ muscle force measurements and subsequent tissue collection were performed on week 12. Bodyweight was recorded prior to each injection. The mice were kept under isoflurane-induced anesthesia (3% in 100% O2) during injections and were continuously monitored until they fully recovered.
2.4. Behavioral and Functional Tests
The open-field behavioral activity of the mice was examined using locomotor activity monitors as described in [33]. The data were acquired and collected by Amon Lite software (version 1.4) every 10 min in a one-hour session per test and were summed up for each mouse. The data reported here were an average of 3 tests. Both the locomotor activity monitors and the software were obtained from Linton Instrumentation (Diss, UK). Treadmill exhaustion tests were performed on a Treadmill Simplex II system (Columbus Instruments, Columbus, OH, USA) with adjusted 15 °C inclination as described previously [33]. The total running time was recorded and the time to fatigue was displayed as a percentage of the time recorded in the initial test. Forelimb muscle force was assessed using a commercial grip strength monitor (Linton Instrumentation, Norfolk, UK) as described in [38]. Briefly, mice were held by the tail, allowed to grasp a metal mesh attached to a force transducer with their forelimbs, and then were gently pulled until they released the grip. Measurements were performed 5 times per mouse, with 30 s elapsed between each measurement. The maximal force recorded was expressed as mN per gram of the bodyweight recorded at the assessment. Tibialis anterior (TA) muscle strength was assessed by in situ muscle force measurements, as detailed in [33], while mice were under terminal anesthesia induced by a mixture of 10 mg/mL dolethal (Vetoquinol, Towcester, UK) and 15 µg/mL buprenodale (Dechra, Shrewsbury, UK) at 6 times of their bodyweight. Data were recorded and analyzed using Dynamic Muscle Control and Analysis Software (Aurora Scientific, Aurora, Canada). All isometric measurements were obtained at an initial length at which the maximal tension was recorded during the tetanus. The responses to tetanic stimulations at increased pulse frequencies from 10 Hz to 180 Hz were recorded and the maximal force was determined. The specific force was subsequently calculated based on a ratio of the maximal force and the muscle cross-sectional area (CSA), which was mathematically approximated by dividing the muscle mass by the optimum fiber length and the density of mammalian muscle as described in TREAT-NMD SOP DMD_M.2.2.005.
2.5. Post-Mortem Tissue Processing
From each mouse, the diaphragm (DIA), gastrocnemius (GAS), quadriceps (QUAD), TA and triceps (TRI) muscles were dissected and weighed after the fat/connective tissue was trimmed off. The weights of the ipsilateral and contralateral type-matched muscles were then averaged, with the exception of the DIA, and normalized to the corresponding final bodyweight (Table S1). The muscles from one side of the hindlimb were frozen immediately in liquid nitrogen for subsequent RNA extraction. The contralateral muscles were embedded in an optimal cutting temperature (OCT) medium (VWR, Lutterworth, UK) and frozen in liquid-nitrogen-cooled isopentane (Sigma, Dorset, UK) for histological analyses. The DIA muscle was longitudinally divided into 2 halves. One half was frozen immediately in liquid nitrogen for RNA work while the other half was prepared as described in [39] for histological analyses. OCT-embedded frozen muscles were cryosectioned on an OTF5000 cryostat (Bright, Huntingdon, UK) at a 10 µm thickness for 10 serial levels through the muscle length, and the transverse sections were collected onto SuperFrost slides (VWR, Lutterworth, UK).
2.6. RT-qPCR Quantification for DUX4 and Relevant Genes
The total RNA from liquid-nitrogen-snap-frozen muscles was isolated using RNeasy Fibrous Tissue kit (QIAgen, Manchester, UK) by following the manufacturer’s instructions. The samples were homogenized in the lysis buffer provided with the kit on a TissueLyser II (QIAgen, Manchester, UK) at 25 Hz for 2–4 min. One microgram of RNA was reverse transcribed using a QuantiTect reverse transcription kit (QIAgen, Manchester, UK). Ten nanograms of diluted cDNA in qPCR water (Roche, Burgess Hill, UK) were then amplified using a LightCycler480 SYBR Green Master I kit (Roche, Burgess Hill, UK) according to the manufacturer’s instructions, with each sample prepared in triplicates. qPCR reactions were run on LightCycler480 System, initialized at 95 °C for 5 min and followed by 45 cycles at 95 °C for 15 s, 58–60 °C for 15 s, and 72 °C for 15 s. The relative quantification for DUX4 and DUX4-related genes was performed against the corresponding housekeeping gene, Gapdh, using Pfaffl’s method as described in [40]. Data are shown as fold-changes compared to the SCR values obtained in the same way. The primers were purchased from Integrated DNA Technologies (Belgium) and are detailed in Table 1.

Table 1.
Details of qRT-PCR primers.
2.7. Immunohistochemistry
For DUX4 and laminin co-immunostaining, frozen muscle sections were fixed in 4% (w/v) paraformaldehyde for 15 min, permeabilized in 0.3% (v/v) Triton X-100 with 1× PBS for 10 min and blocked in a buffer containing 2% (w/v) BSA, 5% (v/v) goat serum, 0.1% (v/v) Triton X-100 and 1× PBS for 30 min. An incubation with rabbit anti-DUX4 [E5.5] (1:100, Abcam, UK) and rat anti-laminin (1:1000) antibodies was performed overnight at 4 °C, and then an incubation with goat anti-rabbit AlexaFluor568 and goat anti-rat AlexaFluor488 antibodies (1:400, Life Technologies, Paisley, UK) was performed for 1 h at room temperature. Nuclei were stained with 1 µg/mL DAPI in 1× PBS for 15 min. For collagen immunostaining, the muscle sections were fixed in ice-cold acetone for 10 min and blocked in 1% (w/v) BSA, 1% (v/v) goat serum, 0.1% (v/v) Triton X-100 and 1× PBS for 1 h. A subsequent incubation with rabbit anti-collagen VI antibodies (1:300, Abcam, Cambridge, UK) was carried out overnight at 4 °C, followed by a 1 h incubation with goat anti-rabbit AlexaFluor488 antibodies (1:1000, Life Technologies, Paisley, UK). The slides were mounted in Mowiol 4-88. Ten random images from each muscle section were captured at a magnification of ×200 for DUX4-positive nuclei quantification. For other analyses, multiple images from the largest mid-belly muscle section were captured at a magnification of x100 and were automatically stitched together by ZEN Imaging software (Zeiss, Cambridge, UK), generating an image of the whole transverse muscle section. Images were acquired on an Axio Observer D1 fluorescence microscope by an AxioCam MR3 (Zeiss, Cambridge, UK). For hematoxylin and eosin (H&E) analysis, frozen muscle sections were fixed in 100% (v/v) ice-cold methanol for 10 min and then submerged in hematoxylin (Vector Laboratories, Peterborough, UK) and eosin solutions. Sections were dehydrated in a series of 50%, 80%, 90% and 100% (v/v) ethanol washes, for 1 min/wash, and were then cleared twice in 100% (v/v) xylene for 5 min/wash. The slides were mounted in DPX Mountant. Images were acquired using an Eclipse Ni-E upright microscope and compatible software (Nikon Instruments Inc., New York, NY, USA). The reagents were purchased from Sigma (Dorset, UK) unless stated otherwise.
