Human Serum Proteins and Susceptibility of Acinetobacter baumannii to Cefiderocol: Role of Iron Transport
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. RNA Extraction and Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
2.3. Antimicrobial Susceptibility Testing
3. Results
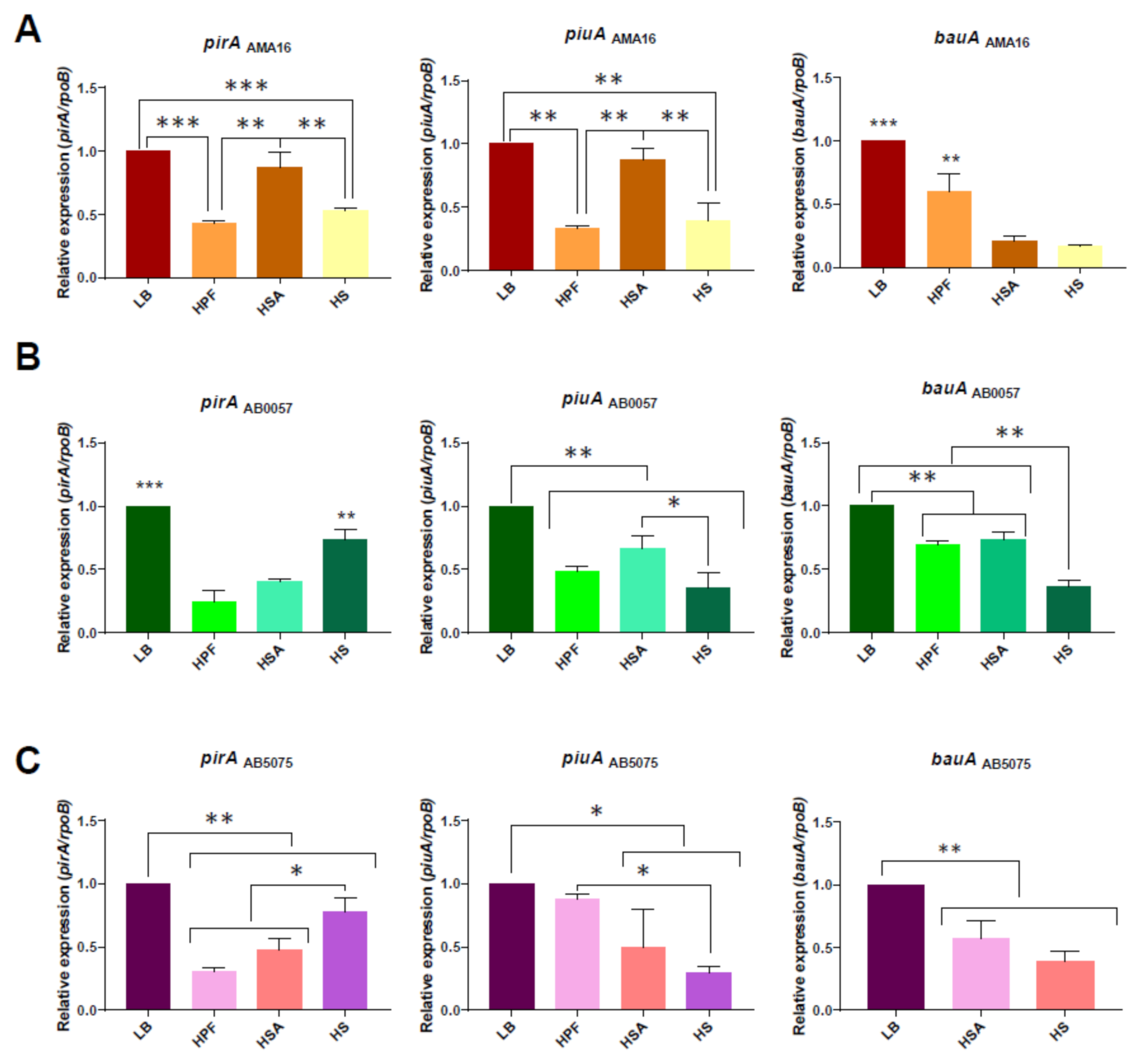
3.1. Changes in the Expression Levels of Genes Associated with Iron Uptake Systems in the Presence of Human Fluids
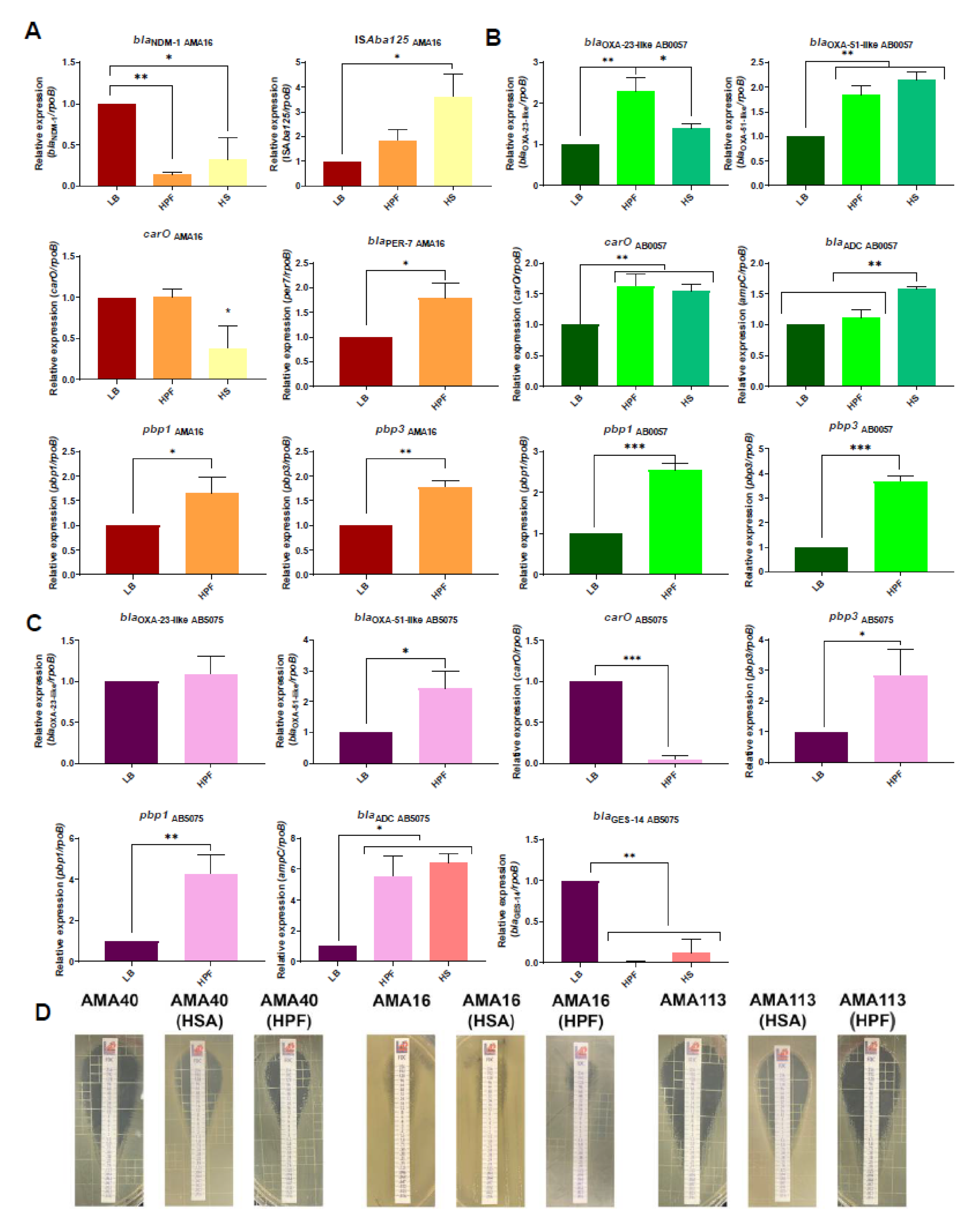
3.2. Modulation of the Expression of β-Lactam Resistance Genes by Human Fluids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eichenberger, E.M.; Thaden, J.T. Epidemiology and Mechanisms of Resistance of Extensively Drug Resistant Gram-Negative Bacteria. Antibiotics 2019, 8, 37. [Google Scholar] [CrossRef] [Green Version]
- Holmes, C.L.; Anderson, M.T.; Mobley, H.L.T.; Bachman, M.A. Pathogenesis of Gram-negative bacteremia. Clin. Microbiol. Rev. 2021, 34, e00234-e20. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ortega, L.; Arch, O.; Perez-Canosa, C.; Lupion, C.; Gonzalez, C.; Rodriguez-Bano, J.; Spanish Study Group of Nosocomial Infections. Control measures for Acinetobacter baumannii: A survey of Spanish hospitals. Enferm. Infecc. Microbiol. Clín. 2011, 29, 36–38. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, M.S.; Bonomo, R.A.; Tolmasky, M.E. Carbapenemases: Transforming Acinetobacter baumannii into a Yet More Dangerous Menace. Biomolecules 2020, 10, 720. [Google Scholar] [CrossRef] [PubMed]
- CDC. Antibiotic Resistance Threats in the United States; US Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019. [Google Scholar]
- Karaiskos, I.; Lagou, S.; Pontikis, K.; Rapti, V.; Poulakou, G. The “Old” and the “New” Antibiotics for MDR Gram-Negative Pathogens: For Whom, When, and How. Front. Public Health 2019, 7, 151. [Google Scholar] [CrossRef] [Green Version]
- Bassetti, M.; Echols, R.; Matsunaga, Y.; Ariyasu, M.; Doi, Y.; Ferrer, R.; Lodise, T.P.; Naas, T.; Niki, Y.; Paterson, D.L.; et al. Efficacy and safety of cefiderocol or best available therapy for the treatment of serious infections caused by carbapenem-resistant Gram-negative bacteria (CREDIBLE-CR): A randomised, open-label, multicentre, pathogen-focused, descriptive, phase 3 trial. Lancet Infect. Dis. 2021, 21, 226–240. [Google Scholar] [CrossRef]
- Bassetti, M.; Peghin, M.; Vena, A.; Giacobbe, D.R. Treatment of Infections Due to MDR Gram-Negative Bacteria. Front. Med. 2019, 6, 74. [Google Scholar] [CrossRef]
- Isler, B.; Doi, Y.; Bonomo, R.A.; Paterson, D.L. New treatment options against carbapenem-resistant Acinetobacter baumannii infections. Antimicrob. Agents Chemother. 2019, 63, e01110-e18. [Google Scholar] [CrossRef] [Green Version]
- Bonomo, R.A.; Burd, E.M.; Conly, J.; Limbago, B.M.; Poirel, L.; Segre, J.A.; Westblade, L.F. Carbapenemase-Producing Organisms: A Global Scourge. Clin. Infect. Dis. 2018, 66, 1290–1297. [Google Scholar] [CrossRef]
- Spellberg, B.; Bonomo, R.A. The deadly impact of extreme drug resistance in Acinetobacter baumannii. Crit. Care Med. 2014, 42, 1289–1291. [Google Scholar] [CrossRef] [Green Version]
