A Comparative Study of Cell Culture Conditions during Conversion from Primed to Naive Human Pluripotent Stem Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Primed hESCs and hiPSCs Culture
2.2. Conversion of Primed hESCs and hiPSCs to Naive Status
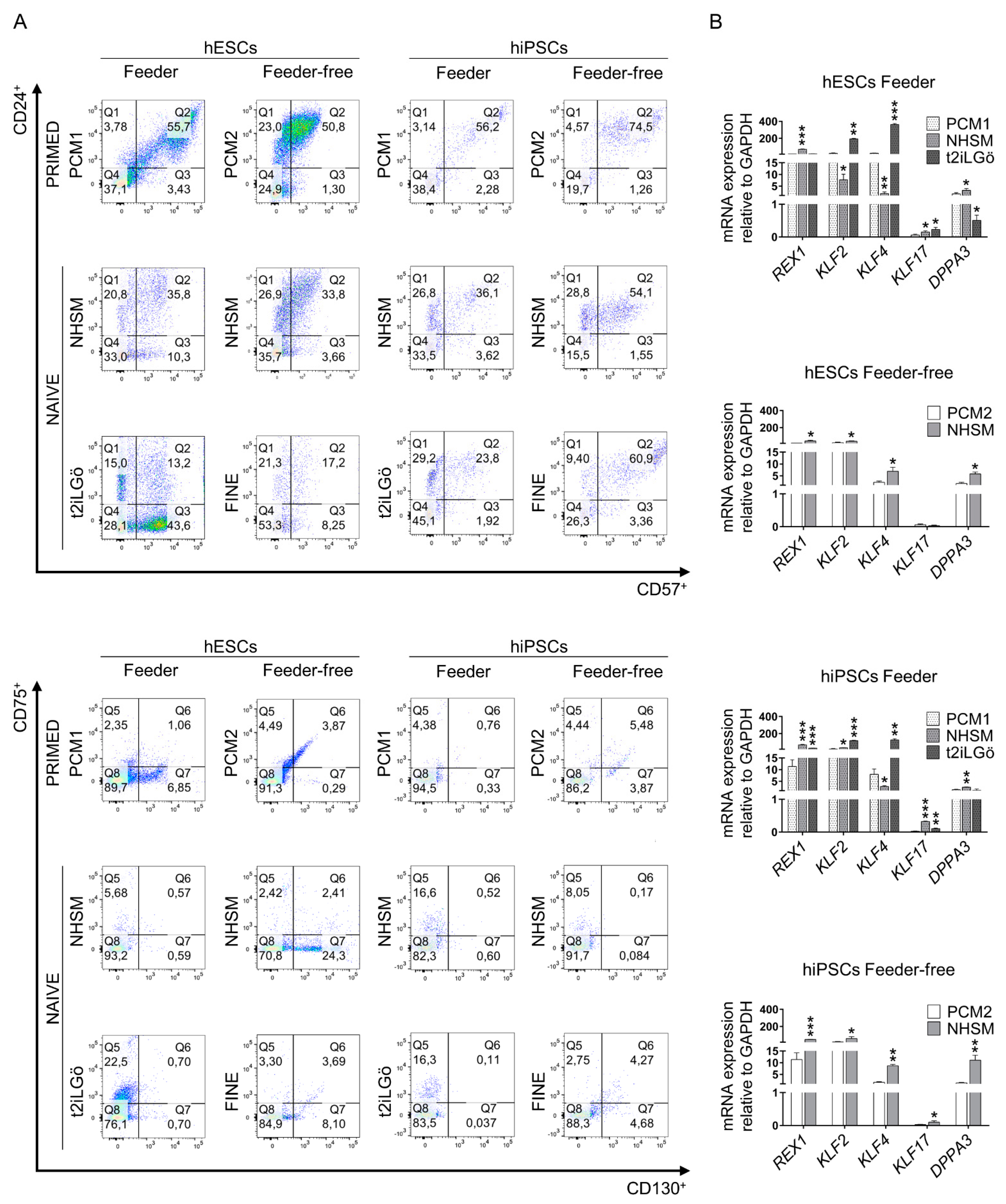
2.3. Flow Cytometry Analysis
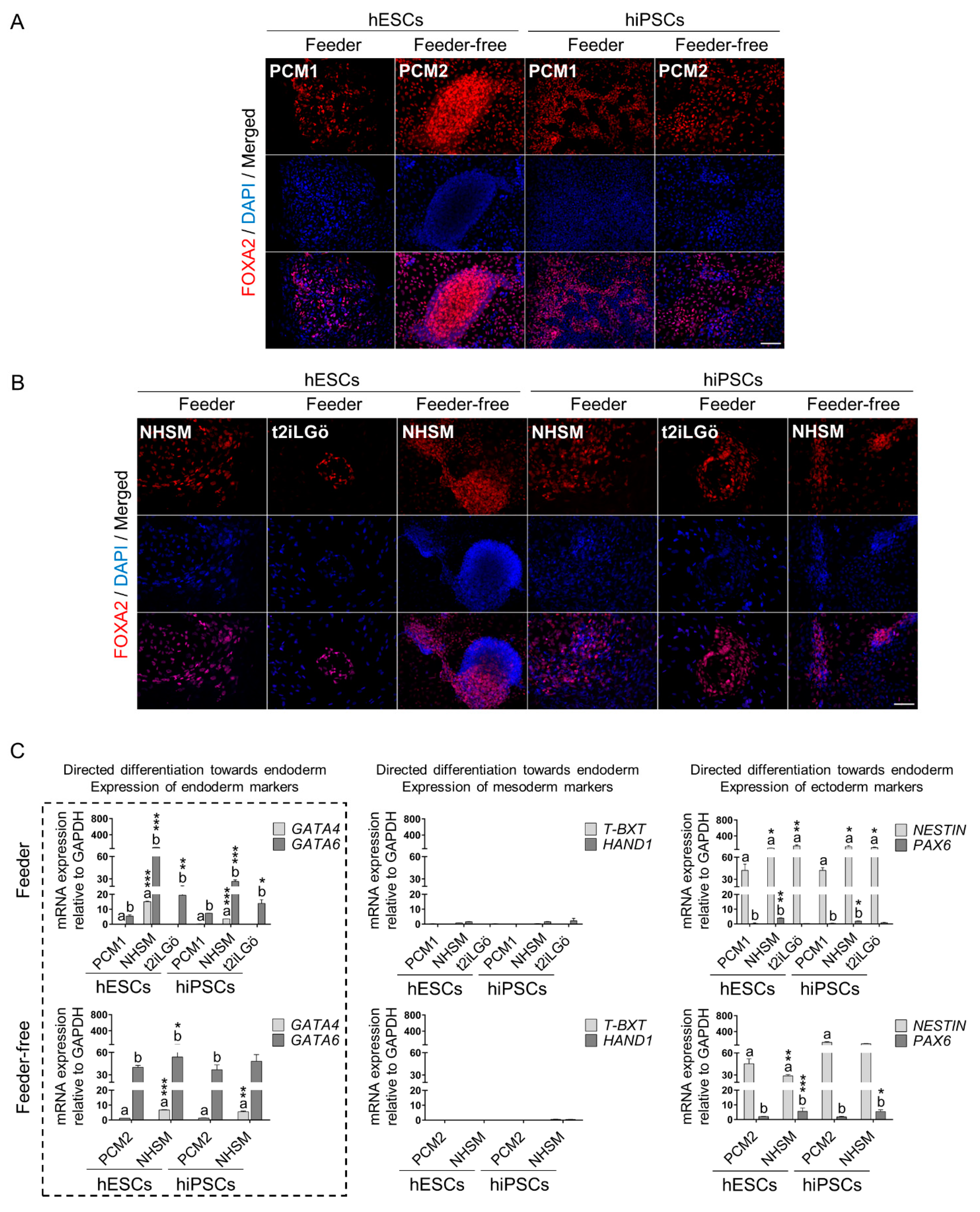
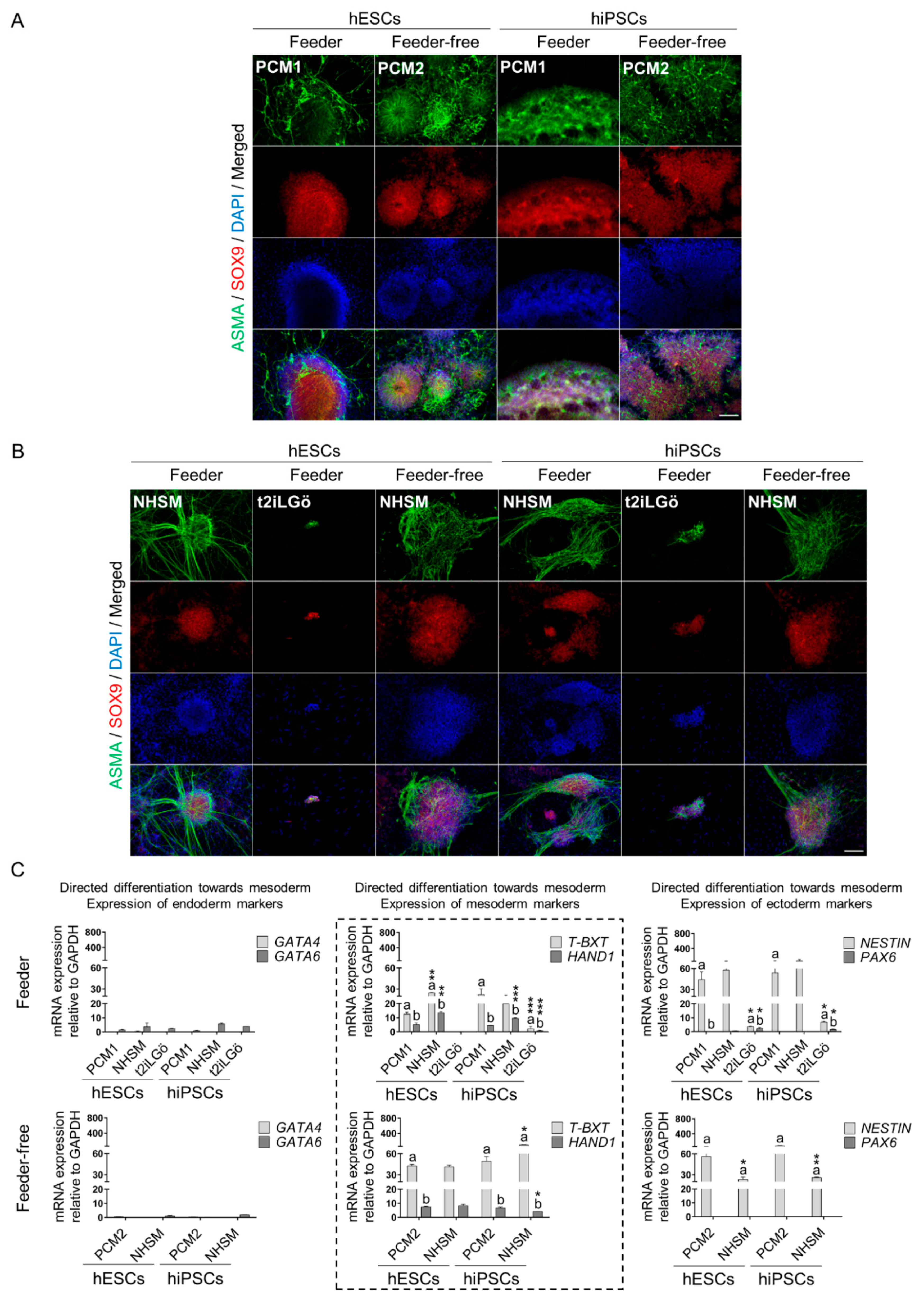
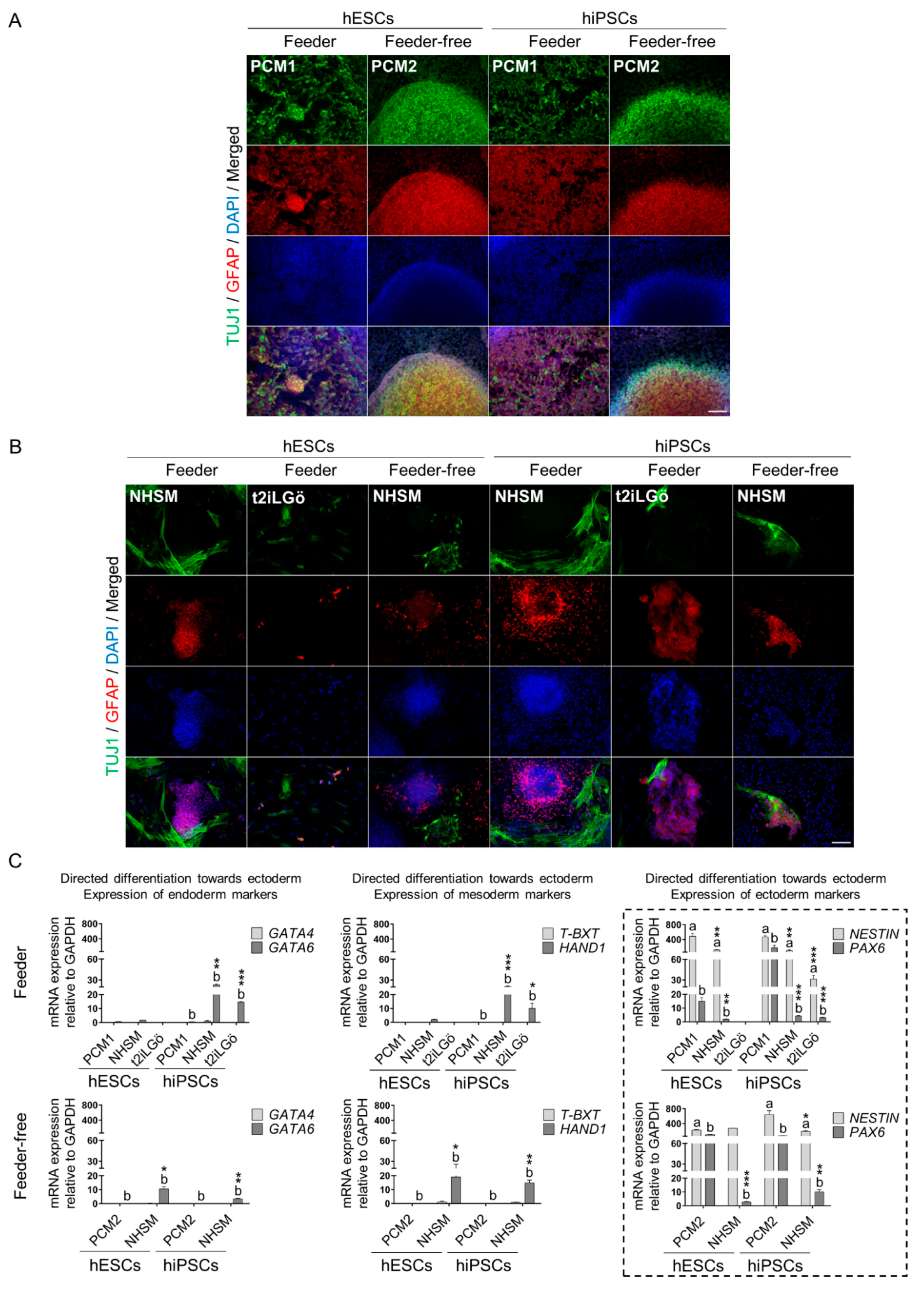
2.4. Directed Differentiation towards the Three Germ Layers
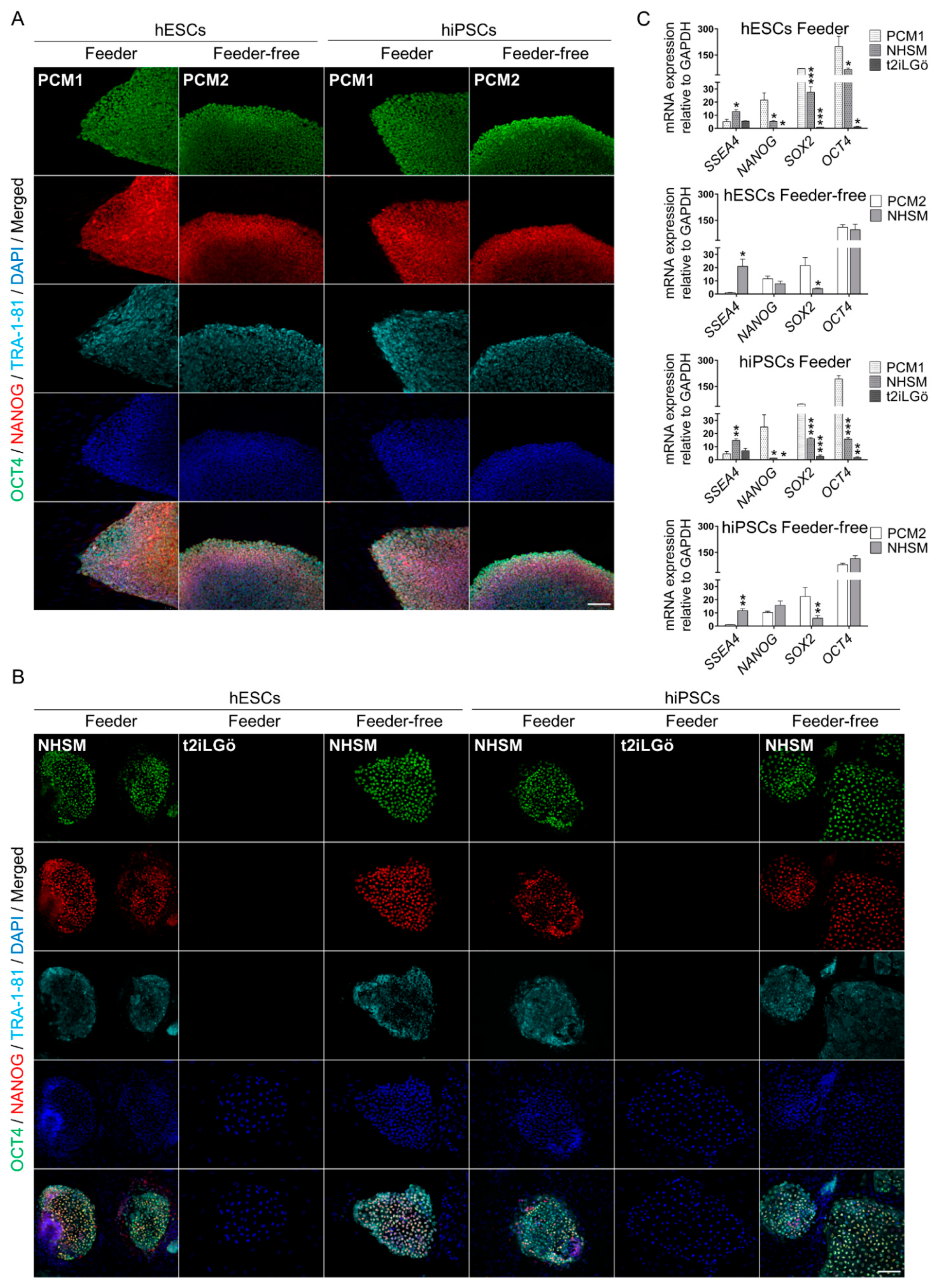
2.5. Immunofluorescence and Imaging
2.6. RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.7. Statistical Analysis
3. Results
3.1. Converted hESCs and hiPSCs Present Naive Morphology and Markers
3.2. Pluripotency Evaluation of the Naive hESCs and hiPSCs
3.3. Naive hESCs and hiPSCs Differentiate Readily into All Three Primary Germ Layers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, J.; Izpisua Belmonte, J.C. Dynamic Pluripotent Stem Cell States and Their Applications. Cell Stem Cell 2015, 17, 509–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, X.; Xiuwen, Z.; Sik, H.H.; Wanguo, W.; Ergeng, H.; Alberto, H.; Sheng, D. Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc. Natl. Acad. Sci. USA 2010, 107, 8129–8134. [Google Scholar] [CrossRef] [Green Version]
- Evans, M.J.; Kaufman, M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981, 292, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic Stem Cell Lines Derived from Human Blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef] [Green Version]
- Nichols, J.; Smith, A. Naive and Primed Pluripotent States. Cell Stem Cell 2009, 4, 487–492. [Google Scholar] [CrossRef] [Green Version]
- Tesar, P.J.; Chenoweth, J.G.; Brook, F.A.; Davies, T.J.; Evans, E.P.; Mack, D.L.; Gardner, R.L.; McKay, R.D.G. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 2007, 448, 196–199. [Google Scholar] [CrossRef]
- Hackett, J.A.; Surani, M.A. Regulatory Principles of Pluripotency: From the Ground State Up. Cell Stem Cell 2014, 15, 416–430. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [Green Version]
