Challenges and Practical Solutions to MRI and Histology Matching and Measurements Using Available ImageJ Software Tools
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dataset Description
2.2. Image Processing
2.3. Statistical Analysis
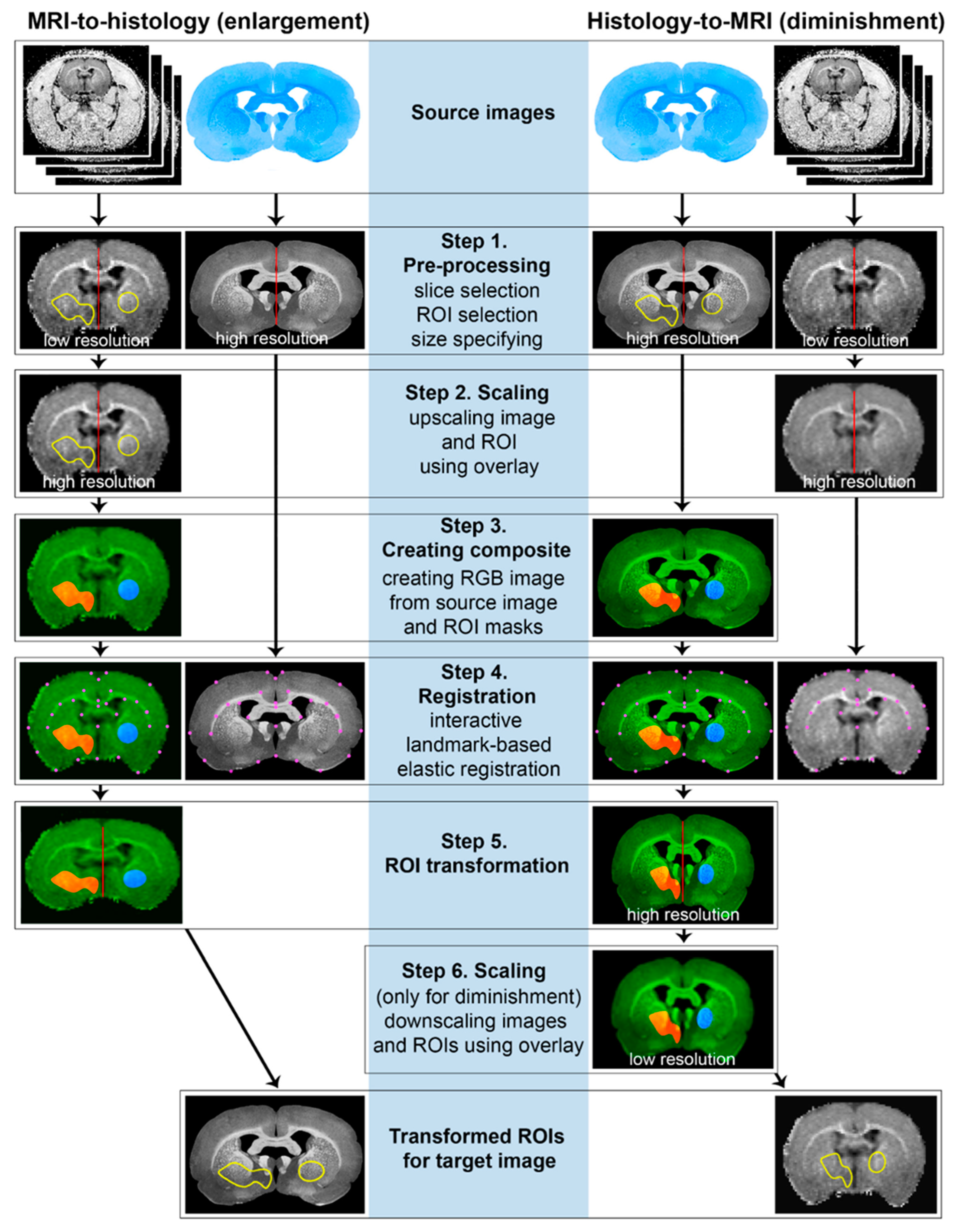
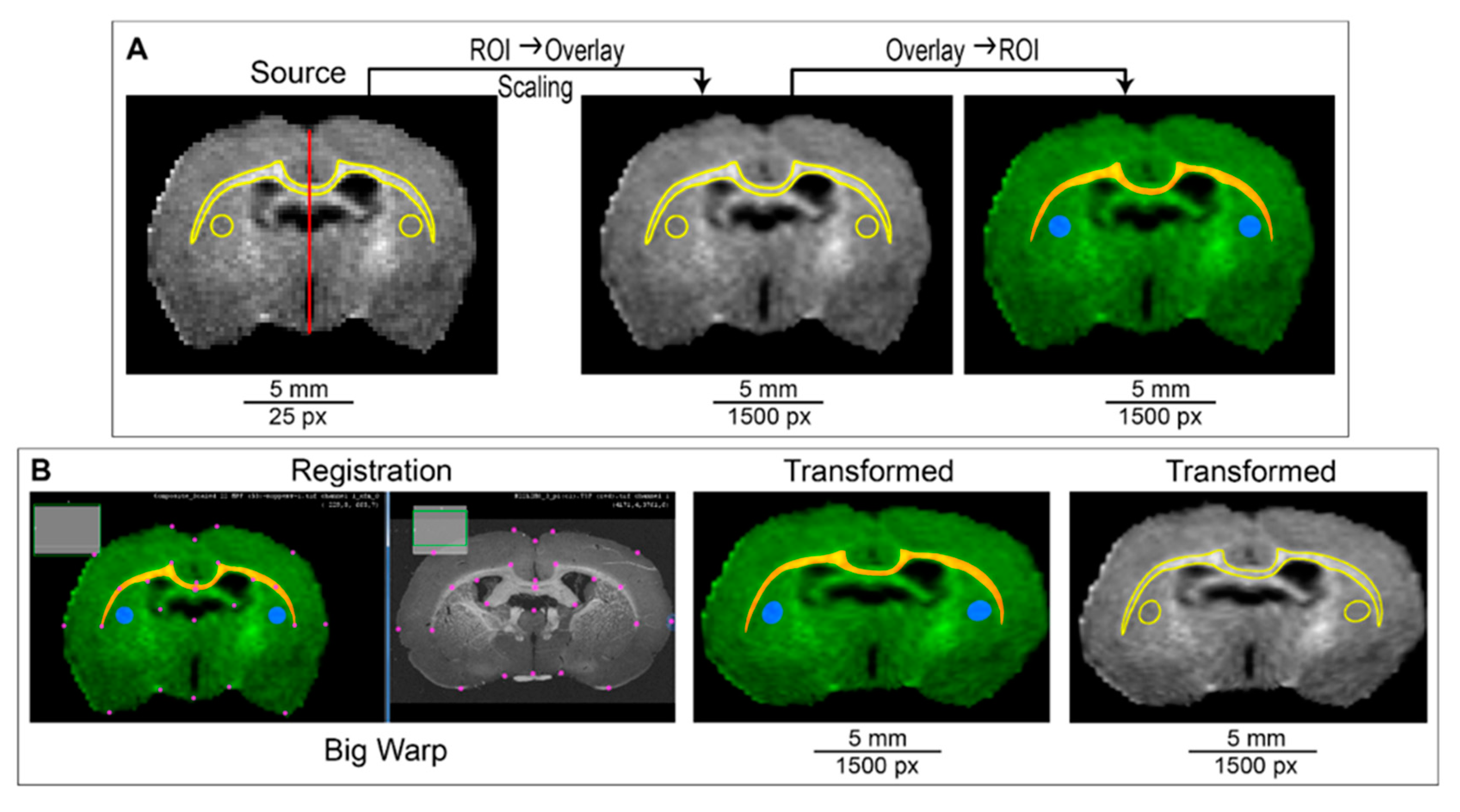
2.4. ROI Transformation Protocol (ROIT Method)
3. Results
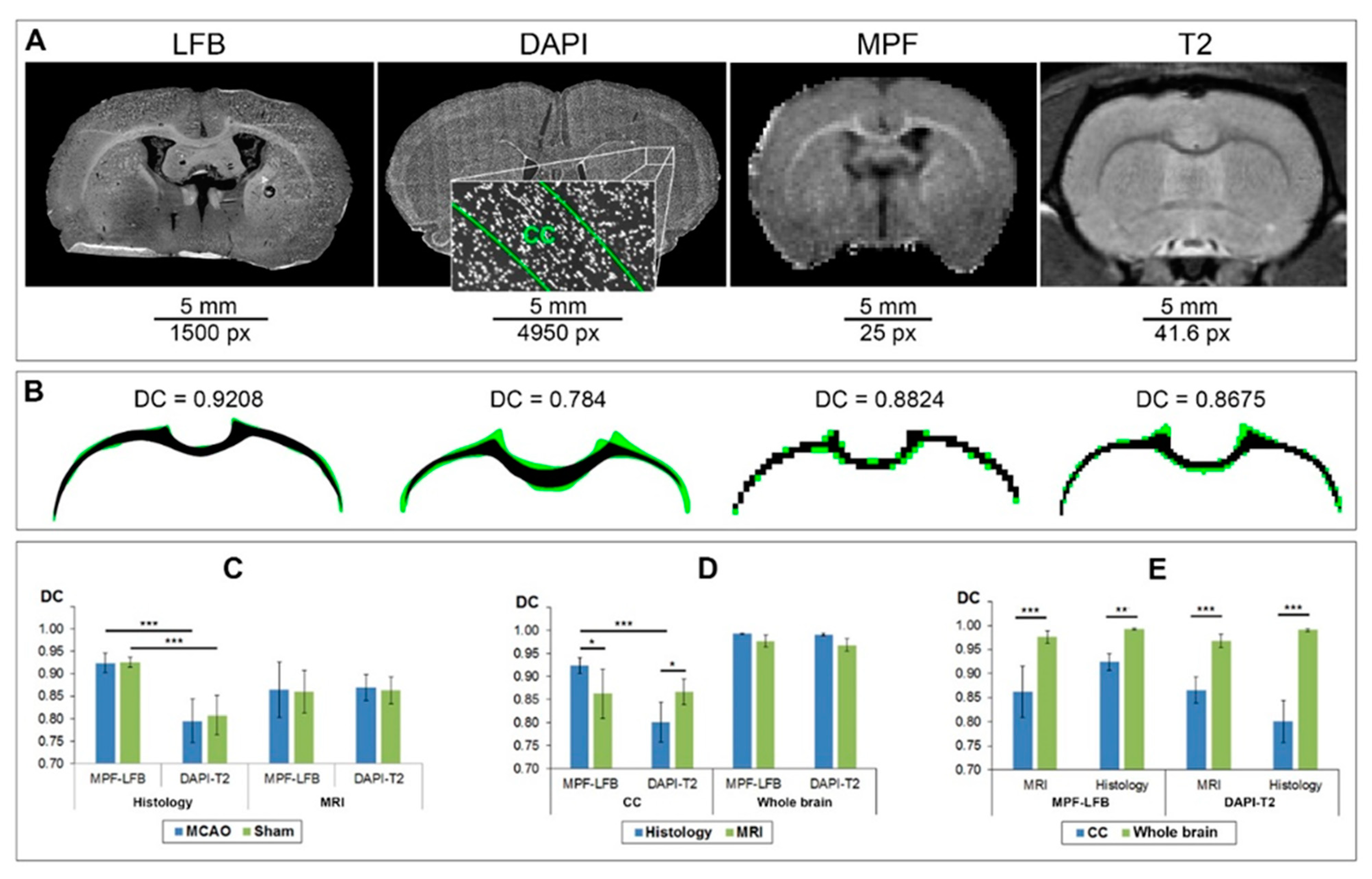
3.1. Correspondence of Manual Delineation between Two Operators
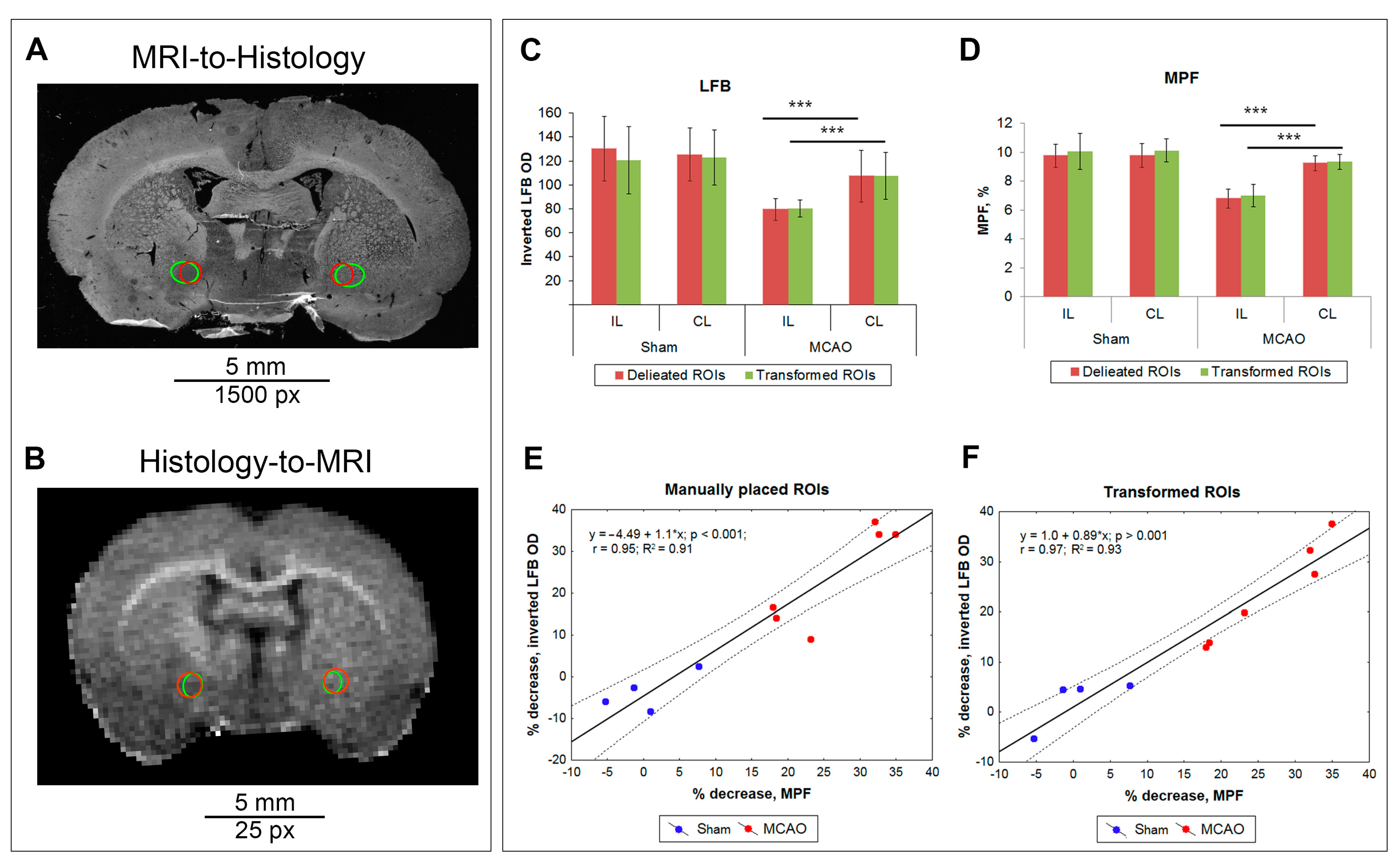
3.2. Correspondence of Transformed and Manually Delineated ROIs
3.3. Comparison of Measurements Performed by the ROIT Method and Manual Delineation
4. Discussion
4.1. Validation of the ROIT Method
4.2. Troubleshooting, Advantages, and Limitations of ROIT Method Application
4.2.1. Troubleshooting
4.2.2. Advantages
4.2.3. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mackay, A.; Whittall, K.; Adler, J.; Li, D.; Paty, D.; Graeb, D. In vivo visualization of myelin water in brain by magnetic resonance. Magn. Reson. Med. 1994, 31, 673–677. [Google Scholar] [CrossRef] [PubMed]
- Hwang, D.; Kim, D.H.; Du, Y.P. In vivo multi-slice mapping of myelin water content using T2* decay. Neuroimage 2010, 52, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Deoni, S.C.L.L.; Rutt, B.K.; Arun, T.; Pierpaoli, C.; Jones, D.K. Gleaning multicomponent T 1 and T 2 information from steady-state imaging data. Magn. Reson. Med. 2008, 60, 1372–1387. [Google Scholar] [CrossRef] [PubMed]
- Morrison, C.; Mark Henkelman, R. A model for magnetization transfer in tissues. Magn. Reson. Med. 1995, 33, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Underhill, H.R.; Rostomily, R.C.; Mikheev, A.M.; Yuan, C.; Yarnykh, V.L. Fast bound pool fraction imaging of the in vivo rat brain: Association with myelin content and validation in the C6 glioma model. Neuroimage 