Radiomics-Based Predictive Model of Radiation-Induced Liver Disease in Hepatocellular Carcinoma Patients Receiving Stereo-Tactic Body Radiotherapy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. SBRT
2.3. Patient Follow-Up and Definition of RILD
2.4. Extraction of Radiomic Features
2.5. Selection of Predictive Factors
2.6. Model Construction
2.7. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Predictive Factors
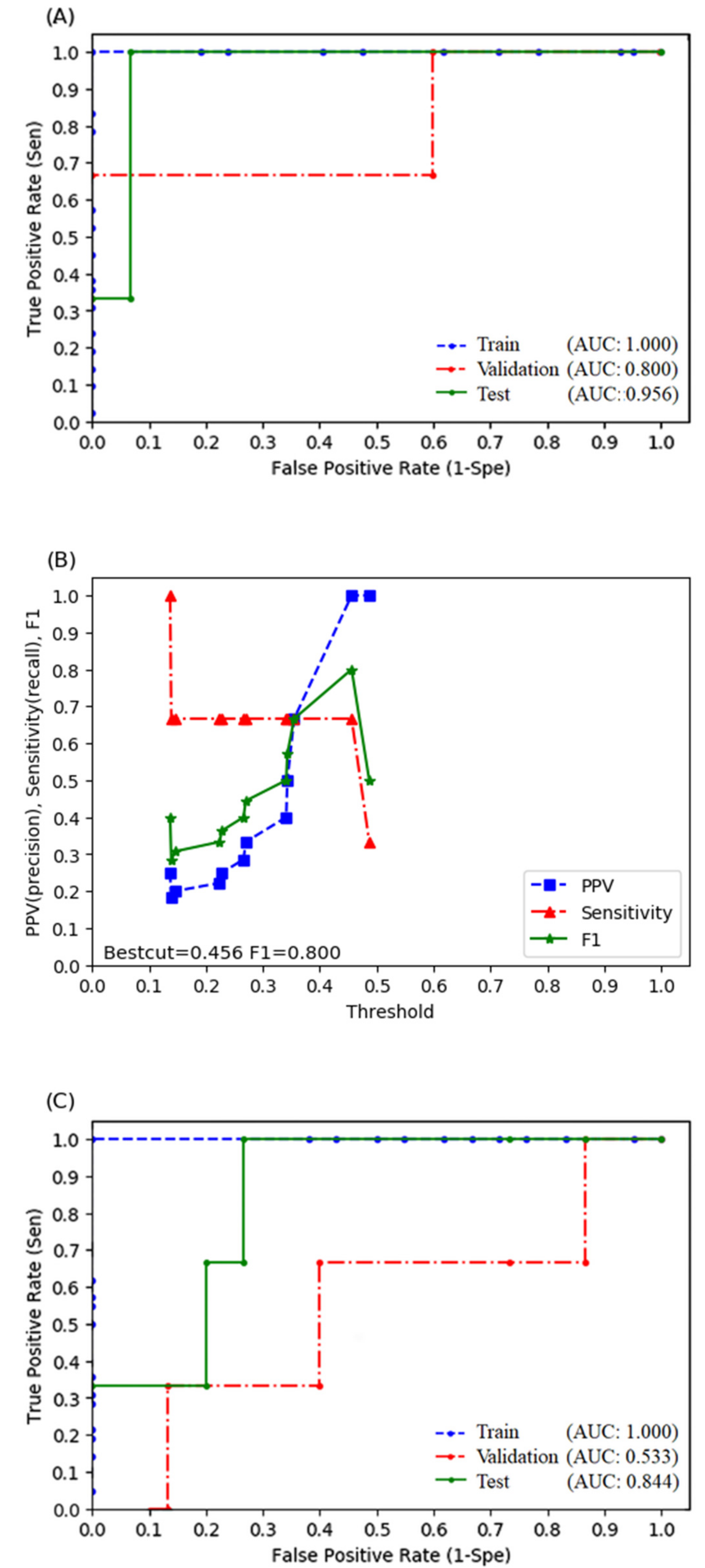
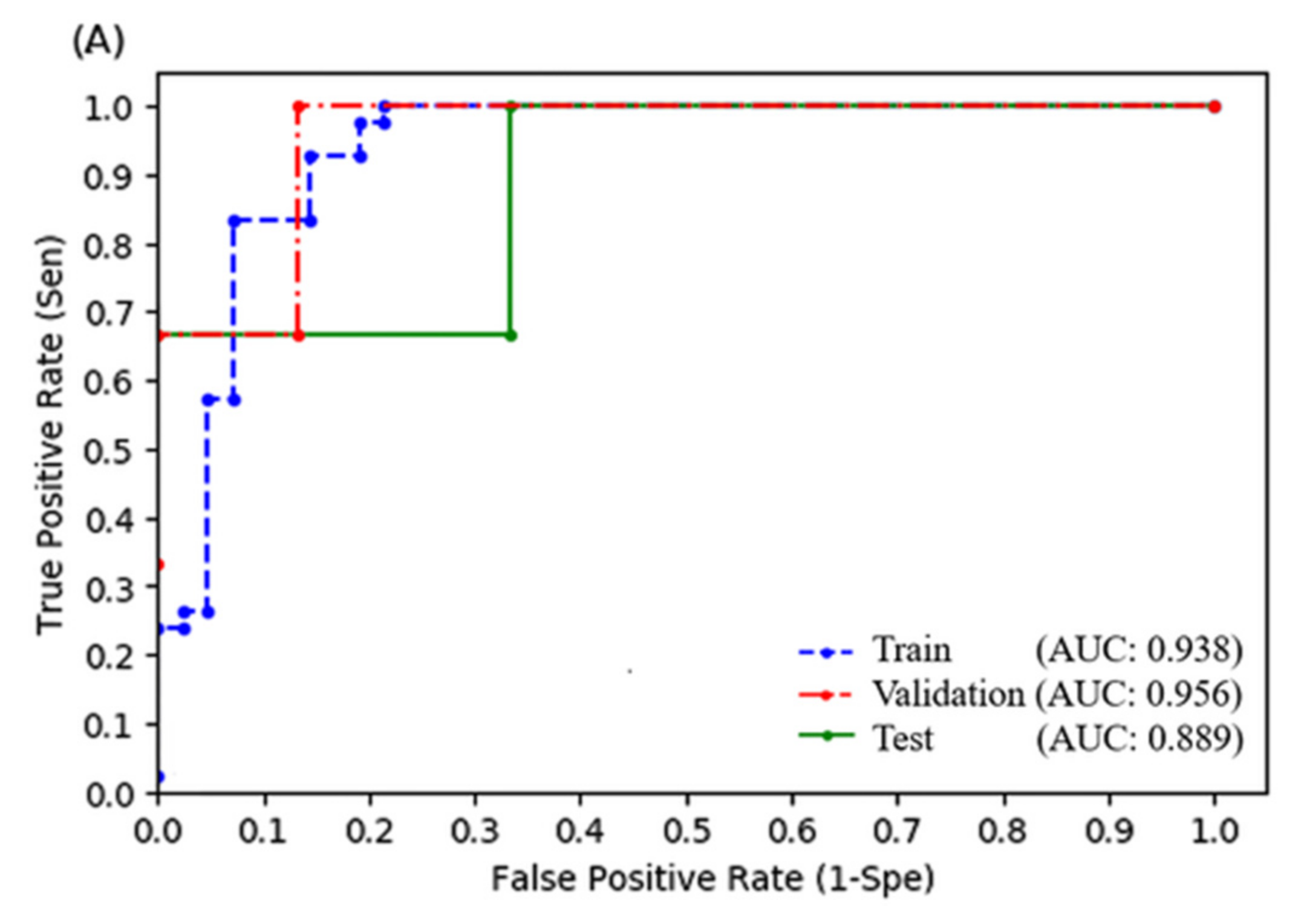
3.3. Random Forest Model
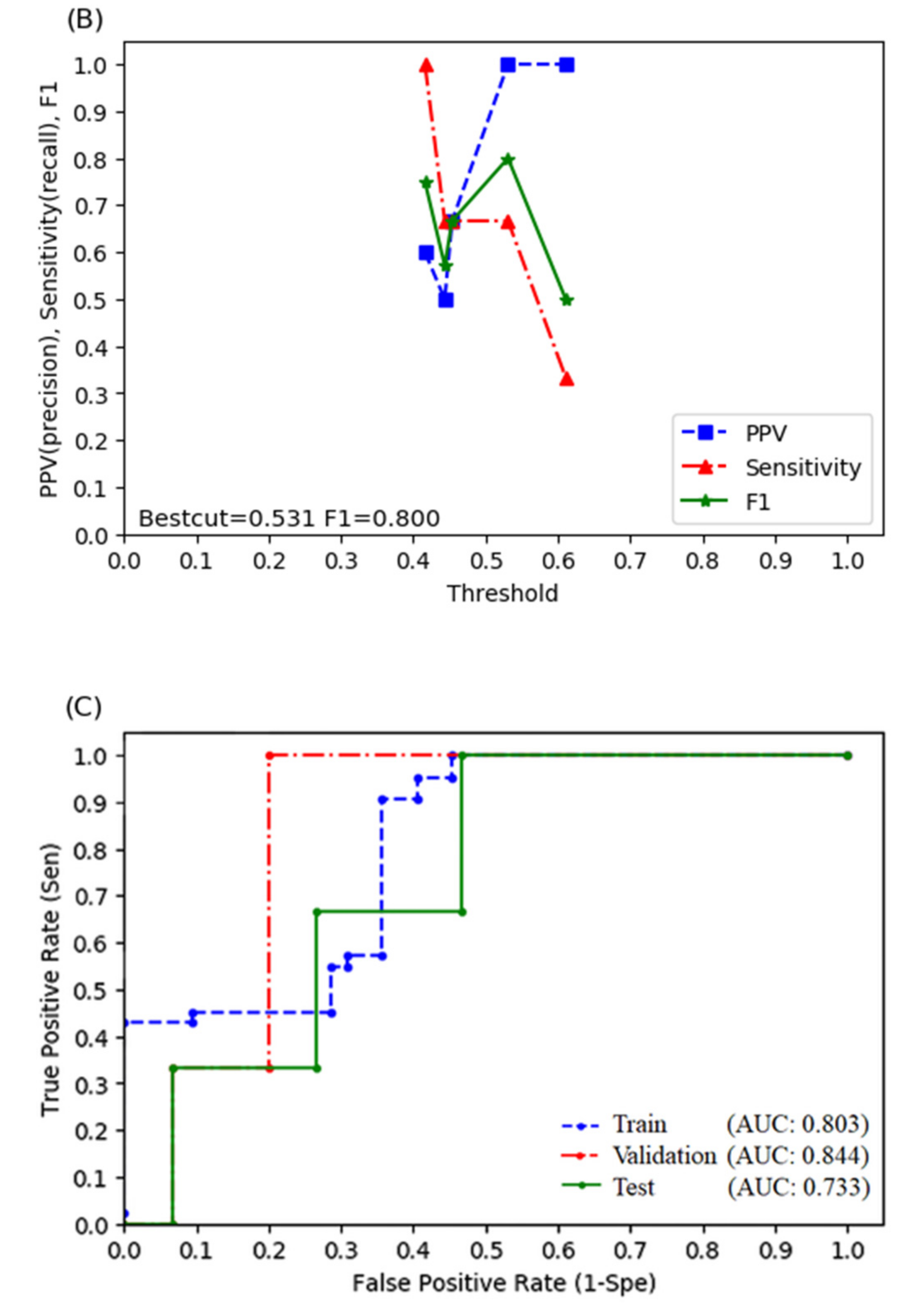
3.4. Logistic Regression Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McGlynn, K.A.; Petrick, J.L.; London, W.T. Global epidemiology of hepatocellular carcinoma: An emphasis on demographic and regional variability. Clin. Liver Dis. 2015, 19, 223–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belghiti, J.; Kianmanesh, R. Surgical treatment of hepatocellular carcinoma. HPB 2005, 7, 42–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, T.C.; Lam, V.W. Surgical management of hepatocellular carcinoma. World J. Hepatol. 2015, 7, 245–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, J.C.-H.; Wu, J.-K.; Lee, P.C.-T.; Liu, H.-S.; Jian, J.J.-M.; Lin, Y.-M.; Sung, J.-L.; Jan, G.-J. Biologic susceptibility of hepatocellular carcinoma patients treated with radiotherapy to radiation-induced liver disease. Int. J. Radiat. Oncol. Biol. Phys. 2004, 60, 1502–1509. [Google Scholar] [CrossRef] [PubMed]