2.8. Histological Analyses
The number of myonuclei that were positive for DUX4 was manually counted using Fiji/MuscleJ software (version 1.53c, National Institutes of Health, Bethesda, MD, USA) and expressed as the total DUX4-positive nuclei per mm2 of muscle section. The myofiber perimeter was identified by laminin staining. The subsequent quantifications of the total fiber number, the number of centrally nucleated fibers (CNFs), the area and the minimal Feret’s diameter of individual myofibers were automatically scored by Fiji/MuscleJ software as described in [41]. Automatic analyses of the frequency distribution and the coefficient of variance of the minimal Feret’s diameter were carried out using GraphPad Prism8 software (San Diego, CA, USA). The CSA of the entire muscle sections or the area positive with collagen VI was semi-automatically measured by Fiji/MuscleJ. The fibrotic area was expressed as the percentage of the total area of the muscle CSA.
2.9. Statistical Analysis
Data were analyzed using the GraphPad Prism8 software (San Diego, CA, USA) and are shown as the means ± SEM. Error bars represent the SEM; “n” refers to the number of mice per group. All data passed the normality Shapiro–Wilk test, which is the most powerful test among four common normality tests, especially for small sample sizes (3 ≤ n ≤ 5000) [42]. Comparisons of statistical significance were further assessed by one-way or two-way ANOVA followed by Tukey’s post hoc test, as detailed in figure legends. All functional tests and histological analyses were performed in a blinded manner.
3. Results
3.1. Systemic Treatment with vivoPMO-PACS4 Increases Muscle Mass of ACTA1-MCM/FLExDUX4 Mice
To best mimic the translation of antisense therapy in clinical settings, in this study we investigated the inhibitory effect of vivoPMO-PACS4 on DUX4 in ACTA1-MCM/FLExDUX4 mice when the disease pathology was established. Two groups of ACTA1-MCM/FLExDUX4 mice and one group of ACTA1-MCM healthy control mice (six mice/group) received the first tamoxifen (TMX) dose at 1.5 mg/kg via intraperitoneal (IP) injection at 10 weeks of age, and then every two weeks afterwards. Three weeks after the first injection of TMX, the ACTA1-MCM/FLExDUX4 mice were injected (IP) with 10 mg/kg of either vivoPMO-PACS4 (PACS4) or vivoPMO-SCR (SCR), considered as a negative control, while the ACTA1-MCM mice—the positive control (CTRL)—received volume-matched saline. The dosing with vivoPMOs or saline was repeated either once or twice per week, depending on the week and the TMX administration, for a total of 12 injections. Details of the experimental design are shown in Figure 1a. The bodyweight recorded during the study and normalized to the initial weight indicated the successful induction and maintenance of detrimental DUX4 expression and the related pathology. CTRL mice gained weight gradually up to 130% by the end of the 12-week experiment, whereas the weight gain in the ACTA1-MCM/FLExDUX4 mice fluctuated between 99 and 105% in the SCR group, and between 100 and 108% in the PACS4 group (Figure 1b). Although the PACS4 treatment appeared to have a marginal effect on the bodyweight, we observed a significant improvement in the mass of the five examined skeletal muscles, including the diaphragm (DIA), gastrocnemius (GAS), quadriceps (QUAD), tibialis anterior (TA) and triceps (TRI) (Figure 1c). The muscle mass of the hindlimbs and forelimbs of PACS4-treated mice was significantly increased by up to 18% of the SCR values, achieving the same levels as the CTRL group for the GAS (p = 0.7027) and TRI (p > 0.9999). Interestingly, the DIA mass of SCR mice was 21% heavier than that of CTRL mice (p < 0.0001), and treatment with PACS4 reduced its weight by 8% (p = 0.0351 compared to SCR). Hence, systemic treatment with vivoPMO-PACS4 provided a significant benefit to body-wide muscle mass.
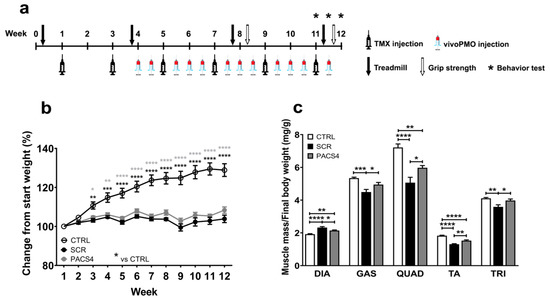
Figure 1.
vivoPMO-PACS4 treatment improved body-wide muscle weight. (a) Diagram of the experiment, including timepoints of injections of tamoxifen (TMX) to induce DUX4 expression and vivoPMOs, and functional tests. All mice received 6 × 1.5 mg/kg/injections of TMX via intraperitoneal (IP) delivery. ACTA1-MCM/FLExDUX4 mice were further IP injected with 12 × 10 mg/kg/injections of either vivoPMO-PACS4 (PACS4, n = 6) or vivoPMO-SCR (SCR, n = 6), considered as a negative control. ACTA1-MCM mice receiving volume-matched saline were considered as a positive control (CTRL, n = 6). (b) Weekly bodyweight is shown as percentage of the initial weight recorded on the day of the first TMX injection. (c) Animals were sacrificed after 8 weeks of vivoPMO treatment. Muscle mass of the diaphragm (DIA), gastrocnemius (GAS), quadriceps (QUAD), tibialis anterior (TA) and triceps (TRI) was normalized to the corresponding final bodyweight. Data are shown as means ± SEM; n = 6. Statistical analysis was by (b) two-way or (c) one-way ANOVA followed by Tukey’s multiple comparisons test; * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. In (b), black asterisks indicate significance between the SCR and CTRL groups; grey asterisks indicate significance between the PACS4 and CTRL groups.
3.2. Long-Term vivoPMO-PACS4 Treatment Greatly Improves Muscle Function and Animal’s Behavior
We evaluated the impact of chronic DUX4 expression and the long-term effect of PACS4 treatment on whole-body muscle function over a 12-week study using the treadmill exhaustion test. Mice were allowed to run on a treadmill until they were unable to move from the stopper for 10 s. The time to fatigue shown in Figure 2a is the total running time at each timepoint calculated as a percentage of the time recorded at the beginning of the experiment. Following two doses of TMX (week 3), the time to reach exhaustion in the ACTA1-MCM/FLExDUX4 mice decreased to 78% (SCR) and 70% (PACS4) of the initial levels, while no reduction was observed in the CTRL mice, confirming the negative impact of the inducible DUX4 expression in both ACTA1-MCM/FLExDUX4 groups. With continual TMX delivery, the level of resistance to fatigue in the SCR mice decreased further to 69% and 60% on weeks 7 and 11, respectively. However, at the same timepoints, the mice receiving PACS4 displayed an increase in resistance to fatigue, at 75% and 83%, respectively. This means that the fatigue in the PACS4 mice decreased by 23% (p = 0.0030) compared to SCR mice, and that the PACS4-treated mice were as healthy as the CTRL mice (p = 0.3686) by the last vivoPMO delivery. An additional assessment of the strength of the forelimb muscles using the grip strength test (Figure 2b) indicated that the forelimbs of the PACS4 mice were as strong as those of the SCR mice (30.3 ± 0.7 mN/g vs. 28.1 ± 1.0 mN/g, p = 0.6725) on week 8. While the forelimb muscle force in the SCR mice dropped by a further 19% on week 11, the value in the PACS4 mice remained unchanged (30.2 ± 1.9 mN/g) and was similar to the CTRL value (35.9 ± 1.8 mN/g, p = 0.0770). Hence, the vivoPMO-PACS4 treatment prevented a 32% loss of forelimb strength compared to the SCR level (p = 0.0180). Further in situ TA force measurements demonstrated obvious muscle weakness in the ACTA1-MCM/FLExDUX4 mice receiving vivoPMO-SCR when compared to the CTRL group (Figure 2c,d). Treatment with PACS4 significantly improved the maximal tetanic force, from 798.1 ± 46.8 mN measured in SCR mice to 1051.8 ± 57.1 mN (p = 0.0006) at the highest stimulation frequency of 180 Hz. Notably, mice receiving PACS4 exhibited a specific maximal force comparable to CTRL mice, with 193.4 ± 4.3 mN/mm2 and 209.9 ± 9.9 mN/mm2 at 180 Hz, respectively (p = 0.1566), which corresponded to a 19% increase compared to the strength of the SCR group’s muscles (p = 0.0025).