2011, 54, 2052–2065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stikov, N.; Perry, L.M.; Mezer, A.; Rykhlevskaia, E.; Wandell, B.A.; Pauly, J.M.; Dougherty, R.F. Bound pool fractions complement diffusion measures to describe white matter micro and macrostructure. Neuroimage 2011, 54, 1112–1121. [Google Scholar] [CrossRef] [Green Version]
- Khodanovich, M.Y.; Sorokina, I.V.; Glazacheva, V.Y.; Akulov, A.E.; Nemirovich-Danchenko, N.M.; Romashchenko, A.V.; Tolstikova, T.G.; Mustafina, L.R.; Yarnykh, V.L. Histological validation of fast macromolecular proton fraction mapping as a quantitative myelin imaging method in the cuprizone demyelination model. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Khodanovich, M.Y.; Kisel, A.A.; Akulov, A.E.; Atochin, D.N.; Kudabaeva, M.S.; Glazacheva, V.Y.; Svetlik, M.V.; Medvednikova, Y.A.; Mustafina, L.R.; Yarnykh, V.L. Quantitative assessment of demyelination in ischemic stroke in vivo using macromolecular proton fraction mapping. J. Cereb. Blood Flow Metab. 2018, 38, 919–931. [Google Scholar] [CrossRef]
- Khodanovich, M.Y.; Pishchelko, A.O.; Glazacheva, V.Y.; Pan, E.S.; Akulov, A.E.; Svetlik, M.V.; Tyumentseva, Y.A.; Anan’ina, T.V.; Yarnykh, V.L. Quantitative imaging of white and gray matter remyelination in the cuprizone demyelination model using the macromolecular proton fraction. Cells 2019, 8, 1204. [Google Scholar] [CrossRef] [Green Version]
- Song, S.K.; Sun, S.W.; Ramsbottom, M.J.; Chang, C.; Russell, J.; Cross, A.H. Dysmyelination revealed through MRI as increased radial (but unchanged axial) diffusion of water. Neuroimage 2002, 17, 1429–1436. [Google Scholar] [CrossRef]
- Piredda, G.F.; Hilbert, T.; Thiran, J.P.; Kober, T. Probing myelin content of the human brain with MRI: A review. Magn. Reson. Med. 2021, 85, 627–652. [Google Scholar] [CrossRef] [PubMed]