- Dawson, L.A.; Normolle, D.; Balter, J.M.; McGinn, C.J.; Lawrence, T.S.; Haken, R.T. Analysis of radiation-induced liver disease using the Lyman NTCP model. Int. J. Radiat. Oncol. Biol. Phys. 2002, 53, 810–821. [Google Scholar] [CrossRef]
- Jun, B.; Kim, Y.D.; Cheon, G.J.; Kim, E.S.; Jwa, E.; Kim, S.G.; Kim, B.S.; Jeong, S.W.; Jang, J.Y.; Lee, S.H.; et al. Clinical significance of radiation-induced liver disease after stereotactic body radiation therapy for hepatocellular carcinoma. Korean J. Intern. Med. 2018, 33, 1093–1102. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.; Yoon, S.M.; Kim, S.Y.; Cho, B.; Park, J.-H.; Kim, S.S.; Song, S.Y.; Lee, S.-W.; Ahn, S.D.; Choi, E.K.; et al. Radiation-induced liver disease after stereotactic body radiotherapy for small hepatocellular carcinoma: Clinical and dose-volumetric parameters. Radiat. Oncol. 2013, 8, 249. [Google Scholar] [CrossRef] [Green Version]
- Liang, S.-X.; Zhu, X.-D.; Xu, Z.-Y.; Zhu, J.; Zhao, J.-D.; Lu, H.-J.; Yang, Y.-L.; Chen, L.; Wang, A.-Y.; Fu, X.-L.; et al. Radiation-induced liver disease in three-dimensional conformal radiation therapy for primary liver carcinoma: The risk factors and hepatic radiation tolerance. Int. J. Radiat. Oncol. Biol. Phys. 2006, 65, 426–434. [Google Scholar] [CrossRef]
- Velec, M.; Haddad, C.R.; Craig, T.; Wang, L.; Lindsay, P.; Brierley, J.; Brade, A.; Ringash, J.; Wong, R.K.; Kim, J.; et al. Predictors of Liver Toxicity Following Stereotactic Body Radiation Therapy for Hepatocellular Carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 2017, 97, 939–946. [Google Scholar] [CrossRef]