- Paterson, D.L.; Isler, B.; Stewart, A. New treatment options for multiresistant gram negatives. Curr. Opin. Infect. Dis. 2020, 33, 214–223. [Google Scholar] [CrossRef] [PubMed]
- McLeod, S.M.; Moussa, S.H.; Hackel, M.A.; Miller, A.A. In Vitro Activity of Sulbactam-Durlobactam against Acinetobacter baumannii-calcoaceticus Complex Isolates Collected Globally in 2016 and 2017. Antimicrob. Agents Chemother. 2020, 64, e02534-e19. [Google Scholar] [CrossRef] [PubMed]
- Lomovskaya, O.; Nelson, K.; Rubio-Aparicio, D.; Tsivkovski, R.; Sun, D.; Dudley, M.N. The Impact of Intrinsic Resistance Mechanisms on Potency of QPX7728, a New Ultra-Broad-Spectrum Beta-lactamase Inhibitor of Serine and Metallo Beta-Lactamases in Enterobacteriaceae, Pseudomonas aeruginosa, and Acinetobacter baumannii. Antimicrob. Agents Chemother. 2020, 65, e00552-e20. [Google Scholar]
- Karaiskos, I.; Galani, I.; Souli, M.; Giamarellou, H. Novel beta-lactam-beta-lactamase inhibitor combinations: Expectations for the treatment of carbapenem-resistant Gram-negative pathogens. Expert Opin. Drug Metab. Toxicol. 2019, 15, 133–149. [Google Scholar] [CrossRef] [PubMed]
- Barnes, M.D.; Kumar, V.; Bethel, C.R.; Moussa, S.H.; O’Donnell, J.; Rutter, J.D.; Good, C.E.; Hujer, K.M.; Hujer, A.M.; Marshall, S.H.; et al. Targeting Multidrug-Resistant Acinetobacter spp.: Sulbactam and the Diazabicyclooctenone beta-Lactamase Inhibitor ETX2514 as a Novel Therapeutic Agent. mBio 2019, 10, e00159-e19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhanel, G.G.; Lawrence, C.K.; Adam, H.; Schweizer, F.; Zelenitsky, S.; Zhanel, M.; Lagace-Wiens, P.R.S.; Walkty, A.; Denisuik, A.; Golden, A.; et al. Imipenem-Relebactam and Meropenem-Vaborbactam: Two Novel Carbapenem-beta-Lactamase Inhibitor Combinations. Drugs 2018, 78, 65–98. [Google Scholar] [CrossRef] [PubMed]
- Wright, H.; Bonomo, R.A.; Paterson, D.L. New agents for the treatment of infections with Gram-negative bacteria: Restoring the miracle or false dawn? Clin. Microbiol. Infect. 2017, 23, 704–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durand-Reville, T.F.; Guler, S.; Comita-Prevoir, J.; Chen, B.; Bifulco, N.; Huynh, H.; Lahiri, S.; Shapiro, A.B.; McLeod, S.M.; Carter, N.M.; et al. ETX2514 is a broad-spectrum beta-lactamase inhibitor for the treatment of drug-resistant Gram-negative bacteria including Acinetobacter baumannii. Nat. Microbiol. 2017, 2, 17104. [Google Scholar] [CrossRef]
- Lee, Y.R.; Yeo, S. Cefiderocol, a New Siderophore Cephalosporin for the Treatment of Complicated Urinary Tract Infections Caused by Multidrug-Resistant Pathogens: Preclinical and Clinical Pharmacokinetics, Pharmacodynamics, Efficacy and Safety. Clin. Drug Investig. 2020, 40, 901–913. [Google Scholar] [CrossRef]
- Parsels, K.A.; Mastro, K.A.; Steele, J.M.; Thomas, S.J.; Kufel, W.D. Cefiderocol: A novel siderophore cephalosporin for multidrug-resistant Gram-negative bacterial infections. J. Antimicrob. Chemother. 2021, 76, 1379–1391. [Google Scholar] [CrossRef]
