1. Introduction
Prostate cancer (PCa) is one of the most common cancers and is the major cause of cancer-related death among aging men worldwide. Statistically, about one out of every 38 men dies from the disease [
1,
2]. PCa tumors usually originate from the glandular tissue of the prostate with uncontrolled proliferation. Androgen has been known to play a vital role in the progression of PCa [
3,
4,
5,
6]. Therefore, hormone therapy, androgen deprivation or androgen ablation, has remained a standard therapy for patients with metastatic PCa for the past several decades [
4]. However, hormone therapy is not curative, and the cancer frequently relapses to a more malignant form. In order to elucidate the mechanism by which PCa relapse from the androgen deprivation therapy, the androgen-responsive genes were investigated extensively. Calreticulin (CRT) is one androgen-responsive gene in prostatic epithelial cells [
7]. It has been demonstrated that both mRNA and protein expression level of CRT are downregulated by castration and upregulated by androgen replacement in the prostate. In addition, the expression level of CRT in the prostate is much higher than that in seminal vesicles, heart, brain, muscle, kidney, and liver. This evidence demonstrates that CRT may play an important role in the androgen-related response, including the regulation of PCa tumorigenesis and progression.
Calreticulin is a multifunctional chaperon protein and participates in a variety of important biological processes. As a molecular chaperone, CRT functions to ensure proper folding of glycoproteins. CRT also possesses high binding affinity to Ca
2+ and is involved in the modulation of intracellular Ca
2+ homeostasis. In addition, several studies have demonstrated that CRT mediates integrin activation, which regulates cell adhesion and tumor cell metastasis [
8,
9,
10]. Recently, several studies further suggest that CRT is an RNA-binding protein and mediates the regulation of RNA stability. In vascular smooth muscle cells, serine dephosphorylation and tyrosine phosphorylation of CRT promoted its binding with AU-rich elements (AREs) at the 3′UTR of mRNA of angiotensin type 1 (AT1) receptor, which increases mRNA stability of AT1 receptor [
11]. Under high-glucose conditions, CRT destabilizes glucose transporter-1 (GLUT-1) mRNA in vascular endothelial and smooth muscle cells [
12].
Integrins are a family of αβ-heterodimeric transmembrane receptors involved in cell–cell and cell–matrix interactions and regulate numerous cellular functions, including cell adhesion, cell migration, tumor invasion, and metastasis [
13]. Integrin complexes are composed of 18 α and 8 β subunits, and 24 distinct types of integrins were discovered [
14]. They were served as receptors of ECM ligands with different affinities [
14]. The functions of integrin are regulated by a wide variety of molecular events, such as glycosylation. Glycosylation is the most common post-translational modifications for proteins and lipids [
15]. Aberrant glycosylation of integrin usually leads to impaired cell–cell adhesion, activation of oncogenic signaling pathways, and induction of pro-metastatic phenotypes [
16].
The previous study has demonstrated that CRT stabilized the mRNA of fucosyltransferase1 (FUT1), resulting in the activation of β1-integrin by fucosylation, thereby enhancing cell adhesion and cell migration in bladder cancer cells [
10]. However, the effects of CRT on the regulation of β1-integrin and the underlying mechanism in PCa cells remains elusive. In this study, PC-3 cell was used as a PCa model to investigate whether β1-integrin activation or expression was regulated by CRT with its RNA-binding activity.
2. Materials and Methods
2.1. Cell Culture
PC-3 human prostate cancer cell line was purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). PC-3 cells were maintained in RPMI 1640 medium (GE Healthcare Life Sciences, Chicago, IL, USA) supplemented with 10% fetal bovine serum (Gibco, Carlsbad, CA, USA) under a humidified atmosphere of 5% CO2 at 37 °C. For the subcultures, cells were trypsinized with 0.05% EDTA-trypsin (Life Technologies, Carlsbad, CA, USA).
2.2. Transfection of siRNA
The siRNAs of CRT were obtained from Santa Cruz Biotechnology (Dallas, TX, USA). The siRNAs were transfected using Lipofectamine® 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instruction.