- Xu, Z.-Y.; Liang, S.-X.; Zhu, J.; Zhu, X.-D.; Zhao, J.-D.; Lu, H.-J.; Yang, Y.-L.; Chen, L.; Wang, A.-Y.; Fu, X.-L.; et al. Prediction of radiation-induced liver disease by Lyman normal-tissue complication probability model in three-dimensional conformal radiation therapy for primary liver carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 2006, 65, 189–195. [Google Scholar] [CrossRef]
- El Naqa, I.; Grigsby, P.; Apte, A.; Kidd, E.; Donnelly, E.; Khullar, D.; Chaudhari, S.; Yang, D.; Schmitt, M.; Laforest, R.; et al. Exploring feature-based approaches in PET images for predicting cancer treatment outcomes. Pattern Recognit. 2009, 42, 1162–1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, V.; Gu, Y.; Basu, S.; Berglund, A.; Eschrich, S.A.; Schabath, M.B.; Forster, K.; Aerts, H.J.W.L.; Dekker, A.; Fenstermacher, D.; et al. Radiomics: The process and the challenges. Magn. Reson. Imaging 2012, 30, 1234–1248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.J.; Lee, S.S.; Park, B.; Yun, J.; Sung, Y.S.; Shim, W.H.; Shin, Y.M.; Kim, S.Y.; Lee, S.J.; Lee, M.-G. Radiomics Analysis of Gadoxetic Acid–enhanced MRI for Staging Liver Fibrosis. Radiology 2019, 290, 380–387. [Google Scholar] [CrossRef]
- Huang, W.-Y.; Jen, Y.-M.; Lee, M.-S.; Chang, L.-P.; Chen, C.-M.; Ko, K.-H.; Lin, K.-T.; Lin, J.-C.; Chao, H.-L.; Lin, C.-S.; et al. Stereotactic Body Radiation Therapy in Recurrent Hepatocellular Carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 2012, 84, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Parmar, C.; Velazquez, E.R.; Leijenaar, R.; Jermoumi, M.; Carvalho, S.; Mak, R.H.; Mitra, S.; Shankar, B.U.; Kikinis, R.; Haibe-Kains, B.; et al. Robust Radiomics Feature Quantification Using Semiautomatic Volumetric Segmentation. PLoS ONE 2014, 9, e102107. