Different HSP90 Inhibitors Exert Divergent Effect on Myxoid Liposarcoma In Vitro and In Vivo
Abstract
1. Introduction
2. Materials and Methods
2.1. Tumor Cell Lines and Cell Culture
2.2. Chemical Compounds
2.3. Cell Viability Assays
2.4. In Vivo Experiments
2.5. Tumor Sectioning and Morphologic Analysis
2.6. Western Blot
2.7. Flow Cytometry
2.8. Phospho-RTK Array
2.9. Reverse Transcription, PCR and Fragment Analyzer
3. Results
3.1. HSP90 Inhibition Affects Cell Viability of MLS Cell Lines In Vitro
3.2. HSP90 Inhibition Leads to a G2-M Arrest and Apoptosis
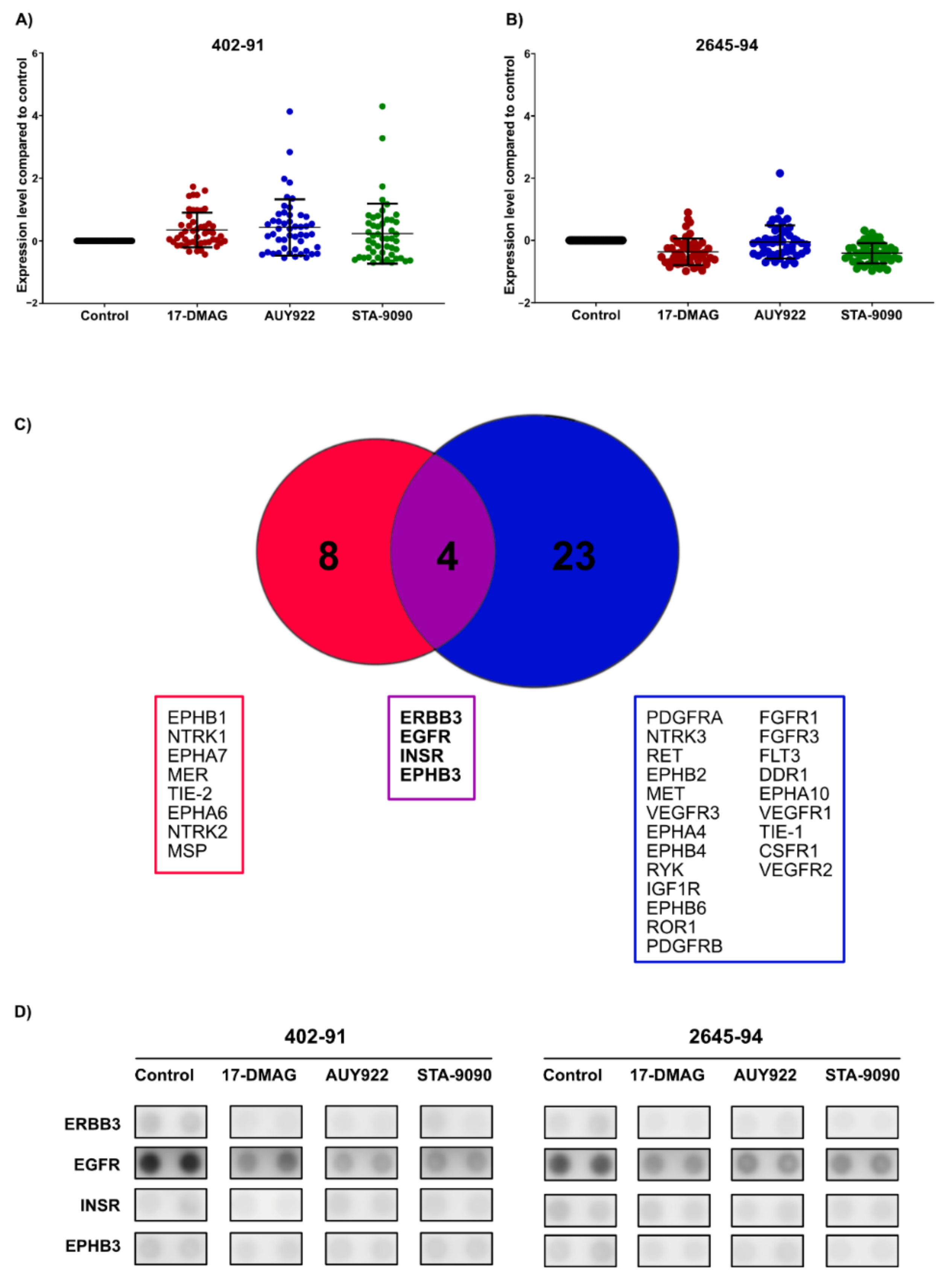
3.3. HSP90 Inhibition Affects Phosphorylation Levels of Receptor Tyrosine Kinases
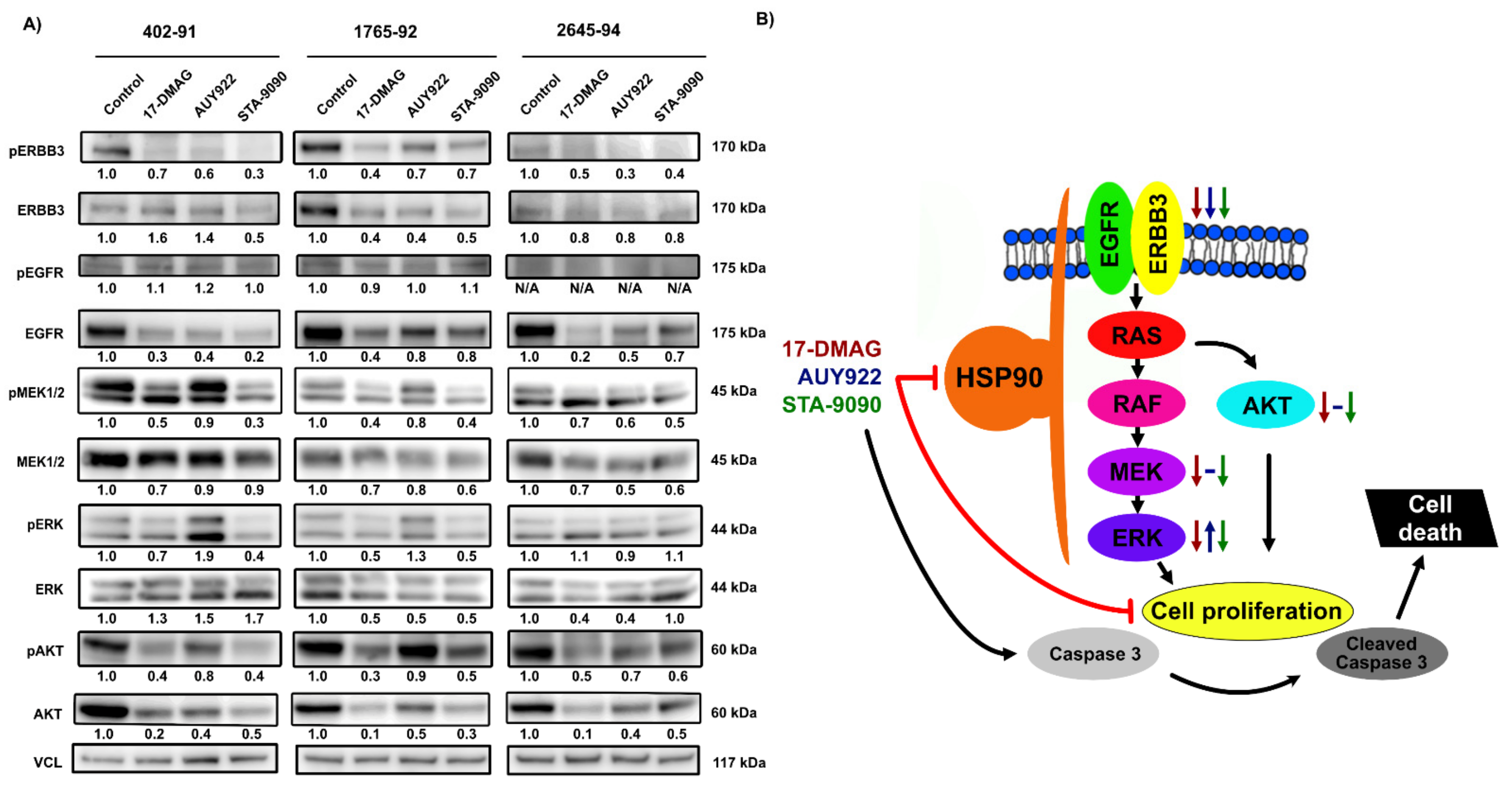
3.4. Effects of HSP90 Inhibition on MAPK and PI3K/AKT Signaling Pathways
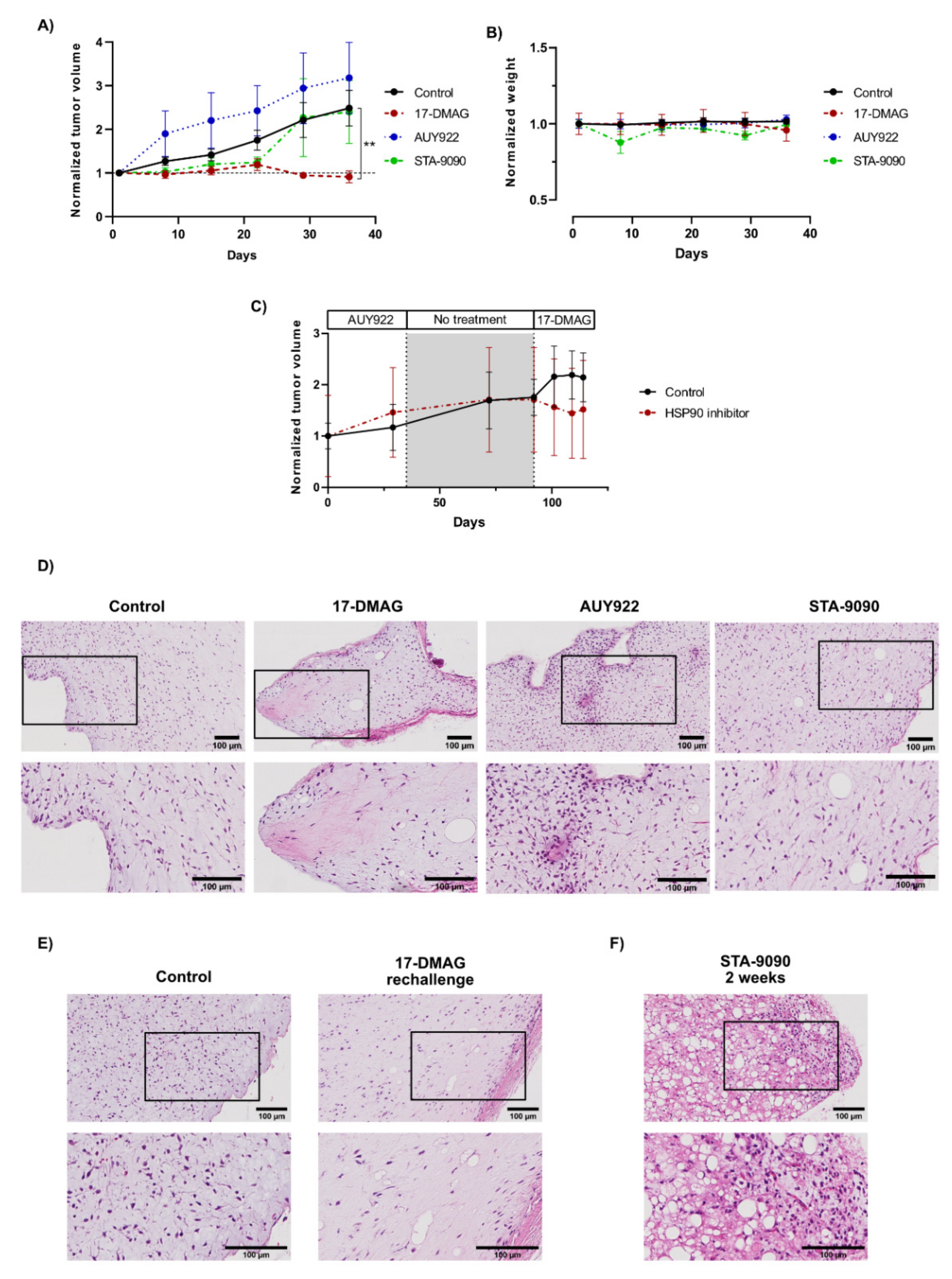
3.5. Effects of HSP90 Inhibitors on Tumor Growth In Vivo
3.6. The Effect and Tolerability of the Different HSP90 Inhibitors Varied Considerably In Vivo
3.7. Morphological Effects of HSP90 Inhibitors In Vivo
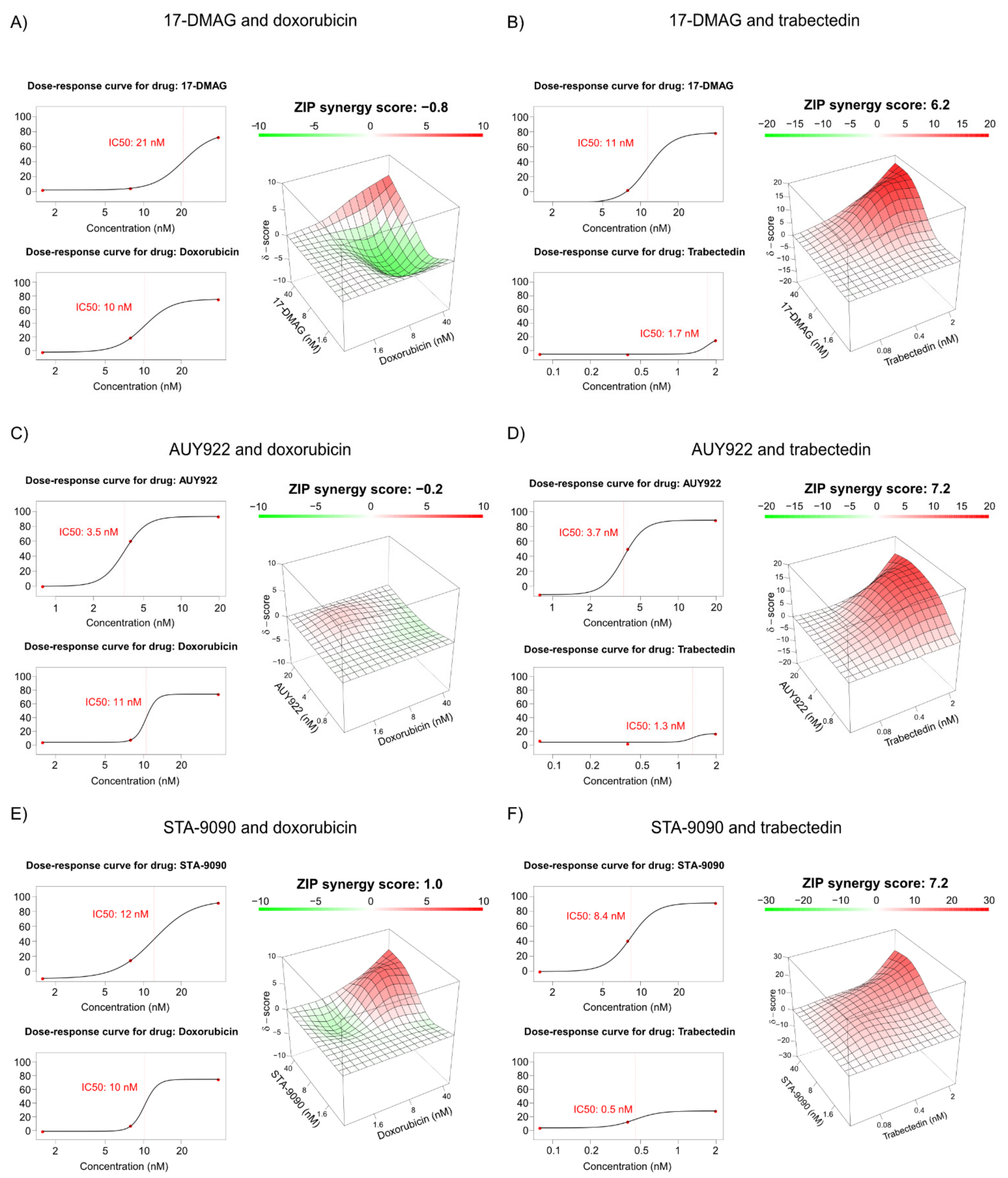
3.8. Combination Treatment of HSP90 Inhibitors with Doxorubicin or Trabectidin
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abaricia, S.; Hirbe, A.C. Diagnosis and treatment of myxoid liposarcomas: Histology matters. Curr. Treat. Options Oncol. 2018, 19, 64. [Google Scholar] [CrossRef] [PubMed]
- Åman, P.; Ron, D.; Mandahl, N.; Fioretos, T.; Heim, S.; Arheden, K.; Willén, H.; Rydholm, A.; Mitelman, F. Rearrangement of the transcription factor geneCHOP in myxoid liposarcomas with t(12;16)(q13;p11). Genes Chromosom. Cancer 1992, 5, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Panagopoulos, I.; Höglund, M.; Mertens, F.; Mandahl, N.; Mitelman, F.; Aman, P. Fusion of the EWS and CHOP genes in myxoid liposarcoma. Oncogene 1996, 12, 489–494. [Google Scholar] [CrossRef]