- Newman, A.M.; Cooper, J.B. Lab-Specific Gene Expression Signatures in Pluripotent Stem Cells. Cell Stem Cell 2010, 7, 258–262. [Google Scholar] [CrossRef] [Green Version]
- Polo, J.M.; Liu, S.; Figueroa, M.E.; Kulalert, W.; Eminli, S.; Tan, K.Y.; Apostolou, E.; Stadtfeld, M.; Li, Y.; Shioda, T.; et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 2010, 28, 848–855. [Google Scholar] [CrossRef] [Green Version]
- Gafni, O.; Weinberger, L.; Mansour, A.A.; Manor, Y.S.; Chomsky, E.; Ben-Yosef, D.; Kalma, Y.; Viukov, S.; Maza, I.; Zviran, A.; et al. Derivation of novel human ground state naive pluripotent stem cells. Nature 2013, 504, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Takashima, Y.; Guo, G.; Loos, R.; Nichols, J.; Ficz, G.; Krueger, F.; Oxley, D.; Santos, F.; Clarke, J.; Mansfield, W.; et al. Resetting Transcription Factor Control Circuitry toward Ground-State Pluripotency in Human. Cell 2014, 158, 1254–1269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szczerbinska, I.; Gonzales, K.A.U.; Cukuroglu, E.; Ramli, M.N.B.; Lee, B.P.G.; Tan, C.P.; Wong, C.K.; Rancati, G.I.; Liang, H.; Göke, J.; et al. A Chemically Defined Feeder-free System for the Establishment and Maintenance of the Human Naive Pluripotent State. Stem Cell Rep. 2019, 13, 612–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiyokawa, Y.; Sato, M.; Noguchi, H.; Inada, E.; Iwase, Y.; Kubota, N.; Sawami, T.; Terunuma, M.; Maeda, T.; Hayasaki, H.; et al. Drug-Induced Naïve iPS Cells Exhibit Better Performance than Primed iPS Cells with Respect to the Ability to Differentiate into Pancreatic β-Cell Lineage. J. Clin. Med. 2020, 9, 2838. [Google Scholar] [CrossRef] [PubMed]
- Guo, G.; Stirparo, G.G.; Strawbridge, S.E.; Spindlow, D.; Yang, J.; Clarke, J.; Dattani, A.; Yanagida, A.; Li, M.A.; Myers, S.; et al. Human naive epiblast cells possess unrestricted lineage potential. Cell Stem Cell 2021, 28, 1040–1056.e6. [Google Scholar] [CrossRef]