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Cheng, J.-Y.; Liu, C.-M.; Wang, Y.-M.; Hsu, H.-C.; Huang, E.-Y.; Huang, T.-T.; Lee, C.-H.; Hung, S.-P.; Huang, B.-S. Proton versus photon radiotherapy for primary hepatocellular carcinoma: A propensity-matched analysis. Radiat. Oncol. 2020, 15, 159. [Google Scholar] [CrossRef]
- Lo, C.-H.; Liu, M.-Y.; Lee, M.-S.; Yang, J.-F.; Jen, Y.-M.; Lin, C.-S.; Chao, H.-L.; Shen, P.-C.; Huang, W.-Y. Comparison Between Child-Turcotte-Pugh and Albumin-Bilirubin Scores in Assessing the Prognosis of Hepatocellular Carcinoma After Stereotactic Ablative Radiation Therapy. Int. J. Radiat. Oncol. Biol. Phys. 2017, 99, 145–152. [Google Scholar] [CrossRef]
- Su, T.-S.; Yang, H.-M.; Zhou, Y.; Huang, Y.; Liang, P.; Cheng, T.; Chen, L.; Li, L.-Q.; Liang, S.-X. Albumin-bilirubin (ALBI) versus Child-Turcotte-Pugh (CTP) in prognosis of HCC after stereotactic body radiation therapy. Radiat. Oncol. 2019, 14, 50. [Google Scholar] [CrossRef]
- Zhu, J.; Zhu, X.D.; Liang, S.X.; Xu, Z.Y.; Zhao, J.D.; Huang, Q.F.; Wang, A.Y.; Chen, L.; Fu, X.L.; Jiang, G.L. Prediction of radiation induced liver disease using artificial neural networks. Jpn. J. Clin. Oncol. 2006, 36, 783–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, T.-S.; Luo, R.; Liang, P.; Cheng, T.; Zhou, Y.; Huang, Y. A prospective cohort study of hepatic toxicity after stereotactic body radiation therapy for hepatocellular carcinoma. Radiother. Oncol. 