Breast Cancer Subtype-Specific miRNAs: Networks, Impacts, and the Potential for Intervention
Abstract
:1. Introduction
2. Breast Cancer Subtypes Defined by Gene and miRNA Expression
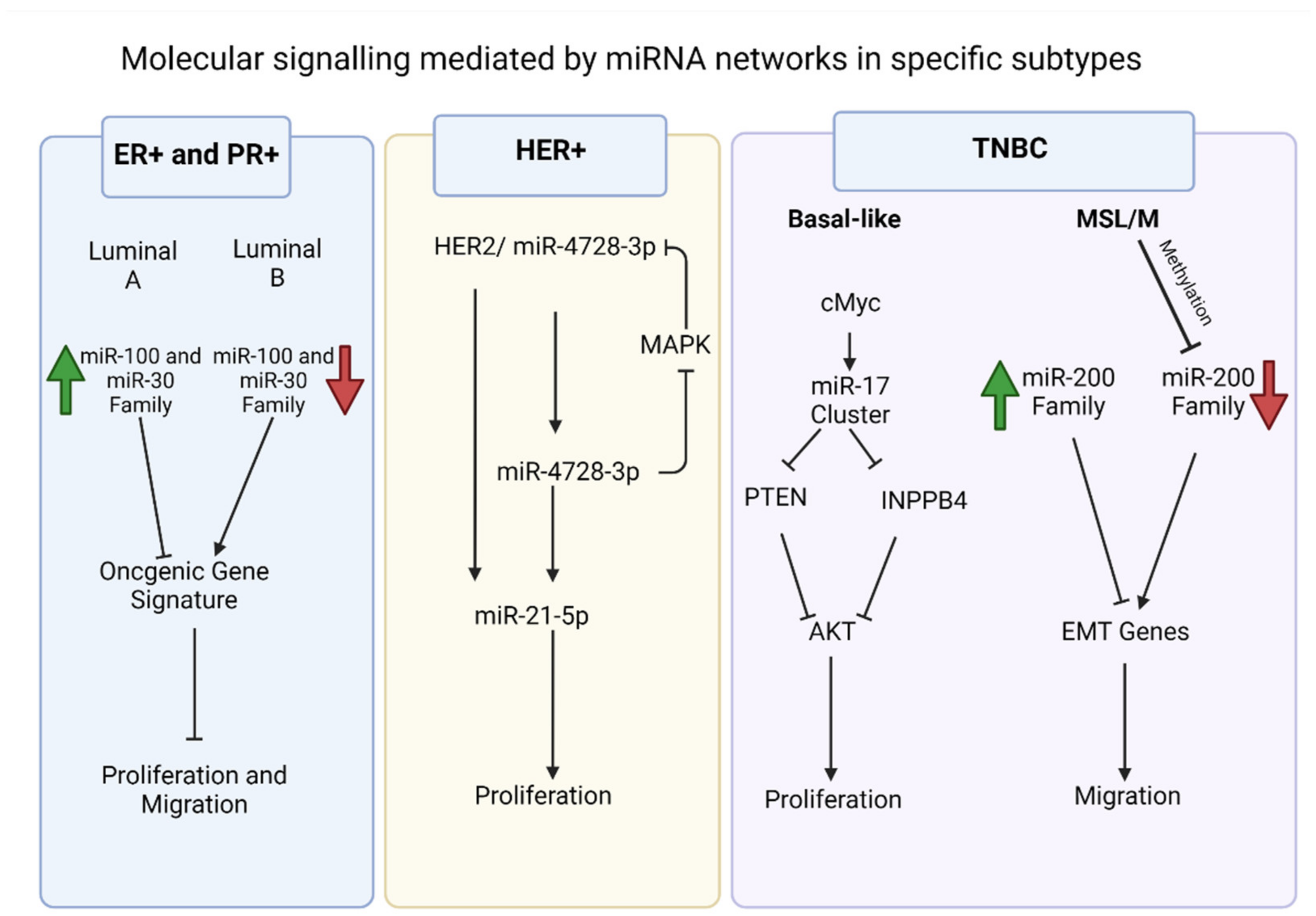
2.1. ER+/PR+ Breast Cancers
2.2. HER2 Overexpressing Breast Cancers
2.3. TNBC
2.4. Therapeutics of miRNA in Breast Cancer
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali Syeda, Z.; Langden, S.S.S.; Munkhzul, C.; Lee, M.; Song, S.J. Regulatory Mechanism of MicroRNA Expression in Cancer. Int. J. Mol. Sci. 2020, 21, 1723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, M.; Wang, H.; Yao, X.; Zhang, D.; Xie, Y.; Cui, R.; Zhang, X. Circulating MicroRNAs in Cancer: Potential and Challenge. Front. Genet. 2019, 10, 626. [Google Scholar] [CrossRef] [Green Version]
- Plotnikova, O.; Baranova, A.; Skoblov, M. Comprehensive Analysis of Human microRNA—mRNA Interactome. Front. Genet. 2019, 10, 933. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Ambros, V.; Bartel, B.; Bartel, D.P.; Burge, C.B.; Carrington, J.C.; Chen, X.; Dreyfuss, G.; Eddy, S.R.; Griffiths-Jones, S.; Marshall, M.; et al. A uniform system for microRNA annotation. RNA 2003, 9, 277–279. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Lee, Y.; Yeom, K.-H.; Nam, J.-W.; Heo, I.; Rhee, J.-K.; Sohn, S.Y.; Cho, Y.; Zhang, B.-T.; Kim, V.N. Molecular Basis for the Recognition of Primary microRNAs by the Drosha-DGCR8 Complex. Cell 2006, 125, 887–901. [Google Scholar] [CrossRef] [Green Version]
- Gregory, R.I.; Yan, K.-P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef]
- Park, J.-E.; Heo, I.; Tian, Y.; Simanshu, D.K.; Chang, H.; Jee, D.; Patel, D.J.; Kim, V.N. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 2011, 475, 201–205. [Google Scholar] [CrossRef]
- Kawamata, T.; Tomari, Y. Making RISC. Trends Biochem. Sci. 2010, 35, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Ørom, U.A.; Nielsen, F.C.; Lund, A.H. MicroRNA-10a Binds the 5′UTR of Ribosomal Protein mRNAs and Enhances Their Translation. Mol. Cell 2008, 30, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Bukhari, S.I.A.; Vasudevan, S. FXR1a-associated microRNP: A driver of specialized non-canonical translation in quiescent conditions. RNA Biol. 2017, 14, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Vidigal, J.A.; Ventura, A. The biological functions of miRNAs: Lessons from in vivo studies. Trends Cell Biol. 2015, 25, 137–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tüfekci, K.U.; Meuwissen, R.L.J.; Genç, Ş. The Role of MicroRNAs in Biological Processes. Methods Pharmacol. Toxicol. 2013, 1107, 15–31. [Google Scholar] [CrossRef]
- Osada, H.; Takahashi, T. MicroRNAs in biological processes and carcinogenesis. Carcinogenesis 2007, 28, 2–12. [Google Scholar] [CrossRef] [Green Version]
- Ebert, M.S.; Sharp, P.A. Roles for MicroRNAs in Conferring Robustness to Biological Processes. Cell 2012, 149, 515–524. [Google Scholar] [CrossRef] [Green Version]
- Bueno, M.J.; Malumbres, M. MicroRNAs and the cell cycle. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2011, 1812, 592–601. [Google Scholar] [CrossRef] [Green Version]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Yi, M.; Xu, L.; Jiao, Y.; Luo, S.; Li, A.; Wu, K. The role of cancer-derived microRNAs in cancer immune escape. J. Hematol. Oncol. 2020, 13, 25. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z. Antisense RNA and Cancer. In Cancer and Noncoding RNAs; Chakrabarti, D.J., Mitra, D.S., Eds.; Translational Epigenetics; Academic Press: Boston, MA, USA, 2018; Volume 1, Chapter 12; pp. 203–227. [Google Scholar]
- Haakensen, V.D.; Nygaard, V.; Greger, L.; Aure, M.R.; Fromm, B.; Bukholm, I.R.; Lüders, T.; Chin, S.-F.; Git, A.; Caldas, C.; et al. Subtype-specific micro-RNA expression signatures in breast cancer progression. Int. J. Cancer 2016, 139, 1117–1128. [Google Scholar] [CrossRef]
