SARS-CoV-2 Dysregulates Neutrophil Degranulation and Reduces Lymphocyte Counts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Virus
2.2. Neutrophil Treatment with Calu-3-Conditioned Media
2.3. SARS-CoV-2 Infection of Neutrophils
2.4. PBL Treatment with Neutrophil Conditioned Media and Coculture
2.5. RNA Extraction and Quantitative Polymerase Chain Reaction (qPCR)
2.6. Immunofluorescence
2.7. Myeloperoxidase (MPO) and Neutrophil Elastase ELISA
2.8. Flow Cytometry
2.9. Statistical Analysis
3. Results
3.1. Factors Secreted by SARS-CoV-2-Infected Epithelial Cells Diminish Myeloperoxidase Release While Modestly Increasing Elastase Release by Human Neutrophils In Vitro
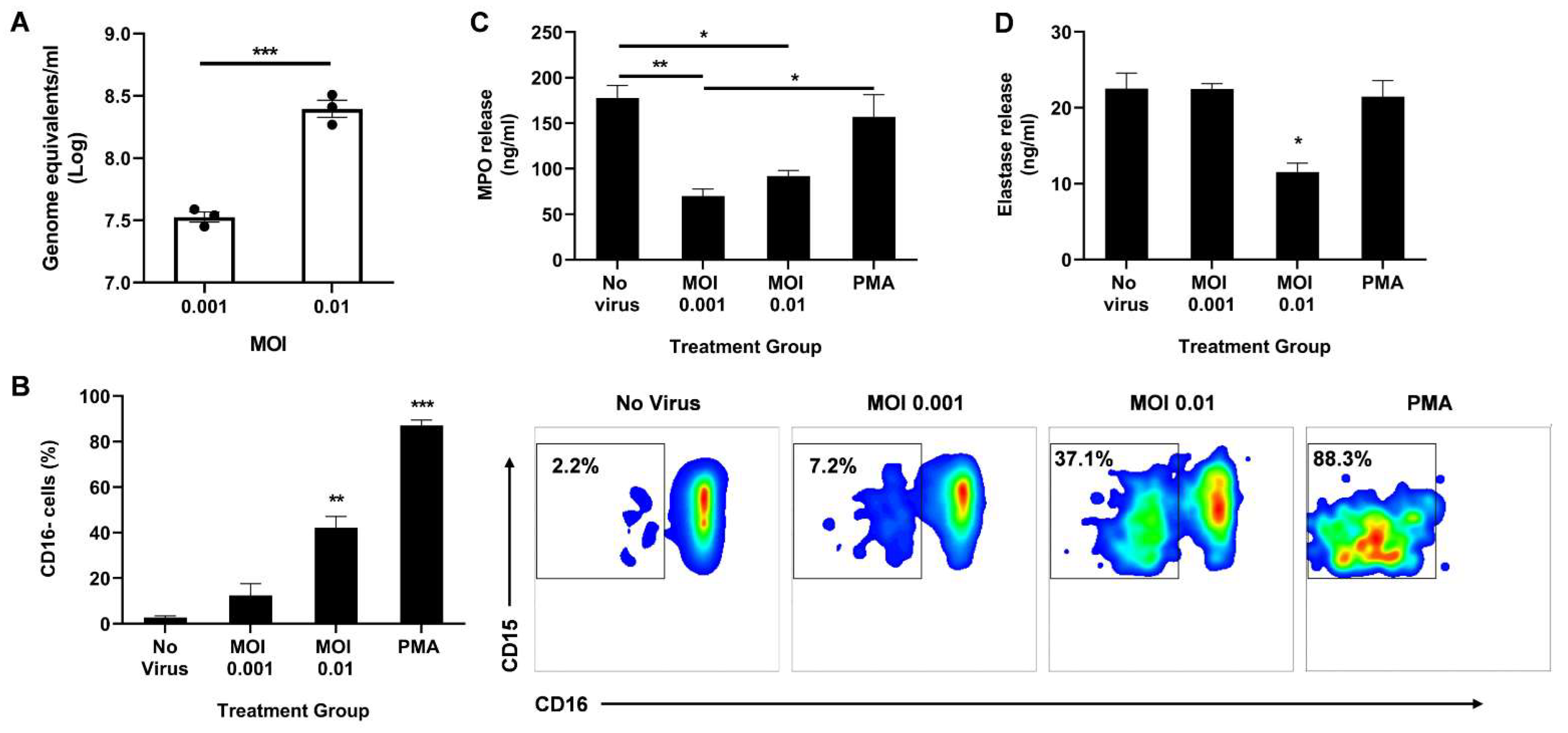
3.2. Direct Infection of Neutrophils with SARS-CoV-2 Promotes CD16 Shedding but Does Not Increase Release of Azurophil Granules
3.3. SARS-CoV-2-Infected Neutrophils Reduce B Cell, CD8+ T Cell, and CD4+ T Cell Counts In Vitro
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morens, D.M.; Breman, J.G.; Calisher, C.H.; Doherty, P.C.; Hahn, B.H.; Keusch, G.T.; Kramer, L.D.; LeDuc, J.W.; Monath, T.P.; Taubenberger, J.K. The Origin of COVID-19 and Why It Matters. Am. J. Trop. Med. Hyg. 2020, 103, 955–959. [Google Scholar] [CrossRef]
- Zhong, N.S.; Zheng, B.J.; Li, Y.M.; Poon, L.L.M.; Xie, Z.H.; Chan, K.H.; Li, P.H.; Tan, S.Y.; Chang, Q.; Xie, J.P.; et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February 2003. Lancet 2003, 362, 1353–1358. [Google Scholar] [CrossRef] [Green Version]
- Ksiazek, T.G.; Erdman, D.; Goldsmith, C.S.; Zaki, S.R.; Peret, T.; Emery, S.; Tong, S.; Urbani, C.; Comer, J.A.; Lim, W.; et al. A Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003, 348, 1953–1966. [Google Scholar] [CrossRef] [PubMed]
- Reina, J.; Reina, N. The Middle East respiratory syndrome coronavirus. Med. Clin. 2015, 145, 529–531. [Google Scholar] [CrossRef] [PubMed]
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, B.; Rajakumar, T.; Malathi, M.; Manikandan, N.; Nagaraj, J.; Santhakumar, A.; Elangovan, A.; Malik, Y.S. Epidemiology and pathobiology of SARS-CoV-2 (COVID-19) in comparison with SARS, MERS: An updated overview of current knowledge and future perspectives. Clin. Epidemiol. Glob. Health 2021, 10, 100694. [Google Scholar] [CrossRef]
- Guan, W.; Ni, Z.; Hu, Y.; Liang, W.; Ou, C.; He, J.; Liu, L.; Shan, H.; Lei, C.; Hui, D.S.C.; et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- He, D.; Gao, D.; Li, Y.; Zhuang, Z.; Cao, P.; Lou, Y.; Yang, L. An Updated Comparison of COVID-19 and Influenza. SSRN Electron. J. 2020, 90, 107233. [Google Scholar] [CrossRef]