- Bonomo, R.A. Cefiderocol: A Novel Siderophore Cephalosporin Defeating Carbapenem-resistant Pathogens. Clin. Infect. Dis. 2019, 69, S519–S520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhanel, G.G.; Golden, A.R.; Zelenitsky, S.; Wiebe, K.; Lawrence, C.K.; Adam, H.J.; Idowu, T.; Domalaon, R.; Schweizer, F.; Zhanel, M.A.; et al. Cefiderocol: A Siderophore Cephalosporin with Activity Against Carbapenem-Resistant and Multidrug-Resistant Gram-Negative Bacilli. Drugs 2019, 79, 271–289. [Google Scholar] [CrossRef] [PubMed]
- Aoki, T.; Yoshizawa, H.; Yamawaki, K.; Yokoo, K.; Sato, J.; Hisakawa, S.; Hasegawa, Y.; Kusano, H.; Sano, M.; Sugimoto, H.; et al. Cefiderocol (S-649266), A new siderophore cephalosporin exhibiting potent activities against Pseudomonas aeruginosa and other gram-negative pathogens including multi-drug resistant bacteria: Structure activity relationship. Eur. J. Med. Chem. 2018, 155, 847–868. [Google Scholar] [CrossRef] [PubMed]
- Ito, A.; Nishikawa, T.; Matsumoto, S.; Yoshizawa, H.; Sato, T.; Nakamura, R.; Tsuji, M.; Yamano, Y. Siderophore Cephalosporin Cefiderocol Utilizes Ferric Iron Transporter Systems for Antibacterial Activity against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2016, 60, 7396–7401. [Google Scholar] [CrossRef] [Green Version]
- Sheldon, J.R.; Skaar, E.P. Acinetobacter baumannii can use multiple siderophores for iron acquisition, but only acinetobactin is required for virulence. PLoS Pathog. 2020, 16, e1008995. [Google Scholar] [CrossRef] [PubMed]
- Antunes, L.C.; Imperi, F.; Towner, K.J.; Visca, P. Genome-assisted identification of putative iron-utilization genes in Acinetobacter baumannii and their distribution among a genotypically diverse collection of clinical isolates. Res. Microbiol. 2011, 162, 279–284. [Google Scholar] [CrossRef]
- Nwugo, C.C.; Arivett, B.A.; Zimbler, D.L.; Gaddy, J.A.; Richards, A.M.; Actis, L.A. Effect of ethanol on differential protein production and expression of potential virulence functions in the opportunistic pathogen Acinetobacter baumannii. PLoS ONE 2012, 7, e51936. [Google Scholar] [CrossRef] [Green Version]
- Zimbler, D.L.; Penwell, W.F.; Gaddy, J.A.; Menke, S.M.; Tomaras, A.P.; Connerly, P.L.; Actis, L.A. Iron acquisition functions expressed by the human pathogen Acinetobacter baumannii. Biometals 2009, 22, 23–32. [Google Scholar] [CrossRef]
- Ramirez, M.S.; Penwell, W.F.; Traglia, G.M.; Zimbler, D.L.; Gaddy, J.A.; Nikolaidis, N.; Arivett, B.A.; Adams, M.D.; Bonomo, R.A.; Actis, L.A.; et al. Identification of potential virulence factors in the model strain Acinetobacter baumannii A118. Front. Microbiol. 2019, 10, 1599. [Google Scholar] [CrossRef] [Green Version]
- Moynié, L.; Serra, I.; Scorciapino, M.A.; Oueis, E.; Page, M.G.; Ceccarelli, M.; Naismith, J.H. Preacinetobactin not acinetobactin is essential for iron uptake by the BauA transporter of the pathogen. eLife 2018, 7, e42270. [Google Scholar] [CrossRef]