- Iorio, M.; Ferracin, M.; Liu, C.-G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA Gene Expression Deregulation in Human Breast Cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viale, G. The current state of breast cancer classification. Ann. Oncol. 2012, 23 (Suppl. S10), x207–x210. [Google Scholar] [CrossRef] [PubMed]
- Althoubaity, F.K. Molecular classification of breast cancer: A retrospective cohort study. Ann. Med. Surg. 2019, 49, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; Van De Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef]
- Van’T Veer, L.J.; Dai, H.; Van De Vijver, M.J.; He, Y.D.; Hart, A.A.M.; Mao, M.; Peterse, H.L.; Van Der Kooy, K.; Marton, M.J.; Witteveen, A.T.; et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef] [Green Version]
- Kandettu, A.; Radhakrishnan, R.; Chakrabarty, S.; Sriharikrishnaa, S.; Kabekkodu, S.P. The emerging role of miRNA clusters in breast cancer progression. Biochim. Biophys. Acta-Rev. Cancer 2020, 1874, 188413. [Google Scholar] [CrossRef]
- Cantini, L.; Bertoli, G.; Cava, C.; Dubois, T.; Zinovyev, A.; Caselle, M.; Castiglioni, I.; Barillot, E.; Martignetti, L. Identification of microRNA clusters cooperatively acting on epithelial to mesenchymal transition in triple negative breast cancer. Nucleic Acids Res. 2019, 47, 2205–2215. [Google Scholar] [CrossRef] [Green Version]
- Amorim, M.; Lobo, J.; Fontes-Sousa, M.; Estevão-Pereira, H.; Salta, S.; Lopes, P.; Coimbra, N.; Antunes, L.; De Sousa, S.P.; Henrique, R.; et al. Predictive and Prognostic Value of Selected MicroRNAs in Luminal Breast Cancer. Front. Genet. 2019, 10, 815. [Google Scholar] [CrossRef] [Green Version]
- Amiruddin, A.; Massi, M.N.; Islam, A.A.; Patellongi, I.; Pratama, M.Y.; Sutandyo, N.; Natzir, R.; Hatta, M.; Latar, N.H.M.; Wahid, S. microRNA-221 and tamoxifen resistance in luminal-subtype breast cancer patients: A case-control study. Ann. Med. Surg. 2021, 73, 103092. [Google Scholar] [CrossRef]
- Søkilde, R.; Persson, H.; Ehinger, A.; Pirona, A.C.; Fernö, M.; Hegardt, C.; Larsson, C.; Loman, N.; Malmberg, M.; Rydén, L.; et al. Refinement of breast cancer molecular classification by miRNA expression profiles. BMC Genom. 2019, 20, 503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDermott, A.M.; Miller, N.; Wall, D.; Martyn, L.M.; Ball, G.; Sweeney, K.J.; Kerin, M. Identification and Validation of Oncologic miRNA Biomarkers for Luminal A-like Breast Cancer. PLoS ONE 2014, 9, e87032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arabkari, V.; Clancy, E.; Dwyer, R.; Kerin, M.; Kalinina, O.; Holian, E.; Newell, J.; Smith, T.J. Relative and Absolute Expression Analysis of MicroRNAs Associated with Luminal a Breast Cancer—A Comparison. Pathol. Oncol. Res. 2019, 26, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Endo, Y.; Toyama, T.; Takahashi, S.; Yoshimoto, N.; Iwasa, M.; Asano, T.; Fujii, Y.; Yamashita, H. miR-1290 and its potential targets are associated with characteristics of estrogen receptor α-positive breast cancer. Endocr.-Relat. Cancer 2012, 20, 91–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Y.; Wang, X.; Niu, W.; Wang, H.; Wen, Q.; Fan, S.; Zhao, R.; Li, Z.; Xiong, W.; Peng, S.; et al. Elevated microRNA-125b levels predict a worse prognosis in HER2-positive breast cancer patients. Oncol. Lett. 2017, 13, 867–874. [Google Scholar] [CrossRef]
- Lowery, A.J.; Miller, N.; Devaney, A.; E McNeill, R.; A Davoren, P.; Lemetre, C.; Benes, V.; Schmidt, S.; Blake, J.; Ball, G.; et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neureceptor status in breast cancer. Breast Cancer Res. 2009, 11, R27. [Google Scholar] [CrossRef]