- Eguizabal, C.; Montserrat, N.; Vassena, R.; Barragan, M.; Garreta, E.; Garcia-Quevedo, L.; Vidal, F.; Giorgetti, A.; Veiga, A.; Belmonte, J.C.I. Complete Meiosis from Human Induced Pluripotent Stem Cells. Stem Cells 2011, 29, 1186–1195. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Tomizawa, D.K.E.-M. States of Pluripotency: Naïve and Primed Pluripotent Stem Cells; IntechOpen: Rijeka, Croatia, 2016; Chapter 3; ISBN 978-953-51-2472-6. [Google Scholar]
- Abu-Dawud, R.; Graffmann, N.; Ferber, S.; Wruck, W.; Adjaye, J. Pluripotent stem cells: Induction and self-renewal. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2018, 373, 20170213. [Google Scholar] [CrossRef] [Green Version]
- De Los Angeles, A.; Loh, Y.-H.; Tesar, P.J.; Daley, G.Q. Accessing naïve human pluripotency. Curr. Opin. Genet. Dev. 2012, 22, 272–282. [Google Scholar] [CrossRef] [Green Version]
- Collier, A.J.; Rugg-Gunn, P.J. Identifying Human Naïve Pluripotent Stem Cells − Evaluating State-Specific Reporter Lines and Cell-Surface Markers. BioEssays 2018, 40, 1700239. [Google Scholar] [CrossRef]
- Nie, Y.; Walsh, P.; Clarke, D.L.; Rowley, J.A.; Fellner, T. Scalable passaging of adherent human pluripotent stem cells. PLoS ONE 2014, 9, e88012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collier, A.J.; Panula, S.P.; Schell, J.P.; Chovanec, P.; Plaza Reyes, A.; Petropoulos, S.; Corcoran, A.E.; Walker, R.; Douagi, I.; Lanner, F.; et al. Comprehensive Cell Surface Protein Profiling Identifies Specific Markers of Human Naive and Primed Pluripotent States. Cell Stem Cell 2017, 20, 874–890.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trusler, O.; Huang, Z.; Goodwin, J.; Laslett, A.L. Cell surface markers for the identification and study of human naive pluripotent stem cells. Stem Cell Res. 2018, 26, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Kilens, S.; Meistermann, D.; Moreno, D.; Chariau, C.; Gaignerie, A.; Reignier, A.; Lelièvre, Y.; Casanova, M.; Vallot, C.; Nedellec, S.; et al. Parallel derivation of isogenic human primed and naive induced pluripotent stem cells. Nat. Commun. 2018, 9, 360. [Google Scholar] [CrossRef] [PubMed]