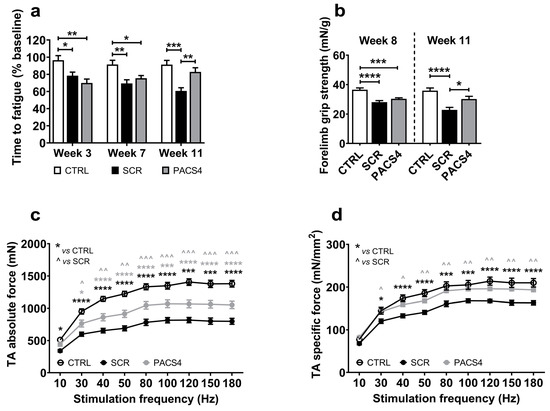
Figure 2.
Significant enhancement in muscle function following vivoPMO-PACS4 treatment. (a) Treadmill exhaustion tests were conducted 1 week before the first TMX injection, and at weeks 3, 7 and 11. The time to fatigue is the total running time in each test, expressed as percentage of the initial time. (b) Forelimb grip strength tests were carried out on weeks 8 and 11. Data are shown as the force corrected to the bodyweight recorded at the assessment. (c) In situ absolute force of TA muscles was measured while animals were under terminal anesthesia on week 12. (d) Specific muscle force is displayed as a ratio of the absolute force and the muscle cross-sectional area (CSA), mathematically approximated by dividing the muscle mass by the optimum fiber length and the density of mammalian muscle. Data are shown as means ± SEM; n = 6. Statistical analysis was by (a,b) one-way or (c,d) two-way ANOVA followed by Tukey’s multiple comparisons test; */^ p < 0.05, **/^^ p < 0.01, ***/^^^ p < 0.001, **** p < 0.0001. In (c,d), black asterisks indicate significance between the SCR and CTRL groups; grey asterisks or carets indicate significance between the PACS4 and CTRL or the PACS4 and SCR groups, respectively.
The efficacy of the vivoPMO treatment was also assessed using open-field activity cage monitors a week before the end of the study. The locomotor behavior of the mice injected with vivoPMO-SCR significantly decreased in 19 of the 22 parameters examined, compared to CTRL mice (Table 2). In contrast, the PACS4-treated animals exhibited a similar behavior to that of the CTRL group in all measurements, and a robust improvement in six parameters as compared to the SCR mice. Notably, the total activity, total rearing counts and rearing time were all increased from 361 ± 51 to 565 ± 52 beam breaks (p = 0.0085), 86 ± 20 to 247 ± 29 beam breaks (p = 0.0050) and 148 ± 31 to 449 ± 53 s (p = 0.0392), respectively. Taken together, these data suggest that chronic vivoPMO-PACS4 treatment has extensive benefits on muscle function and behavior, efficiently stabilizing the disease progression in the treated mice.

Table 2.
Changes in mouse locomotor behavior after longitudinal vivoPMO treatment.
3.3. vivoPMO-PACS4 Robustly Downregulates Expression of DUX4 and Murine DUX4-Related Targets
Among the five muscle types examined here, the TA and DIA muscles of ACTA-MCM/FLExDUX4 mice have been shown to be the most affected by DUX4 expression via systemic TMX-mediated induction [33,34,36]. Hence, our investigation on the effect of vivoPMO-PACS4 against DUX4-mediated pathology at the molecular and histological levels focused on these two muscles. qRT-PCR quantification demonstrated that PACS4 treatment efficiently suppressed the DUX4 mRNA expression by 40% in the TA (p = 0.001) and by 60% in the DIA (p < 0.0001), compared to the corresponding SCR levels (Figure 3a,e). This led to a reduction in the number of DUX4-positive nuclei per mm2 of muscle section from 101.5 ± 6.9 (TA) and 41.4 ± 4.5 (DIA) in the SCR group to 72.8 ± 4.3 (TA, p = 0.0017) and 27.3 ± 3.7 (DIA, p = 0.0249) in the PACS4 group (Figure 3b,f,i). As a consequence, the mRNA expression of a murine DUX4-related target, Wfdc3 [36], in PACS4 muscle was downregulated by 32% in the TA (p = 0.0131) and by 35% in the DIA (p = 0.0008), compared to the SCR group’s levels (Figure 3c,g). Moreover, in agreement with previous findings suggesting that DUX4 interferes with mitochondrial biogenesis via suppressing the expression of a key mediator, Pgc1α [40], we detected a significantly lowered Pgc1α level in the SCR group by 52% (TA, p < 0.0001) and by 69% (DIA, p < 0.0001), compared to the CTRL values. Treatment with PACS4 significantly improved the Pgc1α expression by 31% in the TA muscle (p = 0.0251), but had a minimal effect on the expression in the DIA (p = 0.1958) compared to the SCR treatment (Figure 3d,h). These results confirm the efficacy of the systemic delivery of vivoPMO-PACS4 on DUX4 at both the mRNA and protein levels, as well as on the DUX4 downstream target genes.

Figure 3.
vivoPMO-PACS4 treatment downregulated the expression of DUX4 and murine DUX4-related genes. Following chronic TMX-induced DUX4 expression and an 8-week treatment with vivoPMOs, mRNA levels of DUX4 and DUX4-related genes, and DUX4 protein levels in (a–d) the tibialis anterior and (e–h) the diaphragm muscles were evaluated. mRNA expression of (a,e) DUX4 and (c,d,g,h) DUX4-related genes was quantified by qRT-PCR as relative to the corresponding Gapdh level and expressed as a fold-change of the SCR value obtained by the same way. (b,f) Muscle sections were immunostained with DUX4 and laminin, and nuclei were stained with DAPI. The number of myonuclei positive with DUX4 was manually counted and expressed as the total DUX4+ nuclei per mm2 of the transverse muscle section. Data are shown as means ± SEM; n = 6. Statistical comparison was by one-way ANOVA followed by Tukey’s multiple comparisons test; * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. (i) Representative images are shown at a magnification of ×200; scale bars = 50 µm; white arrows indicate DUX4+ nuclei.