- Estevão-Pereira, H.; Lobo, J.; Salta, S.; Amorim, M.; Lopes, P.; Cantante, M.; Reis, B.; Antunes, L.; Castro, F.; De Sousa, S.P.; et al. Overexpression of circulating MiR-30b-5p identifies advanced breast cancer. J. Transl. Med. 2019, 17, 435. [Google Scholar] [CrossRef] [Green Version]
- Bailey, S.T.; Westerling, T.; Brown, M. Loss of estrogen-regulated microRNA expression increases HER2 signaling and is prognostic of poor outcome in luminal breast cancer. Cancer Res. 2014, 75, 436–445. [Google Scholar] [CrossRef] [Green Version]
- Metzger-Filho, O.; Tutt, A.M.; de Azambuja, E.; Saini, K.S.; Viale, G.; Loi, S.; Bradbury, I.; Bliss, J.; Azim, H.H.A.; Ellis, P.; et al. Dissecting the Heterogeneity of Triple-Negative Breast Cancer. J. Clin. Oncol. 2012, 30, 1879–1887. [Google Scholar] [CrossRef] [Green Version]
- Lü, L.; Mao, X.; Shi, P.; He, B.; Xu, K.; Zhang, S.; Wang, J. MicroRNAs in the Prognosis of Triple-Negative Breast Cancer: A Systematic Review and Meta-Analysis. Medicine 2017, 96, e7085. [Google Scholar] [CrossRef]
- Gasparini, P.; Cascione, L.; Fassan, M.; Lovat, F.; Guler, G.; Balci, S.; Irkkan, C.; Morrison, C.; Croce, C.M.; Shapiro, C.L.; et al. microRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget 2014, 5, 1174–1184. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Wei, N.; Ma, R.; Jiang, S.; Song, D. A miR-210-3p regulon that controls the Warburg effect by modulating HIF-1α and p53 activity in triple-negative breast cancer. Cell Death Dis. 2020, 11, 731. [Google Scholar] [CrossRef] [PubMed]
- Sanges, F.; Floris, M.; Cossu-Rocca, P.; Muroni, M.R.; Pira, G.; Urru, S.A.M.; Barrocu, R.; Gallus, S.; Bosetti, C.; D’Incalci, M.; et al. Histologic subtyping affecting outcome of triple negative breast cancer: A large Sardinian population-based analysis. BMC Cancer 2020, 20, 491. [Google Scholar] [CrossRef] [PubMed]
- Moi, L.; Braaten, T.; Al-Shibli, K.; Lund, E.; Busund, L.-T.R. Differential expression of the miR-17-92 cluster and miR-17 family in breast cancer according to tumor type; results from the Norwegian Women and Cancer (NOWAC) study. J. Transl. Med. 2019, 17, 334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Q.; Ma, D.; Gao, R.-F.; Yu, K.-D. Effect of Ki-67 Expression Levels and Histological Grade on Breast Cancer Early Relapse in Patients with Different Immunohistochemical-based Subtypes. Sci. Rep. 2020, 10, 7648. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Khoshbakht, T.; Hussen, B.M.; Taheri, M.; Samadian, M. A Review on the Role of miR-1290 in Cell Proliferation, Apoptosis and Invasion. Front. Mol. Biosci. 2021, 8, 8. [Google Scholar] [CrossRef]
- Croset, M.; Pantano, F.; Kan, C.W.S.; Bonnelye, E.; Descotes, F.; Alix-Panabières, C.; Lecellier, C.-H.; Bachelier, R.; Allioli, N.; Hong, S.-S.; et al. miRNA-30 Family Members Inhibit Breast Cancer Invasion, Osteomimicry, and Bone Destruction by Directly Targeting Multiple Bone Metastasis—Associated Genes. Cancer Res. 2018, 78, 5259–5273. [Google Scholar] [CrossRef] [Green Version]
- Yin, H.; Wang, Y.; Wu, Y.; Zhang, X.; Zhang, X.; Liu, J.; Wang, T.; Fan, J.; Sun, J.; Yang, A.; et al. EZH2-mediated Epigenetic Silencing of miR-29/miR-30 targets LOXL4 and contributes to Tumorigenesis, Metastasis, and Immune Microenvironment Remodeling in Breast Cancer. Theranostics 2020, 10, 8494–8512. [Google Scholar] [CrossRef]
- Poudel, S.; Song, J.; Jin, E.-J.; Song, K. Sulfuretin-induced miR-30C selectively downregulates cyclin D1 and D2 and triggers cell death in human cancer cell lines. Biochem. Biophys. Res. Commun. 2013, 431, 572–578. [Google Scholar] [CrossRef]