- Aman, P. Fusion genes in solid tumors. Semin. Cancer Biol. 1999, 9, 303–318. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.; Tornin, J.; Suarez, C.; Astudillo, A.; Rubio, R.; Yauk, C.; Williams, A.; Rosu-Myles, M.; Funes, J.M.; Boshoff, C.; et al. Expression of FUS-CHOP fusion protein in immortalized/transformed human mesenchymal stem cells drives mixoid liposarcoma formation. Stem Cells 2013, 31, 2061–2072. [Google Scholar] [CrossRef] [PubMed]
- Engström, K.; Willén, H.; Kåbjörn-Gustafsson, C.; Andersson, C.; Olsson, M.; Göransson, M.; Järnum, S.; Olofsson, A.; Warnhammar, E.; Åman, P. The myxoid/round cell liposarcoma fusion oncogene FUS-DDIT3 and the normal DDIT3 induce a liposarcoma phenotype in transfected human fibrosarcoma cells. Am. J. Pathol. 2006, 168, 1642–1653. [Google Scholar] [CrossRef] [PubMed]
- Lindén, M.; Thomsen, C.; Grundevik, P.; Jonasson, E.; Andersson, D.; Runnberg, R.; Dolatabadi, S.; Vannas, C.; Santamarίa, M.L.; Fagman, H.; et al. FET family fusion oncoproteins target the SWI / SNF chromatin remodeling complex. EMBO Rep. 2019, 20, e45766. [Google Scholar] [CrossRef]
- Dalal, K.M.; Kattan, M.; Antonescu, C.R.; Brennan, M.; Singer, S. Subtype specific prognostic nomogram for patients with primary liposarcoma of the retroperitoneum, extremity, or trunk. Ann. Surg. 2006, 244, 381–391. [Google Scholar] [CrossRef]
- Lansu, J.; Van Houdt, W.J.; Schaapveld, M.; Walraven, I.; Van De Sande, M.A.J.; Ho, V.K.Y.; Haas, R.L. Time trends and prognostic factors for overall survival in myxoid liposarcomas: A population-based study. Sarcoma 2020, 2020, 2437850. [Google Scholar] [CrossRef]
- Fletcher, C.D.M. Pathology and Genetics of Tumours of Soft Tissue and Bone; World Health Organization Classification of Tumours; IARC: Lyon, France, 2002. [Google Scholar]