2.3. Western Blot Assay
Cells were trypnized and washed once with cold PBS (pH 7.4, 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 and 1.8 mM KH2PO4), and then collected by centrifugation at 2000 rpm for 5 min. Cells were lysed in lysis buffer (pH 8.0, 20 mM Tris, 150 mM NaCl, 1% NP-40, 1 mM Na3VO4, and 10% glycerol) containing 1% protease inhibitor Cocktail (Merck Millipore, Billerica, MA, USA). The cell lysates were incubated on ice for 15 min, followed by centrifugation at 13,000 rpm for 15 min. The supernatants were collected, and the protein concentrations were measured using Bradford assay (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions. The cell lysates were mixed with 1/5 volume of 5× sample buffer and boiled at 100 °C for 10 min. The samples were separated by SDS/PAGE (80 V for 15 min in 4% stacking gel and 120 V for 1.5 h in 10% running gel) and transferred to polyvinylidene fluoride membranes (100 V for 90 min at 4 °C). The membrane was blocked with 5% BSA in TBST (pH 7.4 25 mM Tris, 150 mM NaCl, 2 mM KCl, and 0.1% Tween-20) for 1 h at room temperature, followed by incubation with primary antibody overnight at 4 °C with gentle oscillation. After washing three times with TBST for 10 min, the membranes were incubated with HRP-conjugated secondary antibody for 1 h at room temperature. The membranes were washed three times with TBST for 10 min, and signals were then visualized using ECL reagent (Advansta, Menlo Park, CA, USA) according to the manufacturer’s instructions. The antibodies used were as follows: rabbit polyclonal anti-CRT antibody (PA3-900, Thermo Fisher Scientific, Waltham, MA, USA), rabbit polyclonal anti-GAPDH antibody (GTX100118, Genetex, Irvine, CA, USA), rabbit polyclonal anti-VEGF-A antibody (GTX102643, Genetex, Irvine, CA, USA), rabbit polyclonal anti-ITGB1 antibody (MAB2252, Millipore, Burlington, MA, USA), rabbit polyclonal anti-HuR antibody (3A2, Santa Cruz Biotechnology, Dallas, TX, USA), and rabbit polyclonal anti-TTP antibody (ABE285, Millipore).
2.4. Total RNA Extraction and Reverse Transcription Quantitative PCR (RT-QPCR)
Total RNA was extracted by TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The first strand cDNA was synthesized with 1 μg total RNA using a Toyobo reverse transcription (RT)-polymerase chain reaction (PCR) kit (Toyobo, Osaka, Japan). Quantitative real-time PCR reactions were conducted in a Mini-Opticon Real-Time detection system (Bio-Rad, Hercules, CA, USA) using iQTM SYBR® Green Supermix (Bio-Rad, Hercules, CA, USA). The thermal profiles of real-time PCR was 95 °C for 5 min, followed by 40 cycles of 95 °C for 30 s, and 60 °C for 30 s. The melting curve of each tube was examined to confirm a single peak appearance. The sequences of paired primers for real-time PCR detection are as follows: CRT forward: 5′-CCT CCT CTT TGC GTT TCT TG-3′, CRT Reverse: 5′-CAG ACT CCA AGC CTG AGG AC-3′; β1-integrin 3-UTR forward: 5′-TGC AAC AGC TCT CAC CTA CG-3′, β1-integrin 3-UTR reverse:5′- GAT GGG CAA CTC AAA TGG TGA-3′; GAPDH forward: 5′-GGT GGT CTC CTC TGA CTT CAA C-3′, GAPDH reverse: 5′-TCT CTC TTC CTC TTG TGT TCT TG-3′.
2.5. Determination of mRNA Stability
Cells (1 × 105) were seeded on polystyrene 6-well plate (Corning) and treated with 2.5 µg/mL actinomycin D (Act-D, Sigma, St. Louis, MO, USA) at the indicated time points. Subsequently, total RNAs were extracted and reverse-transcribed to cDNA. The real-time PCR was conducted to quantify the levels of mRNA.