- Chang, T.-C.; Yu, D.; Lee, Y.-S.; Wentzel, E.A.; Arking, D.E.; West, K.M.; Dang, C.V.; Thomas-Tikhonenko, A.; Mendell, J.T. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008, 40, 43–50. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Li, Y.; Qi, W.; Zhang, N.; Sun, M.; Huo, Q.; Cai, C.; Lv, S.; Yang, Q. MicroRNA-99a inhibits tumor aggressive phenotypes through regulating HOXA1 in breast cancer cells. Oncotarget 2015, 6, 32737–32747. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Zhu, Q.; Tang, L. MiR-99a Antitumor Activity in Human Breast Cancer Cells through Targeting of mTOR Expression. PLoS ONE 2014, 9, e92099. [Google Scholar] [CrossRef] [Green Version]
- Xia, M.; Li, H.; Wang, J.-J.; Zeng, H.-J.; Wang, S.-H. MiR-99a Suppress Proliferation, Migration and Invasion through Regu-lating Insulin-like Growth Factor 1 Receptor in Breast Cancer. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 1755–1763. [Google Scholar] [PubMed]
- Long, X.; Shi, Y.; Ye, P.; Guo, J.; Zhou, Q.; Tang, Y. MicroRNA-99a Suppresses Breast Cancer Progression by Targeting FGFR3. Front. Oncol. 2020, 9, 1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loibl, S.; Gianni, L. HER2-positive breast cancer. Lancet 2017, 389, 2415–2429. [Google Scholar] [CrossRef]
- Normann, L.S.; Aure, M.R.; Leivonen, S.-K.; Haugen, M.H.; Hongisto, V.; Kristensen, V.N.; Mælandsmo, G.M.; Sahlberg, K.K. MicroRNA in combination with HER2-targeting drugs reduces breast cancer cell viability in vitro. Sci. Rep. 2021, 11, 10893. [Google Scholar] [CrossRef] [PubMed]
- Baselga, J. Why the Epidermal Growth Factor Receptor? The Rationale for Cancer Therapy. Oncologist 2002, 7 (Suppl. S4), 2–8. [Google Scholar] [CrossRef] [PubMed]
- Baselga, J.; Swain, S.M. Novel anticancer targets: Revisiting ERBB2 and discovering ERBB3. Nat. Cancer 2009, 9, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Persson, H.; Kvist, A.; Rego, N.; Staaf, J.; Vallon-Christersson, J.; Luts, L.; Loman, N.; Jönsson, G.B.; Naya, H.; Hoglund, M.; et al. Identification of New MicroRNAs in Paired Normal and Tumor Breast Tissue Suggests a Dual Role for the ERBB2/Her2 Gene. Cancer Res. 2011, 71, 78–86. [Google Scholar] [CrossRef] [Green Version]
- Tashkandi, H.; Shah, N.; Patel, Y.; Chen, H. Identification of new miRNA biomarkers associated with HER2-positive breast cancers. Oncoscience 2015, 2, 924–929. [Google Scholar] [CrossRef] [Green Version]
- Ferracin, M.; Bassi, C.; Pedriali, M.; Pagotto, S.; D’Abundo, L.; Zagatti, B.; Corrà, F.; Musa, G.; Callegari, E.; Lupini, L.; et al. miR-125b targets erythropoietin and its receptor and their expression correlates with metastatic potential and ERBB2/HER2 expression. Mol. Cancer 2013, 12, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banzhaf-Strathmann, J.; Edbauer, D. Good guy or bad guy: The opposing roles of microRNA 125b in cancer. Cell Commun. Signal. 2014, 12, 30. [Google Scholar] [CrossRef] [Green Version]
- Newie, I.; Søkilde, R.; Persson, H.; Grabau, D.; Rego, N.; Kvist, A.; Von Stedingk, K.; Axelson, H.; Borg, A.; Vallon-Christersson, J.; et al. The HER2-Encoded miR-4728-3p Regulates ESR1 through a Non-Canonical Internal Seed Interaction. PLoS ONE 2014, 9, e97200. [Google Scholar] [CrossRef] [PubMed]
- Floros, K.V.; Lochmann, T.L.; Hu, B.; Monterrubio, C.; Hughes, M.T.; Wells, J.D.; Morales, C.B.; Ghotra, M.S.; Costa, C.; Souers, A.J.; et al. Coamplification of miR-4728 protects HER2 -amplified breast cancers from targeted therapy. Proc. Natl. Acad. Sci. USA 2018, 115, E2594–E2603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newie, I.; Søkilde, R.; Persson, H.; Jacomasso, T.; Gorbatenko, A.; Borg, Å.; De Hoon, M.; Pedersen, S.F.; Rovira, C. HER2-encoded mir-4728 forms a receptor-independent circuit with miR-21-5p through the non-canonical poly(A) polymerase PAPD5. Sci. Rep. 2016, 6, 35664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumarswamy, R.; Volkmann, I.; Thum, T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011, 8, 706–713. [Google Scholar] [CrossRef] [Green Version]