- Heath, F.; Hurley, S.A.; Johansen-Berg, H.; Sampaio-Baptista, C. Advances in noninvasive myelin imaging. Dev. Neurobiol. 2018, 78, 136–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mancini, M.; Karakuzu, A.; Cohen-Adad, J.; Cercignani, M.; Nichols, T.E.; Stikov, N. An interactive meta-analysis of MRI biomarkers of Myelin. Elife 2020, 9, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Nikolova, S.; Bartha, R. Registration of in vivo magnetic resonance T1-weighted brain images to triphenyltetrazolium chloride stained sections in small animals. J. Neurosci. Methods 2006, 156, 368–375. [Google Scholar] [CrossRef]
- Meadowcroft, M.D.; Connor, J.R.; Smith, M.B.; Yang, Q.X. MRI and histological analysis of beta-amyloid plaques in both human alzheimer’s disease and APP/PS1 transgenic mice. J. Magn. Reson. Imaging 2009, 29, 997–1007. [Google Scholar] [CrossRef] [Green Version]
- Al-Mubarak, H.; Vallatos, A.; Gallagher, L.; Birch, J.L.; Gilmour, L.; Foster, J.E.; Chalmers, A.J.; Holmes, W.M. Stacked in-plane histology for quantitative validation of non-invasive imaging biomarkers: Application to an infiltrative brain tumour model. J. Neurosci. Methods 2019, 326, 108372. [Google Scholar] [CrossRef]
- Rusu, M.; Kunder, C.; Fan, R.; Ghanouni, P.; West, R.; Sonn, G.; Brooks, J.D. Framework for the co-registration of MRI and histology images in prostate cancer patients with radical prostatectomy. In Proceedings of the SPIE Medical Imaging 2019, San Diego, CA, USA, 15 March 2019. [Google Scholar] [CrossRef]
- Reynolds, H.M.; Williams, S.; Zhang, A.; Chakravorty, R.; Rawlinson, D.; Ong, C.S.; Esteva, M.; Mitchell, C.; Parameswaran, B.; Finnegan, M.; et al. Development of a registration framework to validate MRI with histology for prostate focal therapy. Med. Phys. 2015, 42, 7078–7089. [Google Scholar] [CrossRef]