- Yi, Y.; Lagniton, P.N.P.; Ye, S.; Li, E.; Xu, R.H. COVID-19: What has been learned and to be learned about the novel coronavirus disease. Int. J. Biol. Sci. 2020, 16, 1753–1766. [Google Scholar] [CrossRef]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- Wu, J.T.; Leung, K.; Bushman, M.; Kishore, N.; Niehus, R.; de Salazar, P.M.; Cowling, B.J.; Lipsitch, M.; Leung, G.M. Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat. Med. 2020, 26, 506–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Liu, S.; Liu, J.; Zhang, Z.; Wan, X.; Huang, B.; Chen, Y.; Zhang, Y. COVID-19: Immunopathogenesis and Immunotherapeutics. Signal Transduct. Target. Ther. 2020, 5, 128. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Giamarellos-Bourboulis, E.J.; Netea, M.G.; Rovina, N.; Akinosoglou, K.; Antoniadou, A.; Antonakos, N.; Damoraki, G.; Gkavogianni, T.; Adami, M.E.; Katsaounou, P.; et al. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe 2020, 27, 992–1000. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Zhou, L.; Hu, Z.; Zhang, S.; Yang, S.; Tao, Y.; Xie, C.; Ma, K.; Shang, K.; Wang, W.; et al. Dysregulation of Immune Response in Patients with Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020, 71, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Soehnlein, O.; Steffens, S.; Hidalgo, A.; Weber, C. Neutrophils as protagonists and targets in chronic inflammation. Nat. Rev. Immunol. 2017, 17, 248–261. [Google Scholar] [CrossRef] [PubMed]
- Tomar, B.; Anders, H.J.; Desai, J.; Mulay, S.R. Neutrophils and Neutrophil Extracellular Traps Drive Necroinflammation in COVID-19. Cells 2020, 9, 1383. [Google Scholar] [CrossRef]
- Galani, I.E.; Andreakos, E. Neutrophils in viral infections: Current concepts and caveats. J. Leukoc. Biol. 2015, 98, 557–564. [Google Scholar] [CrossRef] [Green Version]
- Cavalcante-Silva, L.H.A.; Carvalho, D.C.M.; de Almeida Lima, É.; Galvão, J.G.F.M.; de França da Silva, J.S.; de Sales-Neto, J.M.; Rodrigues-Mascarenhas, S. Neutrophils and COVID-19: The road so far. Int. Immunopharmacol. 2021, 90, 107233. [Google Scholar] [CrossRef]
- Naumenko, V.; Turk, M.; Jenne, C.N.; Kim, S.J. Neutrophils in viral infection. Cell Tissue Res. 2018, 371, 505–516. [Google Scholar] [CrossRef]
- Johansson, C.; Kirsebom, F.C.M. Neutrophils in respiratory viral infections. Mucosal Immunol. 2021, 14, 815–827. [Google Scholar] [CrossRef] [PubMed]