- Jenike, A.E.; Halushka, M.K. miR-21: A non-specific biomarker of all maladies. Biomark. Res. 2021, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Pareja, F.; Geyer, F.C.; Marchiò, C.; Burke, K.A.; Weigelt, B.; Reis-Filho, J.S. Triple-negative breast cancer: The importance of molecular and histologic subtyping, and recognition of low-grade variants. npj Breast Cancer 2016, 2, 16036. [Google Scholar] [CrossRef]
- De Rinaldis, E.; Gazinska, P.; Mera, A.; Modrusan, Z.; Fedorowicz, G.M.; Burford, B.; Gillett, C.; Marra, P.; Grigoriadis, A.; Dornan, D.; et al. Integrated genomic analysis of triple-negative breast cancers reveals novel microRNAs associated with clinical and molecular phenotypes and sheds light on the pathways they control. BMC Genom. 2013, 14, 643. [Google Scholar] [CrossRef] [Green Version]
- Ventura, A.; Young, A.G.; Winslow, M.M.; Lintault, L.; Meissner, A.; Erkeland, S.J.; Newman, J.; Bronson, R.T.; Crowley, D.; Stone, J.R.; et al. Targeted Deletion Reveals Essential and Overlapping Functions of the miR-17~92 Family of miRNA Clusters. Cell 2008, 132, 875–886. [Google Scholar] [CrossRef] [Green Version]
- Mogilyansky, E.; Rigoutsos, I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013, 20, 1603–1614. [Google Scholar] [CrossRef] [PubMed]
- Dews, M.; Homayouni, A.; Yu, D.; Murphy, D.; Sevignani, C.; Wentzel, E.; E Furth, E.; Lee, W.M.; Enders, G.H.; Mendell, J.T.; et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat. Genet. 2006, 38, 1060–1065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Donnell, K.A.; Wentzel, E.A.; Zeller, K.I.; Dang, C.; Mendell, J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005, 435, 839–843. [Google Scholar] [CrossRef] [PubMed]
- Kalecky, K.; Modisette, R.; Pena, S.; Cho, Y.-R.; Taube, J. Integrative analysis of breast cancer profiles in TCGA by TNBC subgrouping reveals novel microRNA-specific clusters, including miR-17-92a, distinguishing basal-like 1 and basal-like 2 TNBC subtypes. BMC Cancer 2020, 20, 141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Paddock, M.N.; Wang, H.; Murphy, C.J.; Geck, R.C.; Navarro, A.J.; Wulf, G.M.; Elemento, O.; Haucke, V.; Cantley, L.C.; et al. The INPP4B Tumor Suppressor Modulates EGFR Trafficking and Promotes Triple-Negative Breast Cancer. Cancer Discov. 2020, 10, 1226–1239. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-Y.; Jiang, Z.; Ben-David, Y.; Woodgett, J.R.; Zacksenhaus, E. Molecular stratification within triple-negative breast cancer subtypes. Sci. Rep. 2019, 9, 19107. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, S.; Krishnan, Y. A structural map of oncomiR-1 at single-nucleotide resolution. Nucleic Acids Res. 2017, 45, 9694–9705. [Google Scholar] [CrossRef] [Green Version]
- Donayo, A.O.; Johnson, R.M.; Tseng, H.-W.; Izreig, S.; Gariepy, A.; Mayya, V.K.; Wu, E.; Alam, R.; Lussier, C.; Jones, R.G.; et al. Oncogenic Biogenesis of pri-miR-17~92 Reveals Hierarchy and Competition among Polycistronic MicroRNAs. Mol. Cell 2019, 75, 340–356. [Google Scholar] [CrossRef]
- Fuziwara, C.S.; Kimura, E.T. Insights into Regulation of the miR-17-92 Cluster of miRNAs in Cancer. Front. Med. 2015, 2, 64. [Google Scholar] [CrossRef] [Green Version]
- Uva, P.; Cossu-Rocca, P.; Loi, F.; Pira, G.; Murgia, L.; Orrù, S.; Floris, M.; Muroni, M.R.; Sanges, F.; Carru, C.; et al. miRNA-135b Contributes to Triple Negative Breast Cancer Molecular Heterogeneity: Different Expression Profile in Basal-like Versus non-Basal-like Phenotypes. Int. J. Med Sci. 2018, 15, 536–548. [Google Scholar] [CrossRef] [Green Version]