- Kalkan, T.; Smith, A. Mapping the route from naive pluripotency to lineage specification. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2014, 369, 20130540. [Google Scholar] [CrossRef] [Green Version]
- Messmer, T.; von Meyenn, F.; Savino, A.; Santos, F.; Mohammed, H.; Lun, A.T.L.; Marioni, J.C.; Reik, W. Transcriptional Heterogeneity in Naive and Primed Human Pluripotent Stem Cells at Single-Cell Resolution. Cell Rep. 2019, 26, 815–824.e4. [Google Scholar] [CrossRef] [Green Version]
- Yamane, M.; Ohtsuka, S.; Matsuura, K.; Nakamura, A.; Niwa, H. Overlapping functions of Krüppel-like factor family members: Targeting multiple transcription factors to maintain the naïve pluripotency of mouse embryonic stem cells. Development 2018, 145, dev162404. [Google Scholar] [CrossRef] [Green Version]
- Blakeley, P.; Fogarty, N.M.E.; del Valle, I.; Wamaitha, S.E.; Hu, T.X.; Elder, K.; Snell, P.; Christie, L.; Robson, P.; Niakan, K.K. Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development 2015, 142, 3151–3165. [Google Scholar] [CrossRef] [Green Version]
- Osnato, A.; Brown, S.; Krueger, C.; Andrews, S.; Collier, A.J.; Nakanoh, S.; Quiroga Londoño, M.; Wesley, B.T.; Muraro, D.; Brumm, A.S.; et al. TGFβ signalling is required to maintain pluripotency of human naïve pluripotent stem cells. Elife 2021, 10, e67259. [Google Scholar] [CrossRef]
- Warrier, S.; Van der Jeught, M.; Duggal, G.; Tilleman, L.; Sutherland, E.; Taelman, J.; Popovic, M.; Lierman, S.; Chuva De Sousa Lopes, S.; Van Soom, A.; et al. Direct comparison of distinct naive pluripotent states in human embryonic stem cells. Nat. Commun. 2017, 8, 15055. [Google Scholar] [CrossRef]
- Ware, C.B. Concise Review: Lessons from Naïve Human Pluripotent Cells. Stem Cells 2017, 35, 35–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ware, C.B.; Nelson, A.M.; Mecham, B.; Hesson, J.; Zhou, W.; Jonlin, E.C.; Jimenez-Caliani, A.J.; Deng, X.; Cavanaugh, C.; Cook, S.; et al. Derivation of naive