- Sandgren, K.; Nilsson, E.; Keeratijarut Lindberg, A.; Strandberg, S.; Blomqvist, L.; Bergh, A.; Friedrich, B.; Axelsson, J.; Ögren, M.; Ögren, M.; et al. Registration of histopathology to magnetic resonance imaging of prostate cancer. Phys. Imaging Radiat. Oncol. 2021, 18, 19–25. [Google Scholar] [CrossRef]
- Xiao, G.; Bloch, B.N.; Chappelow, J.; Genega, E.M.; Rofsky, N.M.; Lenkinski, R.E.; Tomaszewski, J.; Feldman, M.D.; Rosen, M.; Madabhushi, A. Determining histology-MRI slice correspondences for defining MRI-based disease signatures of prostate cancer. Comput. Med. Imaging Graph. 2011, 35, 568–578. [Google Scholar] [CrossRef]
- Humphreys, C.A.; Jansen, M.A.; Muñoz Maniega, S.; González-Castro, V.; Pernet, C.; Deary, I.J.; Al-Shahi Salman, R.; Wardlaw, J.M.; Smith, C. A protocol for precise comparisons of small vessel disease lesions between ex vivo magnetic resonance imaging and histopathology. Int. J. Stroke 2019, 14, 310–320. [Google Scholar] [CrossRef]
- Granot, D.; Scheinost, D.; Markakis, E.A.; Papademetris, X.; Shapiro, E.M. Serial monitoring of endogenous neuroblast migration by cellular MRI. Neuroimage 2011, 57, 817–824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granot, D.; Shapiro, E.M. Accumulation of micron sized iron oxide particles in endothelin-1 induced focal cortical ischemia in rats is independent of cell migration. Magn. Reson. Med. 2014, 71, 1568–1574. [Google Scholar] [CrossRef] [PubMed]
- Guglielmetti, C.; Praet, J.; Rangarajan, J.R.; Vreys, R.; De Vocht, N.; Maes, F.; Verhoye, M.; Ponsaerts, P.; Van der Linden, A. Multimodal imaging of subventricular zone neural stem/progenitor cells in the cuprizone mouse model reveals increased neurogenic potential for the olfactory bulb pathway, but no contribution to remyelination of the corpus callosum. Neuroimage 2014, 86, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Elvira, G.; García, I.; Benito, M.; Gallo, J.; Desco, M.; Penadés, S.; Garcia-Sanz, J.A.; Silva, A. Live imaging of mouse endogenous neural progenitors migrating in response to an induced tumor. PLoS ONE 2012, 7, e44466. [Google Scholar] [CrossRef] [Green Version]