3.4. Long-Term vivoPMO-PACS4 Treatment Greatly Improves Muscle Histopathology
The impact of chronic DUX4 expression on the histology of the TA and DIA muscles was first assessed through hematoxylin and eosin (H&E) staining (Figure 4a,b). The SCR group’s muscles displayed substantial deterioration in the muscle architecture and quality, with increased fiber size variation, central nucleation, fibrosis, and necrosis/inflammation compared to the CTRL group’s muscles. The hallmarks of muscle fibrosis and necrosis/inflammation were particularly more abundant in the DIA muscle than in the TA muscle, which may account for the pseudo-hypertrophy in the mass of the DIA contrary to the atrophy in the TA mass, as presented in Figure 1c. In contrast, the PACS4-treated tissues exhibited a histology similar to that of the CTRL muscles, which was consistent with the improvement in the muscle mass towards a value comparable to the CTRL group’s values. The immunohistochemical detection of laminin in these muscle sections provided additional information about the pathology process. The TA muscle appeared to have undergone a significant process of muscle deterioration and regeneration (Figure 4c–g). We observed a high increase in the number of myofibers (p < 0.0001), but a significant reduction in both the cross-sectional area (CSA) and the myofiber diameter of the SCR-treated muscle (p < 0.0001), as compared to the CTRL group. The PACS4 treatment significantly reduced the myofiber number by 25% (p = 0.0248) while increasing the CSA by 23% (p = 0.0158) and the fiber diameter by 10% (p = 0.0410) compared to the SCR values (Figure 4c–e). Accordingly, this led to an 8% reduction in the coefficient of variation for the fiber diameter in the SCR muscle (p = 0.0097), completely normalizing the fiber diameter to the level of the CTRL muscles (p = 0.1422) (Figure 4f,g).
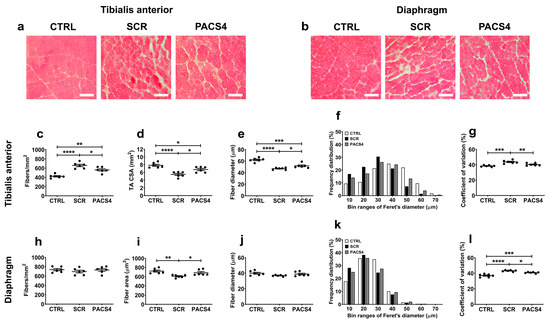
Figure 4.
Improved muscle histopathology following longitudinal vivoPMO-PACS4 treatment. Histopathological changes in (a) the tibialis anterior (TA) and (b) the diaphragm (DIA) muscles were initially assessed by hematoxylin and eosin analysis. Representative images are shown at a magnification of ×100, scale bars = 100 µm. Both muscles were further stained for laminin to identify the myofiber sarcolemma. The total number and size of (c–g) TA and (h–l) DIA myofibers were automatically scored by Fiji/MuscleJ software (version 1.53c, National Institutes of Health, Bethesda, MD, USA). (c,h) The fiber number per mm2 of the muscle CSA, (d) the area of whole transverse TA muscle or (i) area of DIA fibers, (e,j) the mean of minimal Feret’s diameter of fibers, (f,k) a histogram of the frequency distribution of the fiber diameter and (g,l) the coefficient of variance of the fiber diameter are shown for the TA and DIA, respectively. Data are shown as means ± SEM; n = 6. Statistical comparison was by one-way ANOVA followed by Tukey’s multiple comparisons test; * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Surprisingly, in contrast to the remarkable changes observed through H&E staining, we detected only a minor decrease in both the number and the diameter of the DIA myofibers of the SCR group, compared to the CTRL and PACS4 values (Figure 4h,j). However, the area of myofibers in the SCR group’s DIA was significantly smaller than that of the CTRL group’s myofibers by 16% (p = 0.0046, Figure 4i), while the coefficient of variation increased from 36.9 ± 0.8% to 43.1 ± 0.4% (p < 0.0001, Figure 4k,l). The PACS4 treatment successfully restored the myofiber area to a normal level (p = 0.4684) and increased the coefficient of variation to 40.7 ± 0.4% compared to the SCR group (p = 0.0232). These results clearly demonstrate that the PACS4 treatment was effective in counteracting DUX4 pathology in both the TA and DIA muscles of treated ACTA-MCM/FLExDUX4 mice.
3.5. vivoPMO-PACS4 Treatment Significantly Reduces Muscle Turnover and Muscle Fibrosis
Since muscle regeneration in FSHD is correlated with DUX4 pathological severity [43], we further quantified the number of centrally nucleated fibers (CNFs) in both the TA and DIA muscles (Figure 5a,d,g). While the CTRL muscles displayed only 2–4% CNFs in both the TA and the DIA, we detected a substantial increase (p < 0.0001) in CNFs for the SCR group’s TA (18.7 ± 0.9%) and DIA (23.8 ± 1.5%). The treatment with PACS4, although it was started 3 weeks after DUX4 activation, prevented the muscle turnover. Indeed, the PACS4-treated muscles displayed a significant reduction in CNFs by up to 29% of the SCR group (13.2 ± 0.7% CNFs in TA and 18.3 ± 1.0% CNFs in DIA, p = 0.0001).
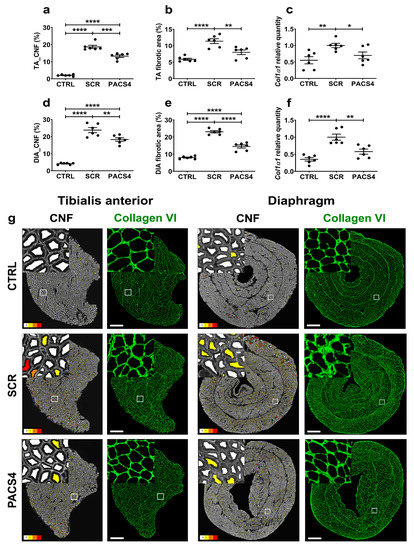
Figure 5.
vivoPMO-PACS4 treatment reduces muscle deterioration and muscle fibrosis. The tibialis anterior (TA) and diaphragm (DIA) muscle sections were stained for laminin or collagen VI; DAPI was used for nuclear staining. The total number of centrally nucleated myofibers (CNFs) of (a) TA and (d) DIA muscles was scored automatically and is expressed as a percentage of the total myofiber number within the same transverse muscle section. Fibrotic area in (b) TA and (e) DIA muscles was semi-automatically evaluated and expressed as percentage of the area positive for collagen VI of the muscle CSA. mRNA expression of Col1α1 indicative for fibrotic response in (c) TA and (f) DIA muscles was quantified by qRT-PCR as relative to Gapdh and shown as a fold-change of SCR values obtained by the same way. Data are shown as means ± SEM; n = 6. Statistical comparison was by one-way ANOVA followed by Tukey’s multiple comparisons test; * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. (g) Representative images of the entire muscle cross-sections analyzed by Fiji/MuscleJ for CNFs, with fibers having 0, 1, 2 and 3+ CNFs color-coded as white, yellow, orange and red, respectively. Images of transverse muscle sections immunostained with collagen VI are shown at a magnification of ×100, scale bar = 500 µm. Corresponding enlarged images at higher magnification are shown in the subsets.