human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2014, 111, 4484–4489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theunissen, T.W.; Powell, B.E.; Wang, H.; Mitalipova, M.; Faddah, D.A.; Reddy, J.; Fan, Z.P.; Maetzel, D.; Ganz, K.; Shi, L.; et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 2014, 15, 471–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Aksoy, I.; Gonnot, F.; Osteil, P.; Aubry, M.; Hamela, C.; Rognard, C.; Hochard, A.; Voisin, S.; Fontaine, E.; et al. Reinforcement of STAT3 activity reprogrammes human embryonic stem cells to naive-like pluripotency. Nat. Commun. 2015, 6, 7095. [Google Scholar] [CrossRef] [Green Version]
- Qin, H.; Hejna, M.; Liu, Y.; Percharde, M.; Wossidlo, M.; Blouin, L.; Durruthy-Durruthy, J.; Wong, P.; Qi, Z.; Yu, J.; et al. YAP Induces Human Naive Pluripotency. Cell Rep. 2016, 14, 2301–2312. [Google Scholar] [CrossRef] [Green Version]
- Chan, Y.-S.; Göke, J.; Ng, J.-H.; Lu, X.; Gonzales, K.A.U.; Tan, C.-P.; Tng, W.-Q.; Hong, Z.-Z.; Lim, Y.-S.; Ng, H.-H. Induction of a Human Pluripotent State with Distinct Regulatory Circuitry that Resembles Preimplantation Epiblast. Cell Stem Cell 2013, 13, 663–675. [Google Scholar] [CrossRef] [Green Version]
- Pastor, W.A.; Chen, D.; Liu, W.; Kim, R.; Sahakyan, A.; Lukianchikov, A.; Plath, K.; Jacobsen, S.E.; Clark, A.T. Naive Human Pluripotent Cells Feature a Methylation Landscape Devoid of Blastocyst or Germline Memory. Cell Stem Cell 2016, 18, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Allegrucci, C.; Young, L.E. Differences between human embryonic stem cell lines. Hum. Reprod. Update 2007, 13, 103–120. [Google Scholar] [CrossRef]
- Ortmann, D.; Vallier, L. Variability of human pluripotent stem cell lines. Curr. Opin. Genet. Dev. 2017, 46, 179–185. [Google Scholar] [CrossRef]
- Zakrzewski, W.; Dobrzyński, M.; Szymonowicz, M.; Rybak, Z. Stem cells: Past, present, and future. Stem Cell Res. Ther. 2019, 10, 68. [Google Scholar] [CrossRef]