- Ghysels, B.; Ochsner, U.; Möllman, U.; Heinisch, L.; Vasil, M.; Cornelis, P.; Matthijs, S. The Pseudomonas aeruginosa pirA gene encodes a second receptor for ferrienterobactin and synthetic catecholate analogues. FEMS Microbiol. Lett. 2005, 246, 167–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moynie, L.; Luscher, A.; Rolo, D.; Pletzer, D.; Tortajada, A.; Weingart, H.; Braun, Y.; Page, M.G.; Naismith, J.H.; Kohler, T. Structure and Function of the PiuA and PirA Siderophore-Drug Receptors from Pseudomonas aeruginosa and Acinetobacter baumannii. Antimicrob. Agents Chemother. 2017, 61, e02531-e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luscher, A.; Moynié, L.; Auguste, P.S.; Bumann, D.; Mazza, L.; Pletzer, D.; Naismith, J.H.; Köhler, T. TonB-Dependent Receptor Repertoire of Pseudomonas aeruginosa for Uptake of Siderophore-Drug Conjugates. Antimicrob. Agents Chemother. 2018, 62, e00097-e18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, T.; Yamawaki, K. Cefiderocol: Discovery, Chemistry, and In Vivo Profiles of a Novel Siderophore Cephalosporin. Clin. Infect. Dis. 2019, 69, S538–S543. [Google Scholar] [CrossRef] [Green Version]
- McCreary, E.K.; Heil, E.L.; Tamma, P.D. New Perspectives on Antimicrobial Agents: Cefiderocol. Antimicrob. Agents Chemother. 2021, 65, e02171-e20. [Google Scholar] [CrossRef]
- Wunderink, R.G.; Matsunaga, Y.; Ariyasu, M.; Clevenbergh, P.; Echols, R.; Kaye, K.S.; Kollef, M.; Menon, A.; Pogue, J.M.; Shorr, A.F.; et al. Cefiderocol versus high-dose, extended-infusion meropenem for the treatment of Gram-negative nosocomial pneumonia (APEKS-NP): A randomised, double-blind, phase 3, non-inferiority trial. Lancet Infect. Dis. 2021, 21, 213–225. [Google Scholar] [CrossRef]
- Jean, S.S.; Gould, I.M.; Lee, W.S.; Hsueh, P.R.; International Society of Antimicrobial Chemotherapy. New Drugs for Multidrug-Resistant Gram-Negative Organisms: Time for Stewardship. Drugs 2019, 79, 705–714. [Google Scholar] [CrossRef]
- Jacobs, M.R.; Abdelhamed, A.M.; Good, C.E.; Rhoads, D.D.; Hujer, K.M.; Hujer, A.M.; Domitrovic, T.N.; Rudin, S.D.; Richter, S.S.; van Duin, D.; et al. ARGONAUT-I: Activity of Cefiderocol (S-649266), a Siderophore Cephalosporin, against Gram-Negative Bacteria, Including Carbapenem-Resistant Nonfermenters and Enterobacteriaceae with Defined Extended-Spectrum beta-Lactamases and Carbapenemases. Antimicrob. Agents Chemother. 2019, 63, e01801-e18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bavaro, D.F.; Belati, A.; Diella, L.; Stufano, M.; Romanelli, F.; Scalone, L.; Stolfa, S.; Ronga, L.; Maurmo, L.; Dell’Aera, M.; et al. Cefiderocol-Based Combination Therapy for “Difficult-to-Treat” Gram-Negative Severe Infections: Real-Life Case Series and Future Perspectives. Antibiotics 2021, 10, 652. [Google Scholar] [CrossRef]
- Oliva, A.; Ceccarelli, G.; De Angelis, M.; Sacco, F.; Miele, M.C.; Mastroianni, C.M.; Venditti, M. Cefiderocol for compassionate use in the treatment of complicated infections caused by extensively and pan-resistant Acinetobacter baumannii. J. Glob. Antimicrob. Resist. 2020, 23, 292–296. [Google Scholar] [CrossRef]
- Simner, P.J.; Beisken, S.; Bergman, Y.; Ante, M.; Posch, A.E.; Tamma, P.D. Defining Baseline Mechanisms of Cefiderocol Resistance in the Enterobacterales. Microb. Drug Resist. 2022, 28, 161–170. [Google Scholar] [CrossRef]
- Van Delden, C.; Page, M.G.; Köhler, T. Involvement of Fe uptake systems and AmpC β-lactamase in susceptibility to the siderophore monosulfactam BAL30072 in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013, 57, 2095–2102. [Google Scholar] [CrossRef] [Green Version]