In addition to muscle deterioration, excessive muscle fibrosis negatively affects muscle function and is an important hallmark of FSHD [44]. We therefore additionally examined the level of fibrosis in TA and DIA muscles using immunostaining for a commonly used marker of fibrosis, collagen VI [45,46] (Figure 5b,e,g). Collagen VI quantification revealed that the DUX4 expression led to an increase in the fibrotic area, from 5.9 ± 0.3% (CTRL) to 11.9 ± 0.7% (SCR, p < 0.0001) in the TA and from 8.0 ± 0.3% (CTRL) to 23.1 ± 0.7% (SCR, p < 0.0001) in the DIA. The PACS4-treated TA and DIA muscles displayed 8.0 ± 0.7% and 14.5 ± 0.9% of their area covered by collagen VI, respectively, which was a remarkable decrease by up to 37% compared to the fibrosis seen in the SCR muscles (p < 0.0001). In agreement with histological analyses, the mRNA level of Col1α1, another indicator of a fibrotic response, was highly elevated in both the SCR-treated TA (p = 0.0053) and DIA (p < 0.0001), compared to the CTRL values. The PACS4 treatment efficiently reduced the Col1α1 expression to the CTRL level by 30% (p = 0.0332) and 42% (p = 0.0026) as compared to the levels seen in the SCR-treated TA and DIA, respectively (Figure 5c,f).
The consistency of our data describing improvements in the muscle mass, muscle function, molecular features and histopathology demonstrates that vivoPMO-PACS4 has significant efficacy in inhibiting DUX4 toxicity and stabilizing the disease progression in a chronic ACTA1-MCM/FLExDUX4 model with established pathology.
4. Discussion
Despite being the third most common muscular dystrophy, no existing disease-modifying treatment is available for FSHD. With remarkable progress over the last two decades in the understanding of FSHD pathogenesis and the contribution of abhorrent DUX4 expression, numerous DUX4-silencing strategies have been extensively developed [14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33]. Surprisingly, although clinical application requires systemic DUX4 inhibition, only few research groups (including ours) have investigated the potential systemic approaches in FSHD disease models [21,30,33,34]. In this work, we verified the inhibitory effect of vivoPMO-PACS4 in the transgenic ACTA1-MCM/FLExDUX4 mouse model of FSHD. We demonstrated for the first time that delivering vivoPMO-PACS4 therapy when the pathology is established, which is more relevant for clinical translation, remains effective in downregulating chronic DUX4 expression at both the mRNA and protein levels in the two most affected skeletal muscles. Furthermore, long-term systemic antisense treatment substantially ameliorates the mass of five muscle types and multiple muscle functions by efficiently stabilizing the disease progression.
ACTA1-MCM/FLExDUX4 is the most used model [21,25,33,34,47] among three transgenic FSHD-like mouse models recently developed [36,48,49]. Despite patients usually being diagnosed with progressive DUX4 pathology, translational research has so far been trialed either in uninduced-DUX4 models [16,21,32], shortly after DUX4 induction [30,33,34] or even before DUX4’s activation [25]. Several groups have suggested that, without TMX-mediated activation, the leaky mosaic levels of DUX4 accumulating over the animal’s lifetime are sufficient enough to cause muscle damage and recapitulate FSHD pathogenesis, potentially more than when DUX4 is forcedly expressed [21,47]. These chronic models indeed provide an advantage for long-term translational studies, offering consistency and reproducibility among research groups. However, mature stages are required in these models (≥3 months old, equivalent to human age ≥20 years) [47] that are unsuitable for investigating the pathological mechanisms involved in infantile FSHD [50]. Recently, the effect of the systemic delivery of gapmer antisense chemistry was evaluated in non-DUX4-induced ACTA1-MCM/FLExDUX4 mice for either 4 or 10 weeks [21]. Although substantial downregulation in the expression of the leaky DUX4 and improvement in some histological features were observed, no changes in muscle mass or muscle function were detected. The authors suggested that this lack of benefit was possibly due to the starting of antisense administration after muscle damage had already occurred, or because females were used, in which the disease is considered more severe. However, using the same mouse model but with continual TMX-mediated DUX4 expression, we [33,34] and others [35] achieved both DUX4 knockdown and amelioration in the muscle/body mass, muscle function and/or muscle histology. In previous studies, we used 16-week-old male mice and inducible DUX4 expression that has been proven to cause more severe pathology than the leaky level, regardless of the animal’s sex [36]. As presented here, even under chronically inducible conditions, our vivoPMO-PACS4 sufficiently counteracted the DUX4 pathogenesis at both the molecular and histological levels, and greatly ameliorated the muscle atrophy and muscle strength. Therefore, our work suggests that inducing DUX4 expression in ACTA1-MCM/FLExDUX4 mice may make the model more suitable for preclinical tests of antisense therapeutics. However, the TMX dosage for inducing DUX4 expression requires careful refinement to avoid a burst-like effect that is highly lethal and/or lasts for a short time due to the effective muscle regeneration in rodents, which does not mimic the chronic situation found in FSHD patients [36]. Hence, the dose regimen based on a low chronic TMX dosage presented in this study is more suitable for allowing researchers to perform experiments at an early stage and possibly for long time periods, for example 12 weeks as we used here. Intriguingly, the low continual TMX dosage of 6 × 1.5 mg/kg/biweekly used in this study seemed to cause more muscle damage than the dosage of 2 × 2.5 mg/kg/biweekly previously used in our 4-week study [33,34]. Although we did not observe major drops in the body weight or fatigue resistance as in the short-term experiments, the histopathological changes in the muscles of vivoPMO-SCR-treated mice, particularly in the DIA, appeared to be more severe in this longitudinal study. This may account for the pseudo-hypertrophy seen in the DIA compared to the atrophy seen in other hind/forelimb muscles. Together with the mRNA quantification of Pgc1α expression, which showed a substantial reduction in the DIA compared to the TA, the greater severity in the former muscle may provide an explanation for the respiratory dysfunction in FSHD patients [6], although this requires further investigation. Interestingly, the number of DUX4-positive nuclei in DIA tissue in this study was 7-fold less than the amount observed previously [34]. We speculate that the low DUX4 level in this chronic model did not cause a burst-like effect, but instead transiently triggered a more severe cumulative pathology. This is consistent with an analysis of FSHD muscle biopsies, where DUX4 protein could be found in some samples, but was never detected in those of severely affected patients [51], emphasizing the impact of long-term vivoPMO-PACS4 treatment in a model that more closely represents the human condition.
RNA-silencing therapies have emerged over the last decade as a hot spot for new drug development. A total of fifteen FDA-approved drugs are on the market and more than 100 are in clinical trials (https://clinicaltrials.gov/; accessed on 20 May 2022). Among these, four drugs (EXONDYS 51®, VYONDYS 53®, VILTEPSO® and SPINRAZA®) receiving FDA conditional approval for neuromuscular dystrophies are all based on PMO chemistry. Despite a high biosafety profile and promising therapeutic efficacy [52], the limited cellular uptake due to the neutral charge of the backbone is a major challenge for clinical applications of PMO, particularly for FSHD. Nevertheless, as demonstrated here, vivoPMO chemistry with a PMO backbone conjugated with a cell-penetrating octaguanidine dendrimer effectively penetrates the skeletal muscles, though the development of safer systems to improve tissue penetration will be necessary for the translation of antisense therapeutics into the clinic. Alternatively, the PMO-PACS4 sequence could be conjugated with other cell-penetrating peptides, such as Sarepta Therapeutics’ PPMO chemistry, which has displayed positive results in a phase 2 clinical trial for Duchenne muscular dystrophy (company’s report: https://investorrelations.sarepta.com/news-releases/news-release-details/sarepta-therapeutics-reports-positive-clinical-results-phase-2; accessed on 20 May 2022), or with monoclonal antibodies targeting the transferrin receptor to enable muscle delivery, such as those being trialed in FSHD animal models by Avidity Biosciences or Dyne Therapeutics [35]. Moreover, a comparison of the antisense efficiency between different systemic delivery routes should be conducted, for example between the intraperitoneal route used here and the intravenous or subcutaneous delivery that has been used in clinical settings, to obtain the best therapeutic benefit with the highest clinical relevance.