2.6. Dual-Luciferase Reporter Assay
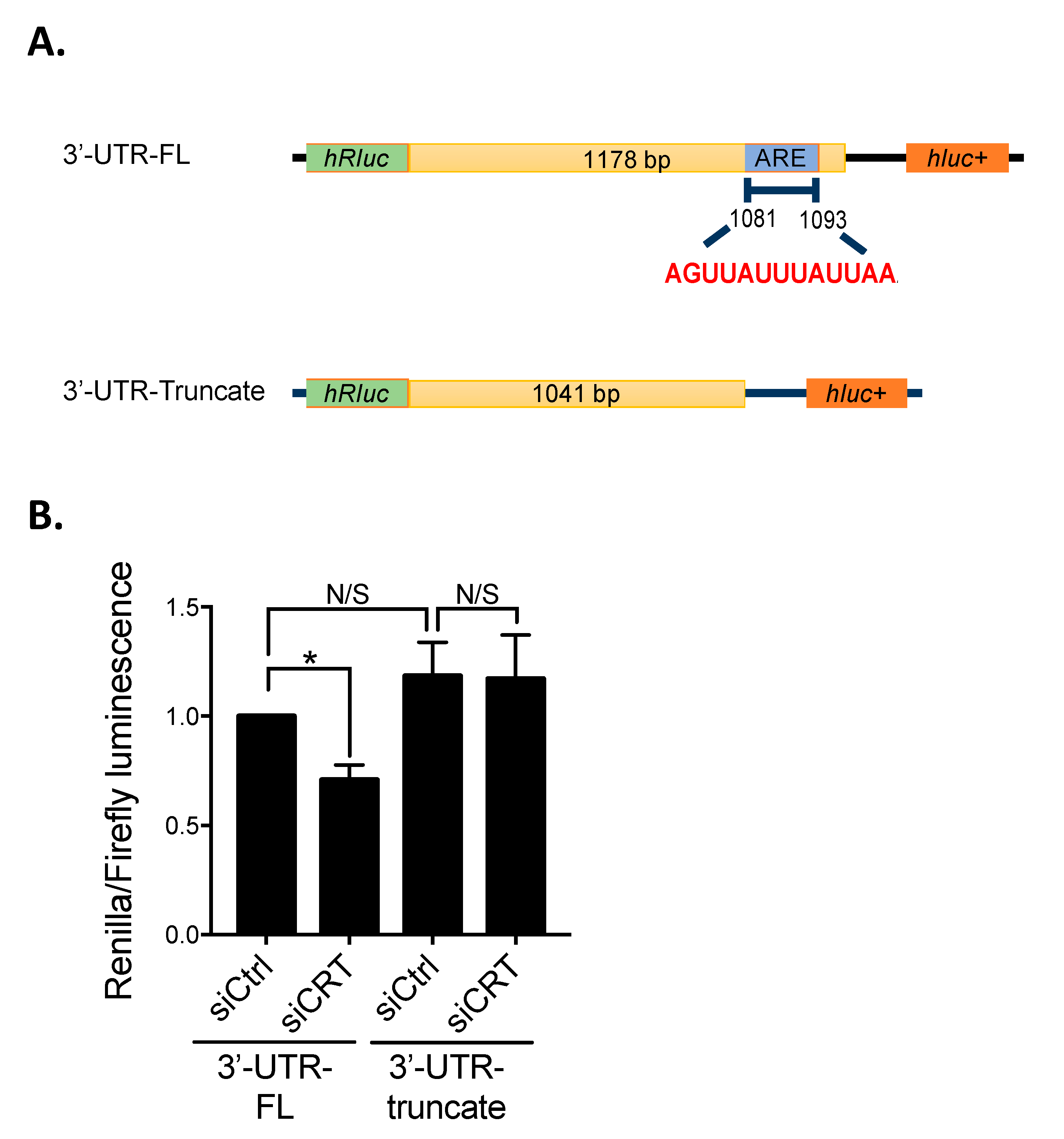
The full-length β1-integrin 3′UTR (β1-integrin-3UTR-FL) and truncated β1 integrin 3′UTR (β1-integrin-3UTR-truncate) were amplified from cDNA of PC-3 cells by PCR with the primers: β1-integrin-3UTR-FL forward: 5′-GGC TCG AGA ATG AGT ACT GCC CGT GCA AAT-3′, β1-integrin-3UTR-FL reverse: 5′-GGG CGG CCG CTC CGA TTT AAG TAT TTT AGG-3′, β1-integrin-3UTR-truncate forward: 5′-AAC TCG AGA ATG AGT ACT GCC CGT GCA AAT CC-3′, β1-integrin-3UTR-truncate reverse: 5′-AAG CGG CCG CAT GGC ACT AAC TCA AAG TAA-3′. The fragments were subsequently inserted into psiCHECK™-2 vector (Promega, Madison, WI, USA) between the XhoI and NotI cutting sites. The psiCHECK™-2 vector contained two reporter genes, Renilla and Firefly luciferases, and is designed for the endpoint lytic assay. Firefly luciferase was used to normalize Renilla luciferase expression.