- Patel, S.R.; Vadhan-Raj, S.; Burgess, M.A.; Plager, C.; Papadopolous, N.; Jenkins, J.; Benjamin, R.S. Results of two consecutive trials of dose-intensive chemotherapy with doxorubicin and ifosfamide in patients with sarcomas. Am. J. Clin. Oncol. 1998, 21, 317–321. [Google Scholar] [CrossRef]
- Reichardt, P.; Tilgner, J.; Hohenberger, P.; Dörken, B. Dose-intensive chemotherapy with ifosfamide, epirubicin, and filgrastim for adult patients with metastatic or locally advanced soft tissue sarcoma: A phase II study. J. Clin. Oncol. 1998, 16, 1438–1443. [Google Scholar] [CrossRef]
- Leyvraz, S.; Zweifel, M.; Jundt, G.; Lissoni, A.; Cerny, T.; Sessa, C.; Fey, M.; Dietrich, D.; Honegger, H.P. Long-term results of a multicenter SAKK trial on high-dose ifosfamide and doxorubicin in advanced or metastatic gynecologic sarcomas. Ann. Oncol. 2006, 17, 646–651. [Google Scholar] [CrossRef]
- Judson, I.; Verweij, J.; Gelderblom, H.; Hartmann, J.T.; Schöffski, P.; Blay, J.-Y.; Kerst, J.M.; Sufliarsky, J.; Whelan, J.; Hohenberger, P.; et al. Doxorubicin alone versus intensified doxorubicin plus ifosfamide for first-line treatment of advanced or metastatic soft-tissue sarcoma: A randomised controlled phase 3 trial. Lancet Oncol. 2014, 15, 415–423. [Google Scholar] [CrossRef]
- Garcia-Carbonero, R.; Supko, J.; Maki, R.; Manola, J.; Ryan, D.; Harmon, D.; Puchalski, T.; Goss, G.; Seiden, M.; Waxman, A.; et al. Ecteinascidin-743 (ET-743) for chemotherapy-naive patients with advanced soft tissue sarcomas: Multicenter phase II and pharmacokinetic study. J. Clin. Oncol. 2005, 23, 5484–5492. [Google Scholar] [CrossRef]
- Garcia-Carbonero, R.; Supko, J.; Manola, J.; Seiden, M.; Harmon, D.; Ryan, D.; Quigley, M.; Merriam, P.; Canniff, J.; Goss, G.; et al. Phase II and pharmacokinetic study of ecteinascidin 743 in patients with progressive sarcomas of soft tissues refractory to chemotherapy. J. Clin. Oncol. 2004, 22, 1480–1490. [Google Scholar] [CrossRef]