- Meizlish, M.L.; Pine, A.B.; Bishai, J.D.; Goshua, G.; Nadelmann, E.R.; Simonov, M.; Chang, C.H.; Zhang, H.; Shallow, M.; Bahel, P.; et al. A neutrophil activation signature predicts critical illness and mortality in COVID-19. Blood Adv. 2021, 5, 1164–1177. [Google Scholar] [CrossRef] [PubMed]
- Dennison, D.; Al Khabori, M.; Al Mamari, S.; Aurelio, A.; Al Hinai, H.; Al Maamari, K.; Alshekaili, J.; Al Khadouri, G. Circulating activated neutrophils in COVID-19: An independent predictor for mechanical ventilation and death. Int. J. Infect. Dis. 2021, 106, 155–159. [Google Scholar] [CrossRef]
- Wang, Y.; Jönsson, F. Expression, Role, and Regulation of Neutrophil Fcγ Receptors. Front. Immunol. 2019, 10, 1958. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Boesen, C.C.; Radaev, S.; Brooks, A.G.; Fridman, W.H.; Sautes-Fridman, C.; Sun, P.D. Crystal Structure of the Extracellular Domain of a Human FcγRIII. Immunity 2000, 13, 387–395. [Google Scholar] [CrossRef] [Green Version]
- Leliefeld, P.H.C.; Koenderman, L.; Pillay, J. How neutrophils shape adaptive immune responses. Front. Immunol. 2015, 6, 471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCracken, J.M.; Allen, L.A.H. Regulation of Human Neutrophil Apoptosis and Lifespan in Health and Disease. J. Cell Death 2014, 7, 15. [Google Scholar] [CrossRef] [Green Version]
- Lawlor, N.; Nehar-Belaid, D.; Grassmann, J.D.S.; Stoeckius, M.; Smibert, P.; Stitzel, M.L.; Pascual, V.; Banchereau, J.; Williams, A.; Ucar, D. Single Cell Analysis of Blood Mononuclear Cells Stimulated Through Either LPS or Anti-CD3 and Anti-CD28. Front. Immunol. 2021, 12, 636720. [Google Scholar] [CrossRef]
- Shafran, N.; Shafran, I.; Ben-Zvi, H.; Sofer, S.; Sheena, L.; Krause, I.; Shlomai, A.; Goldberg, E.; Sklan, E.H. Secondary bacterial infection in COVID-19 patients is a stronger predictor for death compared to influenza patients. Sci. Rep. 2021, 11, 12703. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Farrell, J.M.; Zhao, C.Y.; Tarquinio, K.M.; Brown, S.P. Causes and Consequences of COVID-19-Associated Bacterial Infections. Front. Microbiol. 2021, 12, 682571. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Ling, Y.; Bai, T.; Xie, Y.; Huang, J.; Li, J.; Xiong, W.; Yang, D.; Chen, R.; Lu, F.; et al. COVID-19 with Different Severities: A Multicenter Study of Clinical Features. Am. J. Respir. Crit. Care Med. 2020, 201, 1380–1388. [Google Scholar] [CrossRef]
- Vaillancourt, M.; Jorth, P. The Unrecognized Threat of Secondary Bacterial Infections with COVID-19. MBio 2020, 11, e01806-20. [Google Scholar] [CrossRef] [PubMed]