2018, 129, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Apisarnthanarax, S.; Barry, A.; Cao, M.; Czito, B.; DeMatteo, R.; Drinane, M.; Hallemeier, C.L.; Koay, E.J.; Lasley, F.; Meyer, J.; et al. External Beam Radiation Therapy for Primary Liver Cancers: An ASTRO Clinical Practice Guideline. Pract. Radiat. Oncol. 2022, 12, 28–51. [Google Scholar] [CrossRef]
- Lee, U.E.; Friedman, S.L. Mechanisms of hepatic fibrogenesis. Best Pract. Res. Clin. Gastroenterol. 2011, 25, 195–206. [Google Scholar] [CrossRef]
- Kim, J.; Jung, Y. Radiation-induced liver disease: Current understanding and future perspectives. Exp. Mol. Med. 2017, 49, e359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haddad, M.M.; Merrell, K.W.; Hallemeier, C.L.; Johnson, G.; Mounajjed, T.; Olivier, K.R.; Fidler, J.L.; Venkatesh, S.K. Stereotactic body radiation therapy of liver tumors: Post-treatment appearances and evaluation of treatment response: A pictorial review. Abdom. Radiol. 2016, 41, 2061–2077. [Google Scholar] [CrossRef] [PubMed]
- Takamatsu, S.; Kozaka, K.; Kobayashi, S.; Yoneda, N.; Yoshida, K.; Inoue, D.; Kitao, A.; Ogi, T.; Minami, T.; Kouda, W.; et al. Pathology and images of radiation-induced hepatitis: A review article. Jpn. J. Radiol. 2018, 36, 241–256. [Google Scholar] [CrossRef] [Green Version]
- Guha, C.; Kavanagh, B.D. Hepatic Radiation Toxicity: Avoidance and Amelioration. Semin. Radiat. Oncol. 2011, 21, 256–263. [Google Scholar] [CrossRef] [Green Version]
- Lundberg, S.M.; Lee, S.-I. A unified approach to interpreting model predictions. Adv. Neural Inf. Process. Syst. 2017, 30. Available online: https://www.mdpi.com/2674-1032/1/1/6/htm (accessed on 15 January 2022).