- Aakula, A.; Leivonen, S.-K.; Hintsanen, P.; Aittokallio, T.; Ceder, Y.; Børresen-Dale, A.-L.; Perälä, M.; Östling, P.; Kallioniemi, O. MicroRNA-135b regulates ERα, AR and HIF1AN and affects breast and prostate cancer cell growth. Mol. Oncol. 2015, 9, 1287–1300. [Google Scholar] [CrossRef]
- Hua, K.; Jin, J.; Zhao, J.; Song, J.; Song, H.; Li, D.; Maskey, N.; Zhao, B.; Wu, C.; Xu, H.; et al. miR-135b, upregulated in breast cancer, promotes cell growth and disrupts the cell cycle by regulating LATS2. Int. J. Oncol. 2016, 48, 1997–2006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, Z.; Xin, H.; Yang, Z.; Wang, W.; Dong, J.; Jin, L.; Li, F. miR-135b promotes proliferation and metastasis by targeting APC in triple-negative breast cancer. J. Cell. Physiol. 2019, 234, 10819–10826. [Google Scholar] [CrossRef] [PubMed]
- Pu, T.; Shen, M.; Li, S.; Yang, L.; Gao, H.; Xiao, L.; Zhong, X.; Zheng, H.; Liu, Y.; Ye, F.; et al. Repression of miR-135b-5p promotes metastasis of early-stage breast cancer by regulating downstream target SDCBP. Lab. Investig. 2019, 99, 1296–1308. [Google Scholar] [CrossRef]
- Adams, B.D.; Wali, V.B.; Cheng, C.J.; Inukai, S.; Booth, C.J.; Agarwal, S.; Rimm, D.L.; Győrffy, B.; Santarpia, L.; Pusztai, L.; et al. miR-34a Silences c-SRC to Attenuate Tumor Growth in Triple-Negative Breast Cancer. Cancer Res. 2016, 76, 927–939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, M.; Paranjape, T.; Ullrich, R.; Nallur, S.; Gillespie, E.; Keane, K.; Esquela-Kerscher, A.; Weidhaas, J.B.; Slack, F.J. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene 2009, 28, 2419–2424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, Y.-S.; Tseng, H.-Y.; Chen, Y.-A.; Shen, P.-C.; Al Haq, A.T.; Chen, L.-M.; Tung, Y.-C.; Hsu, H.-L. MCT-1/miR-34a/IL-6/IL-6R signaling axis promotes EMT progression, cancer stemness and M2 macrophage polarization in triple-negative breast cancer. Mol. Cancer 2019, 18, 42. [Google Scholar] [CrossRef] [Green Version]
- Koutsaki, M.; Spandidos, D.; Zaravinos, A. Epithelial—Mesenchymal transition-associated miRNAs in ovarian carcinoma, with highlight on the miR-200 family: Prognostic value and prospective role in ovarian cancer therapeutics. Cancer Lett. 2014, 351, 173–181. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, J.; Li, N.; Liu, Z.; Chen, Z.; Li, Z.; Lai, Y.; Shen, L.; Gao, J. miR-34a increases the sensitivity of colorectal cancer cells to 5-fluorouracil in vitro and in vivo. Am. J. Cancer Res. 2018, 8, 280–290. [Google Scholar]
- Hur, K.; Toiyama, Y.; Takahashi, M.; Balaguer, F.; Nagasaka, T.; Koike, J.; Hemmi, H.; Koi, M.; Boland, C.R.; Goel, A. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut 2013, 62, 1315–1326. [Google Scholar] [CrossRef] [Green Version]
- Senfter, D.; Madlener, S.; Krupitza, G.; Mader, R.M. The microRNA-200 family: Still much to discover. Biomol. Concepts 2016, 7, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Gong, M.; Fang, S.; Hu, C.; Wang, Y.; Zhang, J.; Tang, N.; He, Y. All-trans retinoic acid reverses malignant biological behavior of hepatocarcinoma cells by regulating miR-200 family members. Genes Dis. 2021, 8, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Neves, R.; Scheel, C.; Weinhold, S.; Honisch, E.; Iwaniuk, K.M.; Trompeter, H.-I.; Niederacher, D.; Wernet, P.; Santourlidis, S.; Uhrberg, M. Role of DNA methylation in miR-200c/141 cluster silencing in invasive breast cancer cells. BMC Res. Notes 2010, 3, 219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davalos, V.; Moutinho, C.; Villanueva, A.; Boque, R.; Silva, P.; Carneiro, F.; Esteller, M. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene 2011, 31, 2062–2074. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Cheng, Y.; Wang, Y.; Fan, Y.; Li, C.; Zhang, Y.; Wang, Y.; Dong, Q.; Ma, Y.; Teng, Y.-E.; et al. Tamoxifen reverses epithelial—mesenchymal transition by demethylating miR-200c in triple-negative breast cancer cells. BMC Cancer 2017, 17, 492. [Google Scholar] [CrossRef]