- Malik, S.; Kaminski, M.; Landman, D.; Quale, J. Cefiderocol Resistance in Acinetobacter baumannii: Roles of β-Lactamases, Siderophore Receptors, and Penicillin Binding Protein 3. Antimicrob. Agents Chemother. 2020, 64, e01221-20. [Google Scholar] [CrossRef]
- Poirel, L.; Sadek, M.; Nordmann, P. Contribution of PER-type and NDM-type ss-lactamases to cefiderocol resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2021, 65, e00877-e21. [Google Scholar] [CrossRef]
- Martinez, J.; Razo-Gutierrez, C.; Le, C.; Courville, R.; Pimentel, C.; Liu, C.; Fung, S.E.; Tuttobene, M.R.; Phan, K.; Vila, A.J.; et al. Cerebrospinal fluid (CSF) augments metabolism and virulence expression factors in Acinetobacter baumannii. Sci. Rep. 2021, 11, 4737. [Google Scholar] [CrossRef]
- Pimentel, C.; Le, C.; Tuttobene, M.R.; Subils, T.; Martinez, J.; Sieira, R.; Papp-Wallace, K.M.; Keppetipola, N.; Bonomo, R.A.; Actis, L.A.; et al. Human Pleural Fluid and Human Serum Albumin Modulate the Behavior of a Hypervirulent and Multidrug-Resistant (MDR) Acinetobacter baumannii Representative Strain. Pathogens 2021, 10, 471. [Google Scholar] [CrossRef]
- Martinez, J.; Fernandez, J.S.; Liu, C.; Hoard, A.; Mendoza, A.; Nakanouchi, J.; Rodman, N.; Courville, R.; Tuttobene, M.R.; Lopez, C.; et al. Human pleural fluid triggers global changes in the transcriptional landscape of Acinetobacter baumannii as an adaptive response to stress. Sci. Rep. 2019, 9, 17251. [Google Scholar] [CrossRef]
- Rodman, N.; Martinez, J.; Fung, S.; Nakanouchi, J.; Myers, A.L.; Harris, C.M.; Dang, E.; Fernandez, J.S.; Liu, C.; Mendoza, A.M.; et al. Human Pleural Fluid Elicits Pyruvate and Phenylalanine Metabolism in Acinetobacter baumannii to Enhance Cytotoxicity and Immune Evasion. Front. Microbiol. 2019, 10, 1581. [Google Scholar] [CrossRef] [Green Version]
- Quinn, B.; Rodman, N.; Jara, E.; Fernandez, J.S.; Martinez, J.; Traglia, G.M.; Montana, S.; Cantera, V.; Place, K.; Bonomo, R.A.; et al. Human serum albumin alters specific genes that can play a role in survival and persistence in Acinetobacter baumannii. Sci. Rep. 2018, 8, 14741. [Google Scholar] [CrossRef]
- Le, C.; Pimentel, C.; Tuttobene, M.R.; Subils, T.; Nishimura, B.; Traglia, G.M.; Perez, F.; Papp-Wallace, K.M.; Bonomo, R.A.; Tolmasky, M.E.; et al. Interplay between meropenem and human serum albumin on expression of carbapenem resistance genes and natural competence in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2021, 65, e01019-e21. [Google Scholar] [CrossRef]
- Pimentel, C.; Le, C.; Tuttobene, M.R.; Subils, T.; Papp-Wallace, K.M.; Bonomo, R.A.; Tolmasky, M.E.; Ramirez, M.S. Interaction of Acinetobacter baumannii with Human Serum Albumin: Does the Host Determine the Outcome? Antibiotics 2021, 10, 833. [Google Scholar] [CrossRef]
- Jacobs, A.C.; Thompson, M.G.; Black, C.C.; Kessler, J.L.; Clark, L.P.; McQueary, C.N.; Gancz, H.Y.; Corey, B.W.; Moon, J.K.; Si, Y.; et al. AB5075, a Highly Virulent Isolate of Acinetobacter baumannii, as a Model Strain for the Evaluation of Pathogenesis and Antimicrobial Treatments. mBio 2014, 5, e01076-e14. [Google Scholar] [CrossRef] [Green Version]