- Demetri, G.D.; Schöffski, P.; Grignani, G.; Blay, J.-Y.; Maki, R.G.; Van Tine, B.A.; Alcindor, T.; Jones, R.L.; D’Adamo, D.R.; Guo, M.; et al. Activity of eribulin in patients with advanced liposarcoma demonstrated in a subgroup analysis from a randomized phase III study of eribulin versus dacarbazine. J. Clin. Oncol. 2017, 35, 3433–3439. [Google Scholar] [CrossRef]
- Demetri, G.D.; von Mehren, M.; Jones, R.L.; Hensley, M.J.; Schuetze, S.M.; Staddon, A.; Milhem, M.; Elias, A.; Ganjoo, K.; Tawbi, H.; et al. Efficacy and safety of trabectedin or dacarbazine for metastatic liposarcoma or leiomyosarcoma after failure of conventional chemotherapy: Results of a phase III randomized multicenter clinical trial. J. Clin. Oncol. 2016, 34, 786–793. [Google Scholar] [CrossRef]
- Barone, A.; Chi, D.-C.; Theoret, M.R.; Chen, H.; He, K.; Kufrin, D.; Helms, W.S.; Subramaniam, S.; Zhao, H.; Patel, A.; et al. FDA approval summary: Trabectedin for unresectable or metastatic liposarcoma or leiomyosarcoma following an anthracycline-containing regimen. Clin. Cancer Res. 2017, 23, 7448–7453. [Google Scholar] [CrossRef][Green Version]
- Gronchi, A.; Hindi, N.; Cruz, J.; Blay, J.-Y.; Lopez-Pousa, A.; Italiano, A.; Alvarez, R.; Gutierrez, A.; Rincón, I.; Sangalli, C.; et al. Trabectedin and radiotherapy in soft tissue sarcoma (TRASTS): Results of a phase I study in myxoid liposarcoma from Spanish (GEIS), Italian (ISG), French (FSG) sarcoma groups. eClinicalMedicine 2019, 9, 35–43. [Google Scholar] [CrossRef]
- Negri, T.; Virdis, E.; Brich, S.; Bozzi, F.; Tamborini, E.; Tarantino, E.; Jocollè, G.; Cassinelli, G.; Grosso, F.; Sanfilippo, R.; et al. Functional mapping of receptor tyrosine kinases in myxoid liposarcoma. Clin. Cancer Res. 2010, 16, 3581–3593. [Google Scholar] [CrossRef]
- Safavi, S.; Järnum, S.; Vannas, C.; Udhane, S.S.; Jonasson, E.; Tomić, T.T.; Grundevik, P.; Fagman, H.; Hansson, M.; Kalender, Z.; et al. HSP90 inhibition blocks ERBB3 and RET phosphorylation in myxoid/round cell liposarcoma and causes massive cell death in vitro and in vivo. Oncotarget 2016, 7, 433–445. [Google Scholar] [CrossRef]