- Shapiro, E.M.; Gonzalez-Perez, O.; Manuel García-Verdugo, J.; Alvarez-Buylla, A.; Koretsky, A.P. Magnetic resonance imaging of the migration of neuronal precursors generated in the adult rodent brain. Neuroimage 2006, 32, 1150–1157. [Google Scholar] [CrossRef] [Green Version]
- Panizzo, R.A.; Kyrtatos, P.G.; Price, A.N.; Gadian, D.G.; Ferretti, P.; Lythgoe, M.F. In vivo magnetic resonance imaging of endogenous neuroblasts labelled with a ferumoxide-polycation complex. Neuroimage 2009, 44, 1239–1246. [Google Scholar] [CrossRef]
- Vreys, R.; Soenen, S.J.H.; De Cuyper, M.; Van Der Linden, A. Background migration of USPIO/MLs is a major drawback for in situ labeling of endogenous neural progenitor cells. Contrast Media Mol. Imaging 2011, 6, 1–6. [Google Scholar] [CrossRef]
- Vreys, R.; Velde, G.V.; Krylychkina, O.; Vellema, M.; Verhoye, M.; Timmermans, J.P.; Baekelandt, V.; Van der Linden, A. MRI visualization of endogenous neural progenitor cell migration along the RMS in the adult mouse brain: Validation of various MPIO labeling strategies. Neuroimage 2010, 49, 2094–2103. [Google Scholar] [CrossRef]
- Granot, D.; Nkansah, M.K.; Bennewitz, M.F.; Tang, K.S.; Markakis, E.A.; Shapiro, E.M. Clinically viable magnetic poly(lactide-co-glycolide) particles for MRI-based cell tracking. Magn. Reson. Med. 2014, 71, 1238–1250. [Google Scholar] [CrossRef] [Green Version]
- Pothayee, N.; Cummings, D.M.; Schoenfeld, T.J.; Dodd, S.; Cameron, H.A.; Belluscio, L.; Koretsky, A.P. Magnetic resonance imaging of odorant activity-dependent migration of neural precursor cells and olfactory bulb growth. Neuroimage 2017, 158, 232–241. [Google Scholar] [CrossRef]