- Cohen, D.E.; Melton, D. Turning straw into gold: Directing cell fate for regenerative medicine. Nat. Rev. Genet. 2011, 12, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Doğan, A. Embryonic Stem Cells in Development and Regenerative Medicine BT-Cell Biology and Translational Medicine, Volume 1: Stem Cells in Regenerative Medicine: Advances and Challenges; Turksen, K., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 1–15. ISBN 978-3-319-93867-7. [Google Scholar]
- Cao, S.; Ma, J.; Zhang, J.; Guo, R.; Liu, X.; Li, M. Reprogramming of one human induced pluripotent stem cell line from healthy donor. Stem Cell Res. 2021, 57, 102613. [Google Scholar] [CrossRef] [PubMed]
- Aasen, T.; Raya, A.; Barrero, M.J.; Garreta, E.; Consiglio, A.; Gonzalez, F.; Vassena, R.; Bilić, J.; Pekarik, V.; Tiscornia, G.; et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat. Biotechnol. 2008, 26, 1276–1284. [Google Scholar] [CrossRef] [PubMed]
- Romito, A.; Cobellis, G. Pluripotent Stem Cells: Current Understanding and Future Directions. Stem Cells Int. 2016, 2016, 9451492. [Google Scholar] [CrossRef] [Green Version]
- Carter, M.G.; Smagghe, B.J.; Stewart, A.K.; Rapley, J.A.; Lynch, E.; Bernier, K.J.; Keating, K.W.; Hatziioannou, V.M.; Hartman, E.J.; Bamdad, C.C. A Primitive Growth Factor, NME7AB, Is Sufficient to Induce Stable Naïve State Human Pluripotency; Reprogramming in This Novel Growth Factor Confers Superior Differentiation. Stem Cells 2016, 34, 847–859. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-H.; Laronde, S.; Collins, T.J.; Shapovalova, Z.; Tanasijevic, B.; McNicol, J.D.; Fiebig-Comyn, A.; Benoit, Y.D.; Lee, J.B.; Mitchell, R.R.; et al. Lineage-Specific Differentiation Is Influenced by State of Human Pluripotency. Cell Rep. 2017, 19, 20–35. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Nefzger, C.M.; Rossello, F.J.; Chen, J.; Knaupp, A.S.; Firas, J.; Ford, E.; Pflueger, J.; Paynter, J.M.; Chy, H.S.; et al. Comprehensive characterization of distinct states of human naive pluripotency generated by reprogramming. Nat. Methods 2017, 14, 1055–1062. [Google Scholar] [CrossRef]