- Adams, M.D.; Pasteran, F.; Traglia, G.M.; Martinez, J.; Huang, F.; Liu, C.; Fernandez, J.S.; Lopez, C.; Gonzalez, L.J.; Albornoz, E.; et al. Distinct mechanisms of dissemination of NDM-1 metallo- beta-lactamase in Acinetobacter spp. in Argentina. Antimicrob. Agents Chemother. 2020, 64, e00324-20. [Google Scholar] [CrossRef]
- Rodgers, D.; Pasteran, F.; Calderon, M.; Jaber, S.; Traglia, G.M.; Albornoz, E.; Corso, A.; Vila, A.J.; Bonomo, R.A.; Adams, M.D.; et al. Characterisation of ST25 NDM-1-producing Acinetobacter spp. strains leading the increase in NDM-1 emergence in Argentina. J. Glob. Antimicrob. Resist. 2020, 23, 108–110. [Google Scholar] [CrossRef]
- Hujer, A.M.; Higgins, P.G.; Rudin, S.D.; Buser, G.L.; Marshall, S.H.; Xanthopoulou, K.; Seifert, H.; Rojas, L.J.; Domitrovic, T.N.; Cassidy, P.M.; et al. Nosocomial outbreak of extensively drug-resistant Acinetobacter baumannii isolates containing blaOXA-237 carried on a plasmid. Antimicrob. Agents Chemother. 2017, 61, e00797-e17. [Google Scholar] [CrossRef] [Green Version]
- Adams, M.D.; Chan, E.R.; Molyneaux, N.D.; Bonomo, R.A. Genomewide analysis of divergence of antibiotic resistance determinants in closely related isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2010, 54, 3569–3577. [Google Scholar] [CrossRef] [Green Version]
- Hujer, K.M.; Hujer, A.M.; Hulten, E.A.; Bajaksouzian, S.; Adams, J.M.; Donskey, C.J.; Ecker, D.J.; Massire, C.; Eshoo, M.W.; Sampath, R.; et al. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob. Agents Chemother. 2006, 50, 4114–4123. [Google Scholar] [CrossRef] [Green Version]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 29th ed.; CLSI Document M100; Clinical Lab Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI Document M100; Clinical Lab Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Adams, M.D.; Goglin, K.; Molyneaux, N.; Hujer, K.M.; Lavender, H.; Jamison, J.J.; MacDonald, I.J.; Martin, K.M.; Russo, T.; Campagnari, A.A.; et al. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 2008, 190, 8053–8064. [Google Scholar] [CrossRef] [Green Version]
- Durante-Mangoni, E.; Andini, R.; Zampino, R. Management of carbapenem-resistant Enterobacteriaceae infections. Clin. Microbiol. Infect. 2019, 25, 943–950. [Google Scholar] [CrossRef]
- Martinez-Martinez, L. Carbapenemases: The never-ending story. Enferm. Infecc. Microbiol. Clin. 2019, 37, 73–75. [Google Scholar] [CrossRef]
- Moeck, G.S.; Coulton, J.W. TonB-dependent iron acquisition: Mechanisms of siderophore-mediated active transport. Mol. Microbiol. 1998, 28, 675–681. [Google Scholar] [CrossRef]
- Noinaj, N.; Guillier, M.; Barnard, T.J.; Buchanan, S.K. TonB-dependent transporters: Regulation, structure, and function. Annu. Rev. Microbiol. 2010, 64, 43–60. [Google Scholar] [CrossRef] [Green Version]
- Lau, C.K.; Krewulak, K.D.; Vogel, H.J. Bacterial ferrous iron transport: The Feo system. FEMS Microbiol. Rev. 2016, 40, 273–298. [Google Scholar] [CrossRef]