- Trautmann, M.; Menzel, J.; Bertling, C.; Cyra, M.A.; Isfort, I.; Steinestel, K.; Elges, S.; Grünewald, I.; Altvater, B.; Rossig, C.; et al. FUS–DDIT3 fusion protein-driven IGF-IR signaling is a therapeutic target in myxoid liposarcoma. Clin. Cancer Res. 2017, 23, 6227–6238. [Google Scholar] [CrossRef]
- De Graaff, M.A.; Malu, S.; Guardiola, I.; Kruisselbrink, A.B.; de Jong, Y.; Corver, W.E.; Gelderblom, H.; Hwu, P.; Nielsen, T.O.; Lazar, A.; et al. High-throughput screening of myxoid liposarcoma cell lines: Survivin is essential for tumor growth. Transl. Oncol. 2017, 10, 546–554. [Google Scholar] [CrossRef]
- Steinmann, S.; Gali-Muhtasib, H.; Huebner, K.; Al-Halabi, R.; Merhi, R.A.; Aman, P.; Agaimy, A.; Haller, F.; Schneider-Stock, R. Hsp90 inhibition by AUY922 as an effective treatment strategy against myxoid liposarcoma. Cancer Lett. 2015, 367, 147–156. [Google Scholar] [CrossRef]
- Schopf, F.H.; Biebl, M.M.; Buchner, J. The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 2017, 18, 345–360. [Google Scholar] [CrossRef]
- Hoter, A.; El-Sabban, M.E.; Naim, H.Y. The HSP90 family: Structure, regulation, function, and implications in health and disease. Int. J. Mol. Sci. 2018, 19, 2560. [Google Scholar] [CrossRef]
- Saini, J. Clinical, prognostic and therapeutic significance of heat shock proteins in cancer. Curr. Drug Targets 2018, 19, 1478–1490. [Google Scholar] [CrossRef]
- Li, L.; Chen, N.-N.; You, Q.-D.; Xu, X.-L. An updated patent review of anticancer Hsp90 inhibitors (2013-present). Expert Opin. Ther. Pat. 2021, 31, 67–80. [Google Scholar] [CrossRef]
- Chatterjee, S.; Burns, T.F. Targeting heat shock proteins in cancer: A promising therapeutic approach. Int. J. Mol. Sci. 2017, 18, 1978. [Google Scholar] [CrossRef]