- Linneberg-Agerholm, M.; Wong, Y.F.; Romero Herrera, J.A.; Monteiro, R.S.; Anderson, K.G.V.; Brickman, J.M. Naïve human pluripotent stem cells respond to Wnt, Nodal and LIF signalling to produce expandable naïve extra-embryonic endoderm. Development 2019, 146, dev180620. [Google Scholar] [CrossRef]
- Bredenkamp, N.; Yang, J.; Clarke, J.; Stirparo, G.G.; von Meyenn, F.; Baker, D.; Drummond, R.; Li, D.; Wu, C.; Rostovskaya, M.; et al. Efficient RNA-mediated reprogramming of human somatic cells to naïve pluripotency facilitated by tankyrase inhibition. bioRxiv 2019, 636670. [Google Scholar] [CrossRef] [Green Version]
- Guo, G.; von Meyenn, F.; Santos, F.; Chen, Y.; Reik, W.; Bertone, P.; Smith, A.; Nichols, J. Naive Pluripotent Stem Cells Derived Directly from Isolated Cells of the Human Inner Cell Mass. Stem Cell Rep. 2016, 6, 437–446. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Platero-Luengo, A.; Sakurai, M.; Sugawara, A.; Gil, M.A.; Yamauchi, T.; Suzuki, K.; Bogliotti, Y.S.; Cuello, C.; Morales Valencia, M.; et al. Interspecies Chimerism with Mammalian Pluripotent Stem Cells. Cell 2017, 168, 473–486.e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, A. Formative pluripotency: The executive phase in a developmental continuum. Development 2017, 144, 365–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, G.; von Meyenn, F.; Rostovskaya, M.; Clarke, J.; Dietmann, S.; Baker, D.; Sahakyan, A.; Myers, S.; Bertone, P.; Reik, W.; et al. Epigenetic resetting of human pluripotency. Development 2017, 144, 2748–2763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rostovskaya, M.; Stirparo, G.G.; Smith, A. Capacitation of human naïve pluripotent stem cells for multi-lineage differentiation. Development 2019, 146, dev172916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Cui, W. Sox2, a key factor in the regulation of pluripotency and neural differentiation. World J. Stem Cells 2014, 6, 305–311. [Google Scholar] [CrossRef] [Green Version]