- Mihara, K.; Tanabe, T.; Yamakawa, Y.; Funahashi, T.; Nakao, H.; Narimatsu, S.; Yamamoto, S. Identification and transcriptional organization of a gene cluster involved in biosynthesis and transport of acinetobactin, a siderophore produced by Acinetobacter baumannii ATCC 19606T. Microbiology 2004, 150, 2587–2597. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, V.; Rajeswari, M.R.; Tiwari, M. Proteomic analysis of iron-regulated membrane proteins identify FhuE receptor as a target to inhibit siderophore-mediated iron acquisition in Acinetobacter baumannii. Int. J. Biol. Macromol. 2018, 125, 1156–1167. [Google Scholar] [CrossRef]
- Mussi, M.A.; Limansky, A.S.; Relling, V.; Ravasi, P.; Arakaki, A.; Actis, L.A.; Viale, A.M. Horizontal gene transfer and assortative recombination within the Acinetobacter baumannii clinical population provide genetic diversity at the single carO gene, encoding a major outer membrane protein channel. J. Bacteriol. 2011, 193, 4736–4748. [Google Scholar] [CrossRef] [Green Version]
- Baynes, R.D.; Bezwoda, W.R. Lactoferrin and the inflammatory response. Adv. Exp. Med. Biol. 1994, 357, 133–141. [Google Scholar]
- Fillebeen, C.; Wilkinson, N.; Charlebois, E.; Katsarou, A.; Wagner, J.; Pantopoulos, K. Hepcidin-mediated hypoferremic response to acute inflammation requires a threshold of Bmp6/Hjv/Smad signaling. Blood 2018, 132, 1829–1841. [Google Scholar] [CrossRef] [Green Version]
- Caldwell, M.; Archibald, F. The effect of the hypoferremic response on iron acquisition by and growth of murine lymphoma cells. Biochem. Cell Biol. 1987, 65, 651–657. [Google Scholar] [CrossRef]
- Klein, S.; Boutin, S.; Kocer, K.; Fiedler, M.O.; Störzinger, D.; Weigand, M.A.; Tan, B.; Richter, D.; Rupp, C.; Mieth, M.; et al. Rapid development of cefiderocol resistance in carbapenem-resistant Enterobacter cloacae during therapy is associated with heterogeneous mutations in the catecholate siderophore receptor cira. Clin. Infect Dis. 2021, ciab511. [Google Scholar] [CrossRef]
- Streling, A.P.; Al Obaidi, M.M.; Lainhart, W.D.; Zangeneh, T.; Khan, A.; Dinh, A.Q.; Hanson, B.; Arias, C.A.; Miller, W.R. Evolution of Cefiderocol Non-Susceptibility in Pseudomonas aeruginosa in a Patient without Previous Exposure to the Antibiotic. Clin. Infect. Dis. 2021, 73, e4472–e4474. [Google Scholar] [CrossRef] [PubMed]
- Simner, P.J.; Beisken, S.; Bergman, Y.; Posch, A.E.; Cosgrove, S.E.; Tamma, P.D. Cefiderocol Activity against Clinical Pseudomonas aeruginosa Isolates Exhibiting Ceftolozane-Tazobactam Resistance. Open Forum Infect. Dis. 2021, 8, ofab311. [Google Scholar] [CrossRef] [PubMed]
- Pinsky, M.; Roy, U.; Moshe, S.; Weissman, Z.; Kornitzer, D. Human Serum Albumin Facilitates Heme-Iron Utilization by Fungi. mBio 2020, 11, e00607-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konig, C.; Both, A.; Rohde, H.; Kluge, S.; Frey, O.R.; Rohr, A.C.; Wichmann, D. Cefiderocol in Critically Ill Patients with Multi-Drug Resistant Pathogens: Real-Life Data on Pharmacokinetics and Microbiological Surveillance. Antibiotics 2021, 10, 649. [Google Scholar] [CrossRef]