PC-3 cells (2 × 104) were seeded onto 24-well plates and cultured for 24 h prior to transfection. Cells were transfected with 625 ng plasmid DNA (psiCHECK2-β1-integrin-3UTR-FL or psiCHECK2-β1-integrin-3UTR-truncate) by Lipofectamine® 3000. Four hours after transfection, the medium was replaced with normal culture medium, and cells were cultured for another 48 h. Luciferase assays were performed using the Dual-Glo Luciferase Reporter Assay kit (E2920, Promega, Madison, WI, USA) according to the manufacturer’s procedure. Briefly, cells were lysed with 100 µL Glo lysis buffer (E2661, Promega, Madison, WI, USA). The cell lysates in the plate were oscillated on the orbital shaker for 10 min at room temperature and then transferred into Eppendorf tubes. For each reaction, 25 µL cell lysate was added into a 96-well white plate and mixed with 25 µL Dual-Glo® Reagent followed by incubation for 10 min. The Firefly luciferase activity was first recorded using the SpectraMax M5 (Molecular Devices, San Jose, CA, USA). Then, 25 µL Dual-Glo® Stop & Glo® substrate was added, followed by recording of Renilla luciferase activity.
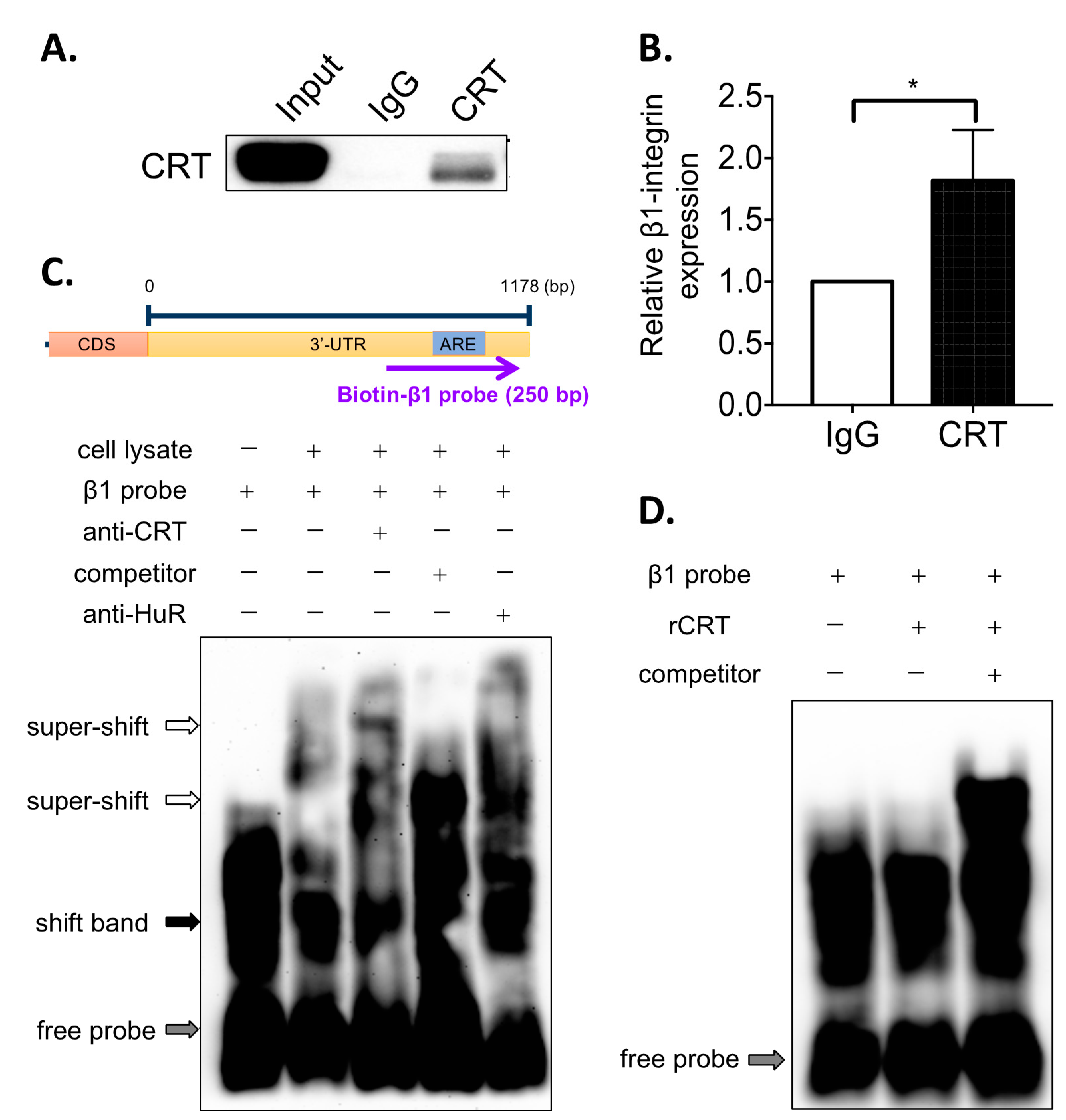
2.7. RNA Immunoprecipitation (RNA-IP)
Cells were lysed by ice-cold RNA-IP lysis buffer (150 mM KCl, pH 7.4 25 mM Tris, 5 mM EDTA and 0.35% Triton X-100) containing 1× protease inhibitor cocktail. Protein concentrations were determined using Bradford assay (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions. Lysates containing 400 µg total protein were pre-cleaned with PureProteome™ Protein A/G Magnetic Beads (Invitrogen, Waltham, MA, USA) at 