- Metzger, D.W.; Sun, K. Immune dysfunction and bacterial coinfections following influenza. J. Immunol. 2013, 191, 2047–2052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCullers, J.A. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat. Rev. Microbiol. 2014, 12, 252–262. [Google Scholar] [CrossRef]
- Reusch, N.; De Domenico, E.; Bonaguro, L.; Schulte-Schrepping, J.; Baßler, K.; Schultze, J.L.; Aschenbrenner, A.C. Neutrophils in COVID-19. Front. Immunol. 2021, 12, 952. [Google Scholar] [CrossRef]
- Chua, R.L.; Lukassen, S.; Trump, S.; Hennig, B.P.; Wendisch, D.; Pott, F.; Debnath, O.; Thürmann, L.; Kurth, F.; Völker, M.T.; et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020, 38, 970–979. [Google Scholar] [CrossRef]
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F.; et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020, 26, 842–844. [Google Scholar] [CrossRef]
- Schulte-Schrepping, J.; Reusch, N.; Paclik, D.; Baßler, K.; Schlickeiser, S.; Zhang, B.; Krämer, B.; Krammer, T.; Brumhard, S.; Bonaguro, L.; et al. Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment. Cell 2020, 182, 1419–1440. [Google Scholar] [CrossRef]
- Wilk, A.J.; Rustagi, A.; Zhao, N.Q.; Roque, J.; Martínez-Colón, G.J.; McKechnie, J.L.; Ivison, G.T.; Ranganath, T.; Vergara, R.; Hollis, T.; et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020, 26, 1070–1076. [Google Scholar] [CrossRef]
- Aschenbrenner, A.C.; Mouktaroudi, M.; Krämer, B.; Oestreich, M.; Antonakos, N.; Nuesch-Germano, M.; Gkizeli, K.; Bonaguro, L.; Reusch, N.; Baßler, K.; et al. Disease severity-specific neutrophil signatures in blood transcriptomes stratify COVID-19 patients. Genome Med. 2021, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Zhao, Y.; Li, J.; Liu, J.; Yang, X.; Guo, X.; Kuang, M.; Xia, H.; Zhang, Z.; Cao, L.; et al. Induction of alarmin S100A8/A9 mediates activation of aberrant neutrophils in the pathogenesis of COVID-19. Cell Host Microbe 2021, 29, 222–235. [Google Scholar] [CrossRef] [PubMed]
- Silvin, A.; Chapuis, N.; Dunsmore, G.; Goubet, A.G.; Dubuisson, A.; Derosa, L.; Almire, C.; Hénon, C.; Kosmider, O.; Droin, N.; et al. Elevated Calprotectin and Abnormal Myeloid Cell Subsets Discriminate Severe from Mild COVID-19. Cell 2020, 182, 1401–1418. [Google Scholar] [CrossRef] [PubMed]
- Saheb Sharif-Askari, N.; Saheb Sharif-Askari, F.; Ahmed, S.B.M.; Hannawi, S.; Hamoudi, R.; Hamid, Q.; Halwani, R. Enhanced Expression of Autoantigens During SARS-CoV-2 Viral Infection. Front. Immunol. 2021, 12, 2271. [Google Scholar] [CrossRef]
- Ye, Q.; Wang, B.; Mao, J. The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID-19. J. Infect. 2020, 80, 607–613. [Google Scholar] [CrossRef]