- Thelin-Järnum, S.; Göransson, M.; Burguete, A.S.; Olofsson, A.; Åman, P. The myxoid liposarcoma specific TLS-CHOP fusion protein localizes to nuclear structures distinct from PML nuclear bodies. Int. J. Cancer 2002, 97, 446–450. [Google Scholar] [CrossRef]
- Ianevski, A.; He, L.; Aittokallio, T.; Tang, J. SynergyFinder: A web application for analyzing drug combination dose–response matrix data. Bioinformatics 2017, 33, 2413–2415. [Google Scholar] [CrossRef] [PubMed]
- Calderwood, S.K. Heat shock proteins and cancer: Intracellular chaperones or extracellular signalling ligands? Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20160524. [Google Scholar] [CrossRef] [PubMed]
- Yamaki, H.; Nakajima, M.; Shimotohno, K.W.; Tanaka, N. Molecular basis for the actions of Hsp90 inhibitors and cancer therapy. J. Antibiot. 2011, 64, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Powers, M.P.; Wang, W.-L.; Hernandez, V.S.; Patel, K.S.; Lev, D.C.; Lazar, A.; López-Terrada, D.H. Detection of myxoid liposarcoma-associated FUS–DDIT3 rearrangement variants including a newly identified breakpoint using an optimized RT-PCR assay. Mod. Pathol. 2010, 23, 1307–1315. [Google Scholar] [CrossRef]
- Åman, P.; Dolatabadi, S.; Svec, D.; Jonasson, E.; Safavi, S.; Andersson, D.; Grundevik, P.; Thomsen, C.; Ståhlberg, A. Regulatory mechanisms, expression levels and proliferation effects of theFUS-DDIT3fusion oncogene in liposarcoma. J. Pathol. 2016, 238, 689–699. [Google Scholar] [CrossRef]
- Yarden, Y. The EGFR family and its ligands in human cancer: Signalling mechanisms and therapeutic opportunities. Eur. J. Cancer 2001, 37, 3–8. [Google Scholar] [CrossRef]
- Taniguchi, C.M.; Emanuelli, B.; Kahn, C.R. Critical nodes in signalling pathways: Insights into insulin action. Nat. Rev. Mol. Cell Biol. 2006, 7, 85–96. [Google Scholar] [CrossRef]