4 °C for 1 h. The pre-cleaned lysates were incubated with rabbit anti-CRT antibody (PA3-900, Thermo Fisher Scientific, Waltham, MA, USA) at 4 °C overnight. The protein–antibody complex was pulled down by magnetic beads at 4 °C for 4 h. Then, all the beads were collected and washed with ice-cold RNA-IP lysis buffer for three times, followed by three washes with NT2 buffer (50 mM pH 7.4 Tris, 150 mM NaCl and 0.05% NP-40). For Western blot, the beads were mixed with sample buffer and then boiled at 100 °C for 15 min for protein elution. For real-time PCR, beads were subjected to RNA extraction by TRIzol reagent.
2.8. RNA Electrophoretic Mobility Shift Assay (RNA EMSA)
Biotinylated RNA probes for β1-integrin-ARE were prepared using in vitro transcription. Briefly, the linearized PCR fragments containing T7 promotor sequence were amplified from psiCHECK2-β1-integrin-3UTR-FL plasmid by PCR. The sequences of the paired primers were as follows: β1-integrin ARE-probe forward: 5′-TAA TAC GAC TCA CTA TAG GGA TAC TGT GGC TAT GCA ACA G-3′, β1-integrin ARE-probe reverse: 5′-CAT CAG AGT CAA GAC ATC CGA T-3′. The PCR fragments were transcribed to RNA by the incubation of T7 polymerases (Promega, Madison, WI, USA) at 37 °C for 4 h, followed by incubation of Recombinant DNase I (Promega, Madison, WI, USA) at 37 °C for 1 h. The transcribed RNAs were purified by phenol–chloroform extraction and resuspended in DEPC-treated water. Biotin-UTP were then added on the 3′-end of RNA probes using Pierce™ RNA 3′ End Biotinylation Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instruction.