5. Conclusions
In summary, the present study provides substantial evidence that long-term systemic vivoPMO-PACS4 treatment in a mouse model of FSHD, with established disease, efficiently stabilizes the pathology progression through suppressing the expression of DUX4 and its downstream targets. The observed improvements in body-wide muscle wasting, muscle weakness and muscle histopathology are particularly important, as this data was generated in a mouse model that closely represents the human condition. Overall, our data support further development of this antisense strategy for FSHD therapy.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biomedicines10071623/s1, Table S1: Muscle and body weight.
Author Contributions
Conceptualization, N.L.-N., G.D., A.M. and L.P.; formal analysis, N.L.-N. and A.M.; investigation, N.L.-N., A.M. and L.P.; methodology, N.L.-N., A.M. and L.P.; resources, G.D. and L.P.; supervision, L.P.; validation, N.L.-N., A.M. and L.P.; writing—original draft, N.L.-N.; writing—review and editing, N.L.-N., A.M. and L.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The animal study was approved by the Animal Welfare Committee of Royal Holloway University of London (approval date: 3 June 2019).
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Takako Jones and Peter Jones, University of Nevada, Nevada, USA, for valuable advice on maintaining the mouse colonies and for the protocol on using tamoxifen to induce DUX4 expression in ACTA1-MCM/FLExDUX4 mice. We also thank Emma Popescu and Nicola Sanderson of the Royal Holloway University of London, UK for the care and maintenance of the animals. The research, though not directly funded by any external body, continued on from previous work supported by Muscular Dystrophy UK.
Conflicts of Interest
A patent named “Antisense oligonucleotides and uses thereof” has been filed by the Institute of Myology, Paris, France. L.P. and G.D. are named as co-inventors. The other authors have declared that no conflict of interest exists.
References
- Mostacciuolo, M.L.; Pastorello, E.; Vazza, G.; Miorin, M.; Angelini, C.; Tomelleri, G.; Galluzzi, G.; Trevisan, C.P. Facioscapulohumeral muscular dystrophy: Epidemiological and molecular study in a north-east Italian population sample. Clin. Genet. 2009, 75, 550–555. [Google Scholar] [CrossRef]
- Deenen, J.C.; Arnts, H.; van der Maarel, S.M.; Padberg, G.W.; Verschuuren, J.J.G.M.; Bakker, E.; Weinreich, S.S.; Verbeek, A.L.; van Engelen, B.G. Population-based incidence and prevalence of facioscapulohumeral dystrophy. Neurology 2014, 83, 1056–1059. [Google Scholar] [CrossRef] [Green Version]
- Statland, J.; Tawil, R. Facioscapulohumeral Muscular Dystrophy. Neurol. Clin. 2014, 32, 721–728. [Google Scholar] [CrossRef] [Green Version]
- Tonini, M.; Passos-Bueno, M.; Cerqueira, A.; Matioli, S.; Pavanello, R.; Zatz, M. Asymptomatic carriers and gender differences in facioscapulohumeral muscular dystrophy (FSHD). Neuromuscul. Disord. 2004, 14, 33–38. [Google Scholar] [CrossRef]
- Hamel, J.; Johnson, N.; Tawil, R.; Martens, W.B.; Dilek, N.; McDermott, M.P.; Heatwole, C. Patient-Reported Symptoms in Facioscapulohumeral Muscular Dystrophy (PRISM-FSHD). Neurology 2019, 93, e1180–e1192. [Google Scholar] [CrossRef]
- Moreira, S.; Wood, L.; Smith, D.; Marini-Bettolo, C.; Guglieri, M.; McMacken, G.; Bailey, G.; Mayhew, A.; Muni-Lofra, R.; Eglon, G.; et al. Respiratory involvement in ambulant and non-ambulant patients with facioscapulohumeral muscular dystrophy. J. Neurol. 2017, 264, 1271–1280. [Google Scholar] [CrossRef]
- Wohlgemuth, M.; Horlings, C.; van der Kooi, E.; Gilhuis, H.; Hendriks, J.; van der Maarel, S.; van Engelen, B.; Heijdra, Y.; Padberg, G. Respiratory function in facioscapulohumeral muscular dystrophy 1. Neuromuscul. Disord. 2017, 27, 526–530. [Google Scholar] [CrossRef]
- Kelly, C.R.; Saw, J.; Thapa, P.; Mandrekar, J.; Naddaf, E. Systemic manifestations and symptom burden of facioscapulohumeral muscular dystrophy in a referral cohort. Muscle Nerve 2022, 65, 415–421. [Google Scholar] [CrossRef]
- Della Marca, G.; Frusciante, R.; Dittoni, S.; Vollono, C.; Buccarella, C.; Iannaccone, E.; Rossi, M.; Scarano, E.; Pirronti, T.; Cianfoni, A.; et al. Sleep disordered breathing in facioscapulohumeral muscular dystrophy. J. Neurol. Sci. 2009, 285, 54–58. [Google Scholar] [CrossRef]
- Scully, M.A.; Eichinger, K.J.; Donlin-Smith, C.M.; Tawil, R.; Statland, J.M. Restrictive lung involvement in facioscapulohumeral muscular dystrophy. Muscle Nerve 2014, 50, 739–743. [Google Scholar] [CrossRef] [Green Version]
- Henke, C.; Spiesshoefer, J.; Kabitz, H.-J.; Herkenrath, S.; Randerath, W.; Brix, T.; Görlich, D.; Young, P.; Boentert, M. Respiratory muscle weakness in facioscapulohumeral muscular dystrophy. Muscle Nerve 2019, 60, 679–686. [Google Scholar] [CrossRef]
- Dixit, M.; Ansseau, E.; Tassin, A.; Winokur, S.; Shi, R.; Qian, H.; Sauvage, S.; Mattéotti, C.; van Acker, A.M.; Leo, O.; et al. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc. Natl. Acad. Sci. USA 2007, 104, 18157–18162. [Google Scholar] [CrossRef] [Green Version]
- Ba, L.M.W.; Bs, S.E.G.; Mei, W.; Belayew, A.; Coppee, F.; Ladner, K.J.; Guttridge, D.; Yang, J.; Harper, S.Q. DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann. Neurol. 2010, 69, 540–552. [Google Scholar] [CrossRef] [Green Version]