- Shuboni-Mulligan, D.D.; Chakravarty, S.; Mallett, C.L.; Wolf, A.M.; Forton, S.; Shapiro, E.M. Age-dependent visualization of neural progenitor cells within the rostral migratory stream via MRI and endogenously labeled micron-sized iron oxide particles. bioRxiv 2018, 429787. [Google Scholar] [CrossRef] [Green Version]
- Khodanovich, M.Y.; Akulov, A.E.; Anan’ina, T.V.; Kudabaeva, M.S.; Pishchelko, A.O.; Krutenkova, E.P.; Nemirovich-Danchenko, N.M.; Svetlik, M.V.; Tumentceva, Y.A.; Van den Haute, C.; et al. Tissue-specific ferritin- and GFP-based genetic vectors visualize neurons by MRI in the intact and post-ischemic rat brain. Int. J. Mol. Sci. 2020, 21, 8951. [Google Scholar] [CrossRef] [PubMed]
- Vande Velde, G.; Rangarajan, J.R.; Toelen, J.; Dresselaers, T.; Ibrahimi, A.; Krylychkina, O.; Vreys, R.; Van Der Linden, A.; Maes, F.; Debyser, Z.; et al. Evaluation of the specificity and sensitivity of ferritin as an MRI reporter gene in the mouse brain using lentiviral and adeno-associated viral vectors. Gene Ther. 2011, 18, 594–605. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Su, P.; Liu, Y.; Wang, H.; Nie, B.; Fang, X.; Xu, Y.; Lin, K.; Lv, P.; He, X.; et al. Detection of neural connections with ex vivo MRI using a ferritin-encoding trans-synaptic virus. Neuroimage 2019, 197, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Scarpelli, M.L.; Healey, D.R.; Mehta, S.; Kodibagkar, V.D.; Quarles, C.C. A practical method for multimodal registration and assessment of whole-brain disease burden using PET, MRI, and optical imaging. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carrera, I.; Frise, E.; Verena, K.; Mark, L.; Tobias, P.; Stephan, P.; Curtis, R.; Stephan, S.; Benjamin, S.; et al. Fiji—An Open platform for biological image analysis. Nat. Methods 2019, 9, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yarnykh, V.L. Time-efficient, high-resolution, whole brain three-dimensional macromolecular proton fraction mapping. Magn. Reson. Med. 2016, 75, 2100–2106. [Google Scholar] [CrossRef] [Green Version]