| Characteristic | All Patient (N = 86) | Without RILD (N = 72) | With RILD (N = 14) |
|---|---|---|---|
| Age (mean) | 63.2 ± 12.3 | 62.9 ± 12.4 | 65.2 ± 12.2 |
| Sex | |||
| Female | 21 (24.4%) | 15 (20.8%) | 6 (42.9%) |
| Male | 65 (75.6%) | 57 (79.2%) | 8 (57.1%) |
| Hepatitis B or C | 75 (87.2%) | 61 (84.7%) | 14 (100%) |
| With prior tx before SBRT | 54 (62.8%) | 46 (63.9%) | 8 (57.1%) |
| Tumor size | |||
| Mean, cm | 6.4 ± 3.7 | 6.3 ± 3.9 | 6.6 ± 2.5 |
| Median (range), cm | 5.6 (1–20.1) | 5.5 (1–20.1) | 6.6 (2–13) |
| BCLC stage | |||
| 0-A | 15 (17.4%) | 14 (19.4%) | 1 (7.1%) |
| B | 11 (12.8%) | 9 (12.5%) | 2 (14.3%) |
| C | 59 (68.6%) | 49 (68.1%) | 10 (71.4%) |
| D | 1 (1.2%) | 0 | 1 (7.1%) |
| ECOG | |||
| 0–1 | 75 (87.2%) | 66 (91.7%) | 9 (64.3%) |
| 2–4 | 11 (12.8%) | 6 (8.3%) | 5 (35.7%) |
| PVT | 32 (37.2%) | 24 (33.3%) | 8 (57.1%) |
| CP class | |||
| A | 68 (79%) | 63 (87.5%) | 5 (35.7%) |
| B | 18 (21%) | 9 (12.5%) | 9 (64.3%) |
| ALBI grade | |||
| 1 | 36 (41.9%) | 35 (48.6%) | 1 (7.1%) |
| 2 | 48 (55.8%) | 37 (51.4%) | 11 (78.6%) |
| 3 | 2 (2.3%) | 0 | 2 (14.3%) |
| Dose | 25–60 Gy/2–6 fx | 30–60 Gy/2–6 fx | 25–55/5 fx |
| GTV volume | |||
| Mean, cc | 217.6 ± 305.0 | 221.5 ± 325.0 | 197.9 ± 178.8 |
| Median (range), cc | 106.8 (0.7–1817.2) | 101.7 (0.7–1817.2) | 139.0 (9.4–572.9) |
| Normal liver volume (mean, cc) | 1384.0 ± 522.9 | 1404.0 ± 515.8 | 1281.1 ± 566.1 |
| V5 (mean,%) | 69.4 ± 20.3 | 69.0 ± 21.6 | 71.6 ± 11.9 |
| V10 (mean,%) | 44.8 ± 18.6 | 45.3 ± 19.8 | 42.3 ± 10.6 |
| V15 (mean,%) | 27.1 ± 12.5 | 27.5 ± 13.2 | 25.1 ± 8.5 |
| V20 (mean,%) | 17.5 ± 8.7 | 17.7 ± 9.1 | 16.2 ± 6.5 |
| V25 (mean,%) | 11.7 ± 6.5 | 11.9 ± 6.7 | 10.6 ± 5.6 |
| V30 (mean,%) | 7.7 ± 4.9 | 7.8 ± 4.9 | 7.0 ± 4.9 |
| Median f/u (month) | 14.7 (1–105) | 18.2 (1.7–105) | 2.8 (1–58) |
| Clinical Variable | Multivariate Analysis | ||
|---|---|---|---|
| HR | 95% CI | p Value | |
| Gender | 0.481 | 0.010–23.675 | 0.713 |
| Age | 1.041 | 0.859–1.261 | 0.683 |
| Pretx AFP | 1.000 | 0.999–1.001 | 0.576 |
| Etiology | 0.952 | ||
| HBV vs. no | 2.256 × 1010 | - | 0.998 |