- Vanderplanck, C.; Ansseau, E.; Charron, S.; Stricwant, N.; Tassin, A.; Laoudj-Chenivesse, D.; Wilton, S.; Coppée, F.; Belayew, A. The FSHD Atrophic Myotube Phenotype Is Caused by DUX4 Expression. PLoS ONE 2011, 6, e26820. [Google Scholar] [CrossRef]
- Chen, J.C.; King, O.D.; Zhang, Y.; Clayton, N.P.; Spencer, C.; Wentworth, B.M.; Emerson, C.P., Jr.; Wagner, K.R. Morpholino-mediated Knockdown of DUX4 Toward Facioscapulohumeral Muscular Dystrophy Therapeutics. Mol. Ther. 2016, 24, 1405–1411. [Google Scholar] [CrossRef] [Green Version]
- Lim, K.R.Q.; Maruyama, R.; Echigoya, Y.; Nguyen, Q.; Zhang, A.; Khawaja, H.; Chandra, S.S.; Jones, T.; Jones, P.; Chen, Y.-W.; et al. Inhibition of DUX4 expression with antisense LNA gapmers as a therapy for facioscapulohumeral muscular dystrophy. Proc. Natl. Acad. Sci. USA 2020, 117, 16509–16515. [Google Scholar] [CrossRef]
- Block, G.J.; Narayanan, D.; Amell, A.M.; Petek, L.M.; Davidson, K.C.; Bird, T.D.; Tawil, R.; Moon, R.T.; Miller, D.G. Wnt/β-catenin signaling suppresses DUX4 expression and prevents apoptosis of FSHD muscle cells. Hum. Mol. Genet. 2013, 22, 4661–4672. [Google Scholar] [CrossRef] [Green Version]
- Campbell, A.E.; Oliva, J.; Yates, M.P.; Zhong, J.W.; Shadle, S.C.; Snider, L.; Singh, N.; Tai, S.; Hiramuki, Y.; Tawil, R.; et al. BET bromodomain inhibitors and agonists of the beta-2 adrenergic receptor identified in screens for compounds that inhibit DUX4 expression in FSHD muscle cells. Skelet. Muscle 2017, 7, 1–18. [Google Scholar] [CrossRef]
- Cruz, J.M.; Hupper, N.; Wilson, L.S.; Concannon, J.B.; Wang, Y.; Oberhauser, B.; Patora-Komisarska, K.; Zhang, Y.; Glass, D.J.; Trendelenburg, A.-U.; et al. Protein kinase A activation inhibits DUX4 gene expression in myotubes from patients with facioscapulohumeral muscular dystrophy. J. Biol. Chem. 2018, 293, 11837–11849. [Google Scholar] [CrossRef] [Green Version]
- Wallace, L.M.; Liu, J.; Domire, J.; Garwick-Coppens, S.E.; Guckes, S.M.; Mendell, J.R.; Flanigan, K.; Harper, S.Q. RNA Interference Inhibits DUX4-induced Muscle Toxicity In Vivo: Implications for a Targeted FSHD Therapy. Mol. Ther. 2012, 20, 1417–1423. [Google Scholar] [CrossRef] [Green Version]
- Bouwman, L.F.; Hamer, B.D.; Heuvel, A.V.D.; Franken, M.; Jackson, M.; Dwyer, C.A.; Tapscott, S.J.; Rigo, F.; van der Maarel, S.M.; de Greef, J.C. Systemic delivery of a DUX4-targeting antisense oligonucleotide to treat facioscapulohumeral muscular dystrophy. Mol. Ther. Nucleic Acids 2021, 26, 813–827. [Google Scholar] [CrossRef] [PubMed]
- Oliva, J.; Galasinski, S.; Richey, A.; Campbell, A.E.; Meyers, M.; Modi, N.; Zhong, J.W.; Tawil, R.; Tapscott, S.J.; Sverdrup, F.M. Clinically Advanced p38 Inhibitors Suppress DUX4 Expression in Cellular and Animal Models of Facioscapulohumeral Muscular Dystrophy. J. Pharmacol. Exp. Ther. 2019, 370, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Saad, N.Y.; Al-Kharsan, M.; Garwick-Coppens, S.E.; Chermahini, G.A.; Harper, M.A.; Palo, A.; Boudreau, R.L.; Harper, S.Q. Human miRNA miR-675 inhibits DUX4 expression and may be exploited as a potential treatment for Facioscapulohumeral muscular dystrophy. Nat. Commun. 2021, 12, 1–18. [Google Scholar] [CrossRef]
- Wallace, L.M.; Saad, N.Y.; Pyne, N.K.; Fowler, A.M.; Eidahl, J.O.; Domire, J.S.; Griffin, D.A.; Herman, A.C.; Sahenk, Z.; Rodino-Klapac, L.R.; et al. Pre-clinical Safety and Off-Target Studies to Support Translation of AAV-Mediated RNAi Therapy for FSHD. Mol. Ther. Methods Clin. Dev. 2018, 8, 121–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Himeda, C.L.; Jones, T.I.; Jones, P.L. Targeted epigenetic repression by CRISPR/dSaCas9 suppresses pathogenic DUX4-fl expression in FSHD. Mol. Ther. Methods Clin. Dev. 2021, 20, 298–311. [Google Scholar] [CrossRef]
- Ciszewski, L.; Lu-Nguyen, N.; Slater, A.; Brennan, A.; Williams, H.; Dickson, G.; Searle, M.S.; Popplewell, L. G-quadruplex ligands mediate downregulation of DUX4 expression. Nucleic Acids Res. 2020, 48, 4179–4194. [Google Scholar] [CrossRef] [Green Version]
- Rojas, L.A.; Valentine, E.; Accorsi, A.; Maglio, J.; Shen, N.; Robertson, A.; Kazmirski, S.; Rahl, P.; Tawil, R.; Cadavid, D.; et al. p38α Regulates Expression of DUX4 in a Model of Facioscapulohumeral Muscular Dystrophy. J. Pharmacol. Exp. Ther. 2020, 374, 489–498. [Google Scholar] [CrossRef]
- Mariot, V.; Joubert, R.; Marsollier, A.-C.; Hourdé, C.; Voit, T.; Dumonceaux, J. A Deoxyribonucleic Acid Decoy Trapping DUX4 for the Treatment of Facioscapulohumeral Muscular Dystrophy. Mol. Ther. Nucleic Acids 2020, 22, 1191–1199. [Google Scholar] [CrossRef]
- DeSimone, A.M.; Leszyk, J.; Wagner, K.R.; Emerson, C.P., Jr. Identification of the hyaluronic acid pathway as a therapeutic target for facioscapulohumeral muscular dystrophy. Sci. Adv. 2019, 5, eaaw7099. [Google Scholar] [CrossRef] [Green Version]
- Bosnakovski, D.; da Silva, M.T.; Sunny, S.T.; Ener, E.T.; Toso, E.A.; Yuan, C.; Cui, Z.; Walters, M.A.; Jadhav, A.; Kyba, M. A novel P300 inhibitor reverses DUX4-mediated global histone H3 hyperacetylation, target gene expression, and cell death. Sci. Adv. 2019, 5, eaaw7781. [Google Scholar] [CrossRef] [Green Version]
- Rashnonejad, A.; Amini-Chermahini, G.; Taylor, N.K.; Wein, N.; Harper, S.Q. Designed U7 snRNAs inhibit DUX4 expression and improve FSHD-associated outcomes in DUX4 overexpressing cells and FSHD patient myotubes. Mol. Ther. Nucleic Acids 2021, 23, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.R.Q.; Bittel, A.; Maruyama, R.; Echigoya, Y.; Nguyen, Q.; Huang, Y.; Dzierlega, K.; Zhang, A.; Chen, Y.-W.; Yokota, T. DUX4 Transcript Knockdown with Antisense 2′-O-Methoxyethyl Gapmers for the Treatment of Facioscapulohumeral Muscular Dystrophy. Mol. Ther. 2020, 29, 848–858. [Google Scholar] [CrossRef] [PubMed]