- Naumova, A.V.; Akulov, A.E.; Khodanovich, M.Y.; Yarnykh, V.L. High-resolution three-dimensional macromolecular proton fraction mapping for quantitative neuroanatomical imaging of the rodent brain in ultra-high magnetic fields. Neuroimage 2017, 147, 985–993. [Google Scholar] [CrossRef] [Green Version]
- Paxinos, G.; Watson, C. The Rat Brain in Stereotaxic Coordinates; Academic Press: San Diego, CA, USA, 2007; ISBN 9780125476126. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Bogovic, J.A.; Hanslovsky, P.; Wong, A.; Saalfeld, S. Robust registration of calcium images by learned contrast synthesis. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; pp. 1123–1126. [Google Scholar] [CrossRef] [Green Version]
- Saalfeld, S.; Tomancák, P. Automatic landmark correspondence detection for ImageJ. In Proceedings of the ImageJ User and Developer Conference, Esch-sur-Alzette, Luxembourg, 6–7 November 2008; pp. 128–133. [Google Scholar]
- Sorzano, C.Ó.S.; Thévenaz, P.; Unser, M. Elastic registration of biological images using vector-spline regularization. IEEE Trans. Biomed. Eng. 2005, 52, 652–663. [Google Scholar] [CrossRef] [Green Version]
- Khodanovich, M.Y.; Gubskiy, I.L.; Kudabaeva, M.S.; Namestnikova, D.D.; Kisel, A.A.; Anan’ina, T.V.; Tumentceva, Y.A.; Mustafina, L.R.; Yarnykh, V.L. Long-term monitoring of chronic demyelination and remyelination in a rat ischemic stroke model using macromolecular proton fraction mapping. J. Cereb. Blood Flow Metab. 2021, 41, 2856–2869. [Google Scholar] [CrossRef] [PubMed]