Cell lysates from PC-3 cells were prepared for EMSA. The cells were lysed with ice-cold All Purpose Buffer (APB, 50 mM Tris-HCl pH7.5, 250 mM NaCl, 3 mM EDTA, 3 mM EGTA, 1% Triton X-100, 0.5% NP-40, 10% Glycerol) containing 1× protease inhibitor cocktail (Merck Millipore, Burlington, MA, USA). The cell lysates were incubated on ice for 15 min, followed by centrifugation at 13,000 rpm for 15 min. The supernatants were collected, and the protein concentrations were determined using Bradford assay.
RNA EMSA was performed using LightShift® Chemiluminescent RNA EMSA Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Briefly, 4 μg cell lysates were incubated with 10× binding buffer at room temperature for 10 min. After heating at 95 °C for 5 min, the RNA probes were gradually cooled on ice, followed by adding the biotinylated RNA probes into mixture and incubating at room temperature for 20 min. Then, 5 μL 5× loading dye was added to the mixtures. Additionally, 5% non-denaturing polyacrylamide gel containing 0.5× TBE (pH 8.3, 45 mM Tris-borate, and 1 mM EDTA) was pre-run at 100 V for 1 h, and the reaction mixtures were then separated in the non-denaturing polyacrylamide gel at 100 V for about 1.5 h. The samples were transferred to Nylon membrane at 400 mA at 4 °C for 60 min. The membrane was crosslinked at 120 mJ/cm2 using a UV-light crosslinking instrument equipped with 254 nm bulbs. Signals were visualized using Chemiluminescent Nucleic Acid Detection Module (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions.
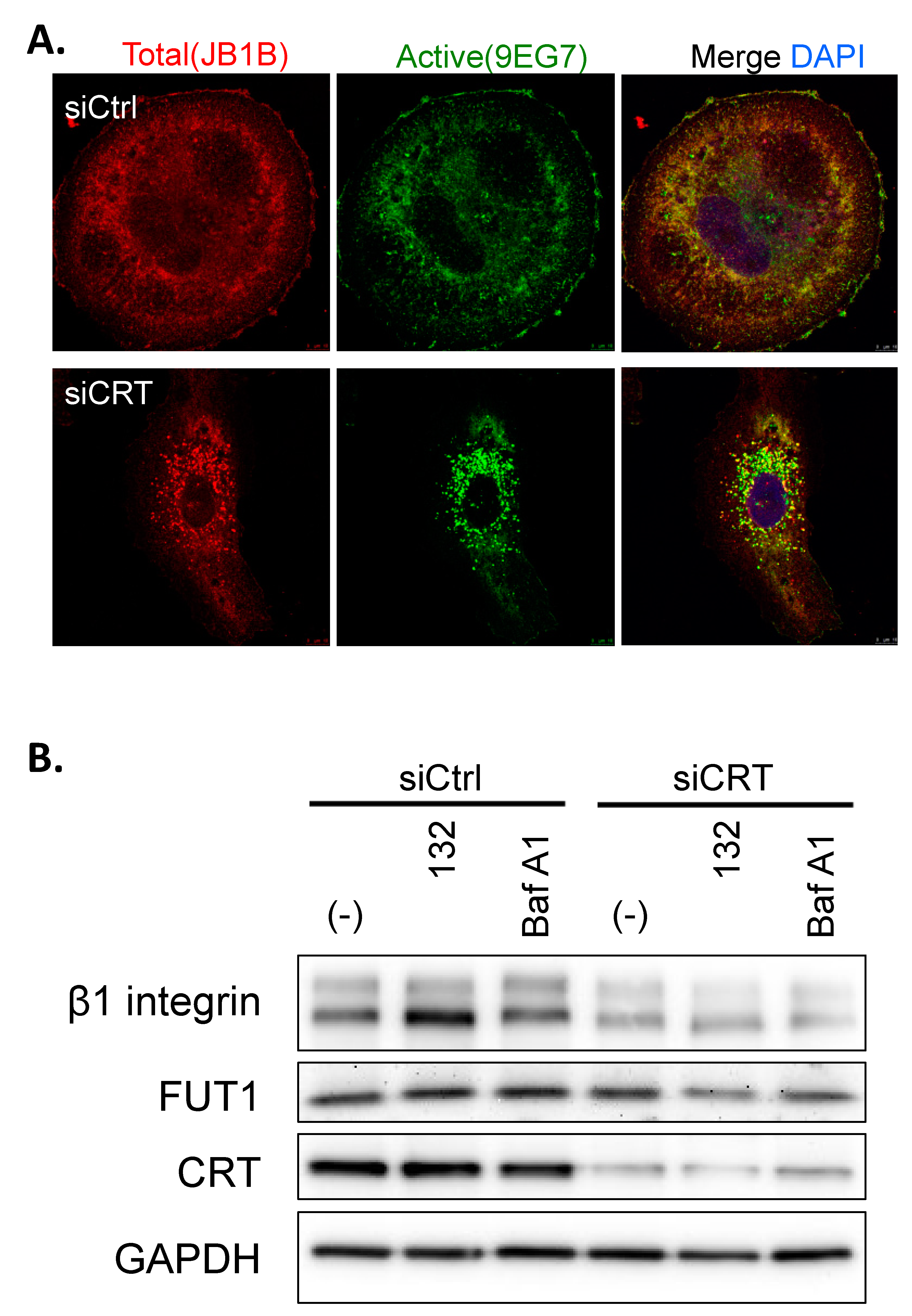
2.9. Immunocytochemistry
Cells were seeded onto poly-l-lysine pre-coated coverslips in 6-well plates. After 24 h incubation, cells were fixed with 4% paraformaldehyde and permeabilized by 0.2% Triton X-100 in PBS at room temperature for 10 min. Detergent-permeabilized cells were blocked with 3% bovine serum albumin in PBST (0.1% Tween 20 in PBS) at room temperature for 30 min and were incubated with mouse anti-β1-integrin (total β1-integrin, Clone