- Miyajima, N.; Tsutsumi, S.; Sourbier, C.; Beebe, K.; Mollapour, M.; Rivas, C.; Yoshida, S.; Trepel, J.B.; Huang, Y.; Tatokoro, M.; et al. The HSP90 inhibitor ganetespib synergizes with the MET kinase inhibitor crizotinib in both crizotinib-sensitive and -resistant MET-driven tumor models. Cancer Res. 2013, 73, 7022–7033. [Google Scholar] [CrossRef]
- Lin, S.-F.; Lin, J.-D.; Hsueh, C.; Chou, T.-C.; Yeh, C.-N.; Chen, M.-H.; Wong, R.J. Efficacy of an HSP90 inhibitor, ganetespib, in preclinical thyroid cancer models. Oncotarget 2017, 8, 41294–41304. [Google Scholar] [CrossRef]
- Bonvalot, S.; Wunder, J.; Gronchi, A.; Broto, J.M.; Turcotte, R.; Rastrelli, M.; Papai, Z.; Radaelli, S.; Lindner, L.H.; Shumelinsky, F.; et al. Complete pathological response to neoadjuvant treatment is associated with better survival outcomes in patients with soft tissue sarcoma: Results of a retrospective multicenter study. Eur. J. Surg. Oncol. (EJSO) 2021, 47, 2166–2172. [Google Scholar] [CrossRef]
- Grosso, F.; Jones, R.; Demetri, G.D.; Judson, I.R.; Blay, J.-Y.; Le Cesne, A.; Sanfilippo, R.; Casieri, P.; Collini, P.; Dileo, P.; et al. Efficacy of trabectedin (ecteinascidin-743) in advanced pretreated myxoid liposarcomas: A retrospective study. Lancet Oncol. 2007, 8, 595–602. [Google Scholar] [CrossRef]
- Wang, W.-L.; Katz, D.; Araujo, D.M.; Ravi, V.; A Ludwig, J.; Trent, J.C.; Patel, S.R.; Lin, P.P.; Guadagnolo, A.; Lòpez-Terrada, D.; et al. Extensive adipocytic maturation can be seen in myxoid liposarcomas treated with neoadjuvant doxorubicin and ifosfamide and pre-operative radiation therapy. Clin. Sarcoma Res. 2012, 2, 25. [Google Scholar] [CrossRef]
- Wang, C.Y.; Guo, S.T.; Wang, J.Y.; Yan, X.G.; Farrelly, M.; Zhang, Y.; Liu, F.; Yari, H.; La, T.; Lei, F.X.; et al. Reactivation of ERK and Akt confers resistance of mutant BRAF colon cancer cells to the HSP90 inhibitor AUY922. Oncotarget 2016, 7, 49597–49610. [Google Scholar] [CrossRef]