- Rudrapatna, U.S.; Wieloch, T.; Beirup, K.; Ruscher, K.; Mol, W.; Yanev, P.; Leemans, A.; van der Toorn, A.; Dijkhuizen, R.M. Can diffusion kurtosis imaging improve the sensitivity and specificity of detecting microstructural alterations in brain tissue chronically after experimental stroke? Comparisons with diffusion tensor imaging and histology. Neuroimage 2014, 97, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Po, C.; Kalthoff, D.; Kim, Y.B.; Nelles, M.; Hoehn, M. White matter reorganization and functional response after focal cerebral ischemia in the rat. PLoS ONE 2012, 7, e45629. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khodanovich, M.Y.; Anan’ina, T.V.; Krutenkova, E.P.; Akulov, A.E.; Kudabaeva, M.S.; Svetlik, M.V.; Tumentceva, Y.A.; Shadrina, M.M.; Naumova, A.V. Challenges and Practical Solutions to MRI and Histology Matching and Measurements Using Available ImageJ Software Tools. Biomedicines 2022, 10, 1556. https://doi.org/10.3390/biomedicines10071556
Khodanovich MY, Anan’ina TV, Krutenkova EP, Akulov AE, Kudabaeva MS, Svetlik MV, Tumentceva YA, Shadrina MM, Naumova AV. Challenges and Practical Solutions to MRI and Histology Matching and Measurements Using Available ImageJ Software Tools. Biomedicines. 2022; 10(7):1556. https://doi.org/10.3390/biomedicines10071556
Chicago/Turabian StyleKhodanovich, Marina Y., Tatyana V. Anan’ina, Elena P. Krutenkova, Andrey E. Akulov, Marina S. Kudabaeva, Mikhail V. Svetlik, Yana A. Tumentceva, Maria M. Shadrina, and Anna V. Naumova. 2022. "Challenges and Practical Solutions to MRI and Histology Matching and Measurements Using Available ImageJ Software Tools" Biomedicines 10, no. 7: 1556. https://doi.org/10.3390/biomedicines10071556
APA StyleKhodanovich, M. Y., Anan’ina, T. V., Krutenkova, E. P., Akulov, A. E., Kudabaeva, M. S., Svetlik, M. V., Tumentceva, Y. A., Shadrina, M. M., & Naumova, A. V. (2022). Challenges and Practical Solutions to MRI and Histology Matching and Measurements Using Available ImageJ Software Tools. Biomedicines, 10(7), 1556. https://doi.org/10.3390/biomedicines10071556