JB1B, Santa Cruz) and rat anti-β1-integrin (active β1-integrin, Clone 9EG7, BD Pharmingen™, Franklin Lakes, NJ, USA) antibodies in PBS/1.5% bovine serum albumin at 4 °C overnight. After extensive washes with PBST, cells were incubated with an Alexa Fluor 546 donkey anti-mouse IgG and Alexa Fluor 488 donkey anti-rat IgG in PBS/1.5% bovine serum albumin 37 °C for 1 h. Images were captured by Leica TCS SP5 confocal microscope.
2.10. Proliferation Assay by Cell Counting
siCtrl- and siCRT-transfected PC-3 cells were seeded in the 24-well plates in the density of 5 × 104 cells/well. After three days of incubation, the cells were trypsinized and stained with 0.4% Trypan Blue. The cell number was counted by the hemocytometer.
2.11. Cell Adhesion Assay
The 96-well culture plates were coated with type I collagen (10 μg/mL) and then incubated at 37 °C overnight, followed by a PBS wash. Cells (5 × 104 cells/100 μL) were seeded in the well and incubated at 37 °C for 10–120 min. After removing the culture medium with non-attached cells, the wells were washed with PBS 3 times. Then, 0.1% crystal violet was applied to stain the attached cell for 10 min. After three PBS washes, 10% acetic acid was added for 20 min. The absorbance of cell lysate at 550 nm was measured using a spectrophotometer.
2.12. Wound Healing Assay
PC-3 cells (3 × 105 cells/well) were seeded in 24-well plates to grow in a monolayer for 24 h. Then, a wound was made in each well by scratching with a sterile 20–200 μL pipette tip. The detached cells were removed by PBS wash. Then, 500 μL of fresh medium was added afterward and incubated for 48 h. The scratch closure was monitored and imaged in 24 h intervals using a BioTek Cytation7 imaging system.
2.13. Statistical Analysis
All statistical comparisons between groups were determined using Student’s t-test by GraphPad Prism (San Diego, CA, USA). Each result was obtained from at least three independent experiments, and a value of * p < 0.05, ** p < 0.01, *** p < 0.001 was considered statistically significant.