- Thomson, M.; Liu, S.J.; Zou, L.-N.; Smith, Z.; Meissner, A.; Ramanathan, S. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell 2011, 145, 875–889. [Google Scholar] [CrossRef] [Green Version]
- Tremble, K.C.; Stirparo, G.G.; Bates, L.E.; Maskalenka, K.; Stuart, H.T.; Jones, K.; Andersson-Rolf, A.; Radzisheuskaya, A.; Koo, B.-K.; Bertone, P.; et al. Sox2 modulation increases naïve pluripotency plasticity. iScience 2021, 24, 102153. [Google Scholar] [CrossRef]
- Kuang, Y.-L.; Munoz, A.; Nalula, G.; Santostefano, K.E.; Sanghez, V.; Sanchez, G.; Terada, N.; Mattis, A.N.; Iacovino, M.; Iribarren, C.; et al. Evaluation of commonly used ectoderm markers in iPSC trilineage differentiation. Stem Cell Res. 2019, 37, 101434. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romayor, I.; Herrera, L.; Burón, M.; Martin-Inaraja, M.; Prieto, L.; Etxaniz, J.; Inglés-Ferrándiz, M.; Pineda, J.R.; Eguizabal, C. A Comparative Study of Cell Culture Conditions during Conversion from Primed to Naive Human Pluripotent Stem Cells. Biomedicines 2022, 10, 1358. https://doi.org/10.3390/biomedicines10061358
Romayor I, Herrera L, Burón M, Martin-Inaraja M, Prieto L, Etxaniz J, Inglés-Ferrándiz M, Pineda JR, Eguizabal C. A Comparative Study of Cell Culture Conditions during Conversion from Primed to Naive Human Pluripotent Stem Cells. Biomedicines. 2022; 10(6):1358. https://doi.org/10.3390/biomedicines10061358
Chicago/Turabian StyleRomayor, Irene, Lara Herrera, Maria Burón, Myriam Martin-Inaraja, Laura Prieto, Jone Etxaniz, Marta Inglés-Ferrándiz, Jose Ramon Pineda, and Cristina Eguizabal. 2022. "A Comparative Study of Cell Culture Conditions during Conversion from Primed to Naive Human Pluripotent Stem Cells" Biomedicines 10, no. 6: 1358. https://doi.org/10.3390/biomedicines10061358
APA StyleRomayor, I., Herrera, L., Burón, M., Martin-Inaraja, M., Prieto, L., Etxaniz, J., Inglés-Ferrándiz, M., Pineda, J. R., & Eguizabal, C. (2022). A Comparative Study of Cell Culture Conditions during Conversion from Primed to Naive Human Pluripotent Stem Cells. Biomedicines, 10(6), 1358. https://doi.org/10.3390/biomedicines10061358