- Lu-Nguyen, N.; Malerba, A.; Herath, S.; Dickson, G.; Popplewell, L. Systemic antisense therapeutics inhibiting DUX4 expression ameliorates FSHD-like pathology in an FSHD mouse model. Hum. Mol. Genet. 2021, 30, 1398–1412. [Google Scholar] [CrossRef] [PubMed]
- Lu-Nguyen, N.; Malerba, A.; Pineda, M.M.A.; Dickson, G.; Popplewell, L.J. Improving Molecular and Histopathology in Diaphragm Muscle of the Double Transgenic ACTA1-MCM/FLExDUX4 Mouse Model of FSHD with Systemic Antisense Therapy. Hum. Gene Ther. 2022; (online ahead of print). [Google Scholar] [CrossRef]
- Jagannathan, S.; de Greef, J.C.; Hayward, L.J.; Yokomori, K.; Gabellini, D.; Mul, K.; Sacconi, S.; Arjomand, J.; Kinoshita, J.; Harper, S.Q. Meeting report: The 2021 FSHD International Research Congress. Skelet. Muscle 2022, 12, 1–10. [Google Scholar] [CrossRef]
- Jones, T.I.; Chew, G.-L.; Barraza-Flores, P.; Schreier, S.; Ramirez, M.; Wuebbles, R.D.; Burkin, D.J.; Bradley, R.K.; Jones, P.L. Transgenic mice expressing tunable levels of DUX4 develop characteristic facioscapulohumeral muscular dystrophy-like pathophysiology ranging in severity. Skelet. Muscle 2020, 10, 1–28. [Google Scholar] [CrossRef] [Green Version]
- Jones, T.; Jones, P.L. A cre-inducible DUX4 transgenic mouse model for investigating facioscapulohumeral muscular dystrophy. PLoS ONE 2018, 13, e0192657. [Google Scholar] [CrossRef] [Green Version]
- Lu-Nguyen, N.; Ferry, A.; Schnell, F.J.; Hanson, G.J.; Popplewell, L.; Dickson, G.; Malerba, A. Functional muscle recovery following dystrophin and myostatin exon splice modulation in aged mdx mice. Hum. Mol. Genet. 2019, 28, 3091–3100. [Google Scholar] [CrossRef]
- Terry, R.L.; Wells, D.J. Histopathological Evaluation of Skeletal Muscle with Specific Reference to Mouse Models of Muscular Dystrophy. Curr. Protoc. Mouse Biol. 2016, 6, 343–363. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Mayeuf-Louchart, A.; Hardy, D.; Thorel, Q.; Roux, P.; Gueniot, L.; Briand, D.; Mazeraud, A.; Bouglé, A.; Shorte, S.L.; Staels, B.; et al. MuscleJ: A high-content analysis method to study skeletal muscle with a new Fiji tool. Skelet. Muscle 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Razali, N.M.; Wah, Y.B. Power Comparisons of Shapiro-Wilk, Kolmogorov-Smirnov, Lilliefors and Anderson-Darling Tests. J. Stat. Model. Anal. 2011, 2, 21–33. [Google Scholar]
- Banerji, C.R.S.; Henderson, D.; Tawil, R.N.; Zammit, P.S. Skeletal muscle regeneration in facioscapulohumeral muscular dystrophy is correlated with pathological severity. Hum. Mol. Genet. 2020, 29, 2746–2760. [Google Scholar] [CrossRef] [PubMed]
- Lassche, S.; Küsters, B.; Heerschap, A.; Schyns, M.V.; Ottenheijm, C.A.; Voermans, N.C.; van Engelen, B.G. Correlation Between Quantitative MRI and Muscle Histopathology in Muscle Biopsies from Healthy Controls and Patients with IBM, FSHD and OPMD. J. Neuromuscul. Dis. 2020, 7, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Boldrin, L.; Ross, J.A.; Whitmore, C.; Doreste, B.; Beaver, C.; Eddaoudi, A.; Pearce, D.J.; Morgan, J.E. The effect of calorie restriction on mouse skeletal muscle is sex, strain and time-dependent. Sci. Rep. 2017, 7, 5160. [Google Scholar] [CrossRef] [Green Version]
- de Greef, J.; Hamlyn, R.; Jensen, B.S.; Landa, R.O.; Levy, J.R.; Kobuke, K.; Campbell, K.P. Collagen VI deficiency reduces muscle pathology, but does not improve muscle function, in the γ-sarcoglycan-null mouse. Hum. Mol. Genet. 2016, 25, 1357–1369. [Google Scholar] [CrossRef] [Green Version]
- Nunes, A.M.; Ramirez, M.; Jones, T.I.; Jones, P.L. Identification of candidate miRNA biomarkers for facioscapulohumeral muscular dystrophy using DUX4-based mouse models. Dis. Model. Mech. 2021, 14, dmm049016. [Google Scholar] [CrossRef]
- Bosnakovski, D.; Chan, S.S.K.; Recht, O.O.; Hartweck, L.M.; Gustafson, C.J.; Athman, L.L.; Lowe, D.A.; Kyba, M. Muscle pathology from stochastic low level DUX4 expression in an FSHD mouse model. Nat. Commun. 2017, 8, 550. [Google Scholar] [CrossRef]
- Giesige, C.R.; Wallace, L.M.; Heller, K.N.; Eidahl, J.O.; Saad, N.Y.; Fowler, A.M.; Pyne, N.K.; Al-Kharsan, M.; Rashnonejad, A.; Chermahini, G.A.; et al. AAV-mediated follistatin gene therapy improves functional outcomes in the TIC-DUX4 mouse model of FSHD. JCI Insight 2018, 3, e123538. [Google Scholar] [CrossRef]
- Chen, T.-H.; Lai, Y.-H.; Lee, P.-L.; Hsu, J.-H.; Goto, K.; Hayashi, Y.K.; Nishino, I.; Lin, C.-W.; Shih, H.-H.; Huang, C.-C.; et al. Infantile facioscapulohumeral muscular dystrophy revisited: Expansion of clinical phenotypes in patients with a very short EcoRI fragment. Neuromuscul. Disord. 2013, 23, 298–305. [Google Scholar] [CrossRef]
- Tassin, A.; Laoudj-Chenivesse, D.; Vanderplanck, C.; Barro, M.; Charron, S.; Ansseau, E.; Chen, Y.-W.; Mercier, J.; Coppée, F.; Belayew, A. DUX4 expression in FSHD muscle cells: How could such a rare protein cause a myopathy? J. Cell. Mol. Med. 2012, 17, 76–89. [Google Scholar] [CrossRef]
- Charleston, J.S.; Schnell, F.J.; Dworzak, J.; Donoghue, C.; Lewis, S.; Chen, L.; Young, G.D.; Milici, A.J.; Voss, J.; Dealwis, U.; et al. Eteplirsen treatment for Duchenne muscular dystrophy. Neurology 2018, 90, e2146–e2154. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).