- Damiano, V.; Brisotto, G.; Borgna, S.; di Gennaro, A.; Armellin, M.; Perin, T.; Guardascione, M.; Maestro, R.; Santarosa, M. Epigenetic silencing of miR-200c in breast cancer is associated with aggressiveness and is modulated by ZEB1. Genes Chromosom. Cancer 2017, 56, 147–158. [Google Scholar] [CrossRef]
- Krützfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef]
- Kaboli, P.J.; Rahmat, A.; Ismail, P.; Ling, K.-H. MicroRNA-based therapy and breast cancer: A comprehensive review of novel therapeutic strategies from diagnosis to treatment. Pharmacol. Res. 2015, 97, 104–121. [Google Scholar] [CrossRef]
- Bader, A.G.; Brown, D.L.; Stoudemire, J.; Lammers, P.J. Developing therapeutic microRNAs for cancer. Gene Ther. 2011, 18, 1121–1126. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Xie, X.; Luo, J.; Liu, M.; Xi, S.; Guo, J.; Kong, Y.; Wu, M.; Gao, J.; Xie, Z.; et al. Targeted Expression of miR-34a Using the T-VISA System Suppresses Breast Cancer Cell Growth and Invasion. Mol. Ther. 2012, 20, 2326–2334. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.; Cao, M.; Zhang, J.; Hu, K.; Yin, Z.; Zhou, Z.; Xiao, X.; Yang, Y.; Sheng, W.; Wu, Y.; et al. Hyaluronic acid-chitosan nanoparticles for co-delivery of MiR-34a and doxorubicin in therapy against triple negative breast cancer. Biomaterials 2014, 35, 4333–4344. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Reinhardt, F.; Pan, E.; Soutschek, J.; Bhat, B.; Marcusson, E.G.; Teruya-Feldstein, J.; Bell, G.W.; A Weinberg, R. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat. Biotechnol. 2010, 28, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Devulapally, R.; Sekar, N.M.; Sekar, T.V.; Foygel, K.; Massoud, T.F.; Willmann, J.K.; Paulmurugan, R. Polymer Nanoparticles Mediated Codelivery of AntimiR-10b and AntimiR-21 for Achieving Triple Negative Breast Cancer Therapy. ACS Nano 2015, 9, 2290–2302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, H.; Xiong, G.; Guo, S.; Xu, C.; Xu, R.; Guo, P.; Shu, D. Delivery of Anti-miRNA for Triple-Negative Breast Cancer Therapy Using RNA Nanoparticles Targeting Stem Cell Marker CD133. Mol. Ther. 2019, 27, 1252–1261. [Google Scholar] [CrossRef] [Green Version]
- Holliday, D.L.; Speirs, V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011, 13, 215. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef] [Green Version]
| Analysis Type | Findings | miRNA Signature | Subtype | Reference |
|---|---|---|---|---|
| microRNA (miRNA)quantitative real-time polymerase chain reaction (qPCR) panel on 139 breast cancer (BRCA) patient tissues compared against 26 normal tissues | patients with miR-182-5p and miR-200b-3p expression show better prognosis | miR-30c-5p, miR-30b-5p, miR-182-5p, and miR-200b-3p | ER/PR + | [30] |
| case control study of estrogen receptor/progesterone receptor positive (ER/PR +) patients with tamoxifen treatment | miR-221 expression is high in ER/PR + patients and is not changed by KI67/PR levels | miR-221 | ER/PR + | [31] |
| clustering analysis of RNA seq from patient cohort of 186 patients | miR-99a/let-7c/miR-125b cluster is high in luminal A compared to luminal B | miR-99a/let-7c/ miR-125b | ER/PR + | [32] |
| qPCR of 54 luminal A type patients against 56 controls | study identified miRNAs specifically downregulated in luminal A type patients | miR-29a, miR-625, miR-181a | ER/PR + | [33] |
| miRNA qPCR of luminal A patients compared against controls | diagnostic markers for luminal A | miR-145, miR-195 and miR-486 | ER/PR+ | [34] |