| HCV vs. no | 4.076 × 1010 | - | 0.998 |
| HBV and HCV vs. no | 1.314 × 1010 | - | 0.998 |
| ECOG | |||
| 2–4 vs. 0–1 | 57.790 | 0.613–5444.863 | 0.080 |
| EQD2 | 1.018 | 0.901–1.151 | 0.770 |
| ALBI score | 91.304 | 2.700–3087.382 | 0.012 |
| Random Forest (With Radiomics) | Random Forest (Without Radiomics) | Logistic Regression (With Radiomics) | Logistic Regression (Without Radiomics) | |
|---|---|---|---|---|
| Sensitivity | 1.000 | 1.000 | 0.667 | 0.667 |
| Specificity | 0.933 | 0.600 | 1.000 | 0.733 |
| Positive predictive rate | 0.750 | 0.333 | 1.000 | 0.333 |
| Accuracy | 0.944 | 0.667 | 0.944 | 0.722 |
| F1 score | 0.857 | 0.500 | 0.800 | 0.444 |
| AUROC | 0.956 | 0.844 | 0.889 | 0.733 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, P.-C.; Huang, W.-Y.; Dai, Y.-H.; Lo, C.-H.; Yang, J.-F.; Su, Y.-F.; Wang, Y.-F.; Lu, C.-F.; Lin, C.-S. Radiomics-Based Predictive Model of Radiation-Induced Liver Disease in Hepatocellular Carcinoma Patients Receiving Stereo-Tactic Body Radiotherapy. Biomedicines 2022, 10, 597. https://doi.org/10.3390/biomedicines10030597
Shen P-C, Huang W-Y, Dai Y-H, Lo C-H, Yang J-F, Su Y-F, Wang Y-F, Lu C-F, Lin C-S. Radiomics-Based Predictive Model of Radiation-Induced Liver Disease in Hepatocellular Carcinoma Patients Receiving Stereo-Tactic Body Radiotherapy. Biomedicines. 2022; 10(3):597. https://doi.org/10.3390/biomedicines10030597
Chicago/Turabian StyleShen, Po-Chien, Wen-Yen Huang, Yang-Hong Dai, Cheng-Hsiang Lo, Jen-Fu Yang, Yu-Fu Su, Ying-Fu Wang, Chia-Feng Lu, and Chun-Shu Lin. 2022. "Radiomics-Based Predictive Model of Radiation-Induced Liver Disease in Hepatocellular Carcinoma Patients Receiving Stereo-Tactic Body Radiotherapy" Biomedicines 10, no. 3: 597. https://doi.org/10.3390/biomedicines10030597
APA StyleShen, P.-C., Huang, W.-Y., Dai, Y.-H., Lo, C.-H., Yang, J.-F., Su, Y.-F., Wang, Y.-F., Lu, C.-F., & Lin, C.-S. (2022). Radiomics-Based Predictive Model of Radiation-Induced Liver Disease in Hepatocellular Carcinoma Patients Receiving Stereo-Tactic Body Radiotherapy. Biomedicines, 10(3), 597. https://doi.org/10.3390/biomedicines10030597