- Kidd, J.M.; Abdelraouf, K.; Nicolau, D.P. Development of Neutropenic Murine Models of Iron Overload and Depletion to Study the Efficacy of Siderophore-Antibiotic Conjugates. Antimicrob. Agents Chemother. 2019, 64, e01961-e19. [Google Scholar] [CrossRef]
- Kidd, J.M.; Abdelraouf, K.; Nicolau, D.P. Efficacy of Humanized Cefiderocol Exposure Is Unaltered by Host Iron Overload in the Thigh Infection Model. Antimicrob. Agents Chemother. 2019, 64, e01767-19. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, S.; Singley, C.M.; Hoover, J.; Nakamura, R.; Echols, R.; Rittenhouse, S.; Tsuji, M.; Yamano, Y. Efficacy of Cefiderocol against Carbapenem-Resistant Gram-Negative Bacilli in Immunocompetent-Rat Respiratory Tract Infection Models Recreating Human Plasma Pharmacokinetics. Antimicrob. Agents Chemother. 2017, 61, e00700-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| CFDC MICs (mg/L) | |||
|---|---|---|---|
| Strain | LB | HPF | 3.5% HSA |
| AB5075 | 0.5 (S) | 1 (S) | 2 (S) |
| ABUH702 | 0.38 (S) | 1.5 (S) | 3 (S) |
| AMA16 | >4.5 * (I) | >256 (R) | 32 * (R) |
| AB0057 | 1 (S) | 8 (I) | 1.5 (S) |
| AMA40 | 0.5 (S) | 16 * (R) | 3 (S) |
| AMA41 | 0.094 (S) | 0.5–0.75 (S) | 2 (S) |
| AMA113 | 0.5 (S) | 1.5 (S) | 1.5 (S) |
| AMA181 | 0.19 (S) | 0.19 (S) | 0.75 (S) |
| AMA3 | 24 (R) | >256 (R) | 32 * (R) |
| AMA4 | 16 * (R) | 48 * (R) | 64 * (R) |
| AMA5 | >256 (R) | >256 (R) | 16 * (R) |
| AMA9 | 32 (R) | 48 (R) | 16 (R) |
| AMA14 | 8 * (I) | 16 * (R) | 12 (I) |
| AMA17 | >256 (R) | >256 (R) | >256 (R) |
| AMA18 | 64 * (R) | 16 * (R) | 16 * (R) |
| AMA19 | 4 (S) | 4 (S) | 48 * (R) |
| AMA28 | 32 * (R) | >256 (R) | 32 * (R) |
| AMA30 | 64 * (R) | 128 * (R) | 12 * (I) |
| AMA31 | >256 (R) | >256 (R) | 96 * (R) |
| AMA33 | 16 * (R) | >256 (R) | >256 (R) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le, C.; Pimentel, C.; Pasteran, F.; Tuttobene, M.R.; Subils, T.; Escalante, J.; Nishimura, B.; Arriaga, S.; Carranza, A.; Mezcord, V.; et al. Human Serum Proteins and Susceptibility of Acinetobacter baumannii to Cefiderocol: Role of Iron Transport. Biomedicines 2022, 10, 600. https://doi.org/10.3390/biomedicines10030600
Le C, Pimentel C, Pasteran F, Tuttobene MR, Subils T, Escalante J, Nishimura B, Arriaga S, Carranza A, Mezcord V, et al. Human Serum Proteins and Susceptibility of Acinetobacter baumannii to Cefiderocol: Role of Iron Transport. Biomedicines. 2022; 10(3):600. https://doi.org/10.3390/biomedicines10030600
Chicago/Turabian StyleLe, Casin, Camila Pimentel, Fernando Pasteran, Marisel R. Tuttobene, Tomás Subils, Jenny Escalante, Brent Nishimura, Susana Arriaga, Aimee Carranza, Vyanka Mezcord, and et al. 2022. "Human Serum Proteins and Susceptibility of Acinetobacter baumannii to Cefiderocol: Role of Iron Transport" Biomedicines 10, no. 3: 600. https://doi.org/10.3390/biomedicines10030600
APA StyleLe, C., Pimentel, C., Pasteran, F., Tuttobene, M. R., Subils, T., Escalante, J., Nishimura, B., Arriaga, S., Carranza, A., Mezcord, V., Vila, A. J., Corso, A., Actis, L. A., Tolmasky, M. E., Bonomo, R. A., & Ramírez, M. S. (2022). Human Serum Proteins and Susceptibility of Acinetobacter baumannii to Cefiderocol: Role of Iron Transport. Biomedicines, 10(3), 600. https://doi.org/10.3390/biomedicines10030600