| immunohistochemistry of miR-1290 targets among 256 ER positive breast cancer | miR-1290 is a prognostic marker for luminal breast cancers | miR-1290 | ER/PR+ | [35] |
| meta-analysis of patient datasets | specific miRNA signature between luminal A and luminal B breast cancer subtypes | miR-30b-5p, miR-30c-5p high in luminal A, miR-182-5p, miR-200b-3p, miR-15b-3p, miR-149-5p, miR-193b-3p and miR-342-3p, 5p high in luminal B | ER/PR+ | [22] |
| mimic transfection, luciferase activity and qPCR | miR-125b functions as a competitive endogenous RNA with EPOR and ERBB2 | miR-125b | HER2+ | [36] |
| in silico and qPCR analysis of 300 miRNAs | upregulated miRNA biomarkers for human epidermal growth factor receptor 2 (HER2) subtype | miR-146a-5p | HER2+ | [37] |
| in silico and qPCR analysis of 300 miRNAs | downregulated miRNA biomarkers for HER2 subtype | miR-181d and miR-195-5p | HER2+ | [38] |
| miRNA screen (1626) in combination with targeted treatments lapatinib and trastuzumab | tumor suppressive miRNA signature identified, treatment with mimics sensitize cells to trastuzumab and lapatinib | miR-101-5p, mir-518a-5p, miR-19b-2-5p, miR-1237-3p, miR-29a-3p, miR-29c-3p, miR-106a-5p, and miR-744-3p | HER2+ | [39] |
| protein expression and The Cancer Genome Atlas (TCGA) data analysis | overexpression of miR-4728 in HER2 minimizes the effect of laptinib | miR-4728 | ||
| triple negative breast cancer (TNBC) vs. non-TNBC patient samples | diagnostic markers of TNBCs | hsa-miR-10a, hsa-miR-18a, hsa-miR-135b and hsa-miR-577 | TNBC | [40] |
| miR arrays from stored tissues | distinct in TNBCs compared to ER negative patients | miR-10a, miR-18a, miR-135b and miR-577 | TNBC | [41] |
| miR arrays from stored tissues | basal-like subtype has overexpression of both clusters, derived from copy number | miR-17-92 and miR-106b-25 cluster overexpression | TNBC | [42] |
| upregulated in TNBCs, prognostic signature | miR-455-3p, miR-107, miR-146b-5p, miR-17-5p, miR-324-5p, miR-20a-5p and miR-142-3p, | |||
| downregulated in TNBCs | miR-139-5p, miR-10b-5p, miR-486-5p | |||
| regression analysis of patient data compared against gene expression omnibus (GEO) datasets | upregulated in TNBCs | miR-455-3p, miR-107, miR-146b-5p, miR-17-5p, miR-324-5p, miR-20a-5p and miR-142-3p | TNBC | [43] |
| clustering of miRNAs significantly different between TNBCs and HER2+ subtypes | miR-139-5p, miR-10b-5p, miR-486-5p | |||
| meta-analysis of published research articles | upregulated in TNBCs, prognostic signature | miR-10b, miR-21, miR-29, miR-9, miR-221/222, miR-373 | TNBC | [44] |
| downregulated in TNBCs | miR-145, miR-199a-5p, miR-200 family, miR-203, miR-205 | |||
| miRNA extraction and microarray from formalin-fixed paraffin-embedded (FFPE) tissues | TNBC-specific four miRNA signature which is reduced in other subtypes | miR-17-5p, miR-20a-5p, miR-92a-3p, miR-106b-5p | TNBC | [45] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arun, R.P.; Cahill, H.F.; Marcato, P. Breast Cancer Subtype-Specific miRNAs: Networks, Impacts, and the Potential for Intervention. Biomedicines 2022, 10, 651. https://doi.org/10.3390/biomedicines10030651
Arun RP, Cahill HF, Marcato P. Breast Cancer Subtype-Specific miRNAs: Networks, Impacts, and the Potential for Intervention. Biomedicines. 2022; 10(3):651. https://doi.org/10.3390/biomedicines10030651
Chicago/Turabian StyleArun, Raj Pranap, Hannah F. Cahill, and Paola Marcato. 2022. "Breast Cancer Subtype-Specific miRNAs: Networks, Impacts, and the Potential for Intervention" Biomedicines 10, no. 3: 651. https://doi.org/10.3390/biomedicines10030651
APA StyleArun, R. P., Cahill, H. F., & Marcato, P. (2022). Breast Cancer Subtype-Specific miRNAs: Networks, Impacts, and the Potential for Intervention. Biomedicines, 10(3), 651. https://doi.org/10.3390/biomedicines10030651