4. Discussion
Gene expression in eukaryotic cells is highly regulated by multiple processes, such as mRNA splicing and mRNA decay. The degradation of mRNA is mediated by the interaction between RNA-binding proteins (RBPs) and mRNA structures, including 5′-capping structure, 5′- and 3′-untranslated region (UTR), and 3′-polyadenylate (polyA) [
17]. AU-rich elements (AREs) in 3′UTR were considered the most common
cis-acting factor that regulates mRNA stability in mammalian cells [
18]. AREs are composed of multiple copies of the AUUUA motif, which bind directly or indirectly with RNA-binding proteins to regulate mRNA stability [
18]. Previous studies have suggested that CRT is a novel RNA-binding protein involved in the regulation of RNA stability. In vascular cells under high-glucose conditions, CRT has been shown to destabilize glucose transporter-1 (GLUT-1) mRNA [
12]. Dephosphorylated and phosphorylated CRT on serine and tyrosine, respectively, resulted in the binding of CRT with ARE at the 3′UTR of mRNA of angiotensin type 1 (AT1) receptor [
11]. This interaction is required for the regulation of mRNA stability of AT1 receptor. In this study, we demonstrate a novel mechanism whereby CRT regulates β1-integrin expression with its RNA binding property in PCa cells. By binding to the ARE at the 3′UTR of β1-integrin mRNA, CRT increases the stability of β1-integrin mRNA, resulting in a higher expression level of β1-integrin (
Figure 2 and
Figure 3). This mechanism is distinct from our previous findings in bladder cancer that CRT stabilized the mRNA of FUT1 resulting in the activation of β1-integrin by fucosylation [
10]. Therefore, CRT regulates β1-integrin in a manner that is dependent on the cell types. However, multiple PCa cell lines should be tested to confirm that this specialized regulation of β1-integrin mRNA stability by CRT is universal in PCa cells.
Our current data suggested that CRT regulates β1-integrin mRNA stability by indirect binding to its 3′UTR (
Figure 3). That is, CRT requires other RNA-binding proteins to interact with AREs of β1-integrin mRNA. Several ARE-binding proteins, such as HuR and TTP, have been identified as regulators for mRNA stability [
19]. From our EMSA results in
Figure 3C, we have found that anti-HuR antibody shows a similar effect to anti-CRT antibody, which induces the upper super-shift of bands. This finding suggest that the ARE-binding protein HuR may be involved in the CRT RNA-binding complex and help CRT to interact with 3′UTR of β1-integrin mRNA. However, it needs more experiments to further confirm this observation. Other ARE-binding proteins may also contribute to the β1-integrin mRNA binding of CRT. Proteomics approaches would help to identify other binding partners of CRT in the future studies.
Although our results showed that binding of ARE at the 3′UTR is required for CRT to regulate β1-integrin mRNA stability, CRT-dependent RNA regulation might be mediated by other post-transcriptional mechanisms as well. It was reported that ARE not only locates at the 3′UTR of mRNA but also at the intron of pre-mRNA [
20]. Intron plays essential roles in gene regulation, such as alternative splicing, modulation of mRNA and protein translation, and mRNA 3′-end processing [
21]. Therefore, CRT may also interact with ARE at the intron of pre-mRNA, and that regulates RNA stability and protein synthesis. Likewise, GU-rich element (GRE) was identified as another highly enriched sequence in 3′UTR of mRNA, which regulates mRNA degradation [
22]. Recently, CRT was reported to interact with a GRE-binding protein, CUG-binding protein 1 (CUGBP1), and suppressed the translation of C/EBPα and C/EBPβ mRNA to protein [
23]. Altogether, these studies suggest that CRT may regulate mRNA stability through participating in not only ARE-binding protein complex but also GRE-binding protein complex. Therefore, whether the regulation of β1-integrin mRNA stability by CRT in PCa cells is also mediated by the GRE-dependent mechanism is worth further investigation.
In conclusion, this study demonstrated that CRT stabilizes mRNA stability through indirectly binding to ARE in the 3′UTR of mRNA. This mechanism expands our insight into the function of CRT in the regulation of mRNA stability.