- Chatterjee, S.; Huang, E.H.-B.; Christie, I.; Kurland, B.; Burns, T.F. Acquired resistance to the Hsp90 inhibitor, ganetespib, in KRAS-mutant NSCLC is mediated via reactivation of the ERK–p90RSK–mTOR signaling network. Mol. Cancer Ther. 2017, 16, 793–804. [Google Scholar] [CrossRef]
- Fogh, J.; Fogh, J.M.; Orfeo, T. One Hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice. JNCI J. Natl. Cancer Inst. 1977, 59, 221–226. [Google Scholar] [CrossRef]
- Grohar, P.J.; Griffin, L.B.; Yeung, C.; Chen, Q.-R.; Pommier, Y.; Khanna, C.; Khan, J.; Helman, L.J. Ecteinascidin 743 interferes with the activity of EWS-FLI1 in ewing sarcoma cells. Neoplasia 2011, 13, 145–153. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vannas, C.; Andersson, L.; Dolatabadi, S.; Ranji, P.; Lindén, M.; Jonasson, E.; Ståhlberg, A.; Fagman, H.; Åman, P. Different HSP90 Inhibitors Exert Divergent Effect on Myxoid Liposarcoma In Vitro and In Vivo. Biomedicines 2022, 10, 624. https://doi.org/10.3390/biomedicines10030624
Vannas C, Andersson L, Dolatabadi S, Ranji P, Lindén M, Jonasson E, Ståhlberg A, Fagman H, Åman P. Different HSP90 Inhibitors Exert Divergent Effect on Myxoid Liposarcoma In Vitro and In Vivo. Biomedicines. 2022; 10(3):624. https://doi.org/10.3390/biomedicines10030624
Chicago/Turabian StyleVannas, Christoffer, Lisa Andersson, Soheila Dolatabadi, Parmida Ranji, Malin Lindén, Emma Jonasson, Anders Ståhlberg, Henrik Fagman, and Pierre Åman. 2022. "Different HSP90 Inhibitors Exert Divergent Effect on Myxoid Liposarcoma In Vitro and In Vivo" Biomedicines 10, no. 3: 624. https://doi.org/10.3390/biomedicines10030624
APA StyleVannas, C., Andersson, L., Dolatabadi, S., Ranji, P., Lindén, M., Jonasson, E., Ståhlberg, A., Fagman, H., & Åman, P. (2022). Different HSP90 Inhibitors Exert Divergent Effect on Myxoid Liposarcoma In Vitro and In Vivo. Biomedicines, 10(3), 624. https://doi.org/10.3390/biomedicines10030624