- Parackova, Z.; Zentsova, I.; Bloomfield, M.; Vrabcova, P.; Smetanova, J.; Klocperk, A.; Mesežnikov, G.; Casas Mendez, L.F.; Vymazal, T.; Sediva, A. Disharmonic Inflammatory Signatures in COVID-19: Augmented Neutrophils’ but Impaired Monocytes’ and Dendritic Cells’ Responsiveness. Cells 2020, 9, 2206. [Google Scholar] [CrossRef]
- Didangelos, A. COVID-19 Hyperinflammation: What about Neutrophils? mSphere 2020, 5, e00367-20. [Google Scholar] [CrossRef]
- Arnhold, J. The Dual Role of Myeloperoxidase in Immune Response. Int. J. Mol. Sci. 2020, 21, 8057. [Google Scholar] [CrossRef]
- Veras, F.P.; Pontelli, M.C.; Silva, C.M.; Toller-Kawahisa, J.E.; de Lima, M.; Nascimento, D.C.; Schneider, A.H.; Caetité, D.; Tavares, L.A.; Paiva, I.M.; et al. SARS-CoV-2–triggered neutrophil extracellular traps mediate COVID-19 pathology. J. Exp. Med. 2020, 217, e20201129. [Google Scholar] [CrossRef]
- Zuo, Y.; Yalavarthi, S.; Shi, H.; Gockman, K.; Zuo, M.; Madison, J.A.; Blair, C.; Weber, A.; Barnes, B.J.; Egeblad, M.; et al. Neutrophil extracellular traps in COVID-19. JCI Insight 2020, 5, e138999. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.Y.; Wang, X.M.; Xing, X.; Xu, Z.; Zhang, C.; Song, J.W.; Fan, X.; Xia, P.; Fu, J.L.; Wang, S.Y.; et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 2020, 21, 1107–1118. [Google Scholar] [CrossRef]
- Arunachalam, P.S.; Wimmers, F.; Mok, C.K.P.; Perera, R.A.P.M.; Scott, M.; Hagan, T.; Sigal, N.; Feng, Y.; Bristow, L.; Tsang, O.T.Y.; et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 2020, 369, 1210–1220. [Google Scholar] [CrossRef]
- Kamenyeva, O.; Boularan, C.; Kabat, J.; Cheung, G.Y.C.; Cicala, C.; Yeh, A.J.; Chan, J.L.; Periasamy, S.; Otto, M.; Kehrl, J.H. Neutrophil recruitment to lymph nodes limits local humoral response to Staphylococcus aureus. PLoS Pathog. 2015, 11, e1004827. [Google Scholar] [CrossRef]
- Gelderman, K.A.; Hultqvist, M.; Holmberg, J.; Olofsson, P.; Holmdahl, R. T cell surface redox levels determine T cell reactivity and arthritis susceptibility. Proc. Natl. Acad. Sci. USA 2006, 103, 12831–12836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malmberg, K.-J.; Arulampalam, V.; Ichihara, F.; Petersson, M.; Seki, K.; Andersson, T.; Lenkei, R.; Masucci, G.; Pettersson, S.; Kiessling, R. Inhibition of activated/memory (CD45RO+) T cells by oxidative stress associated with block of NF-κB activation. J. Immunol. 2001, 167, 2595–2601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagunas-Rangel, F.A. Neutrophil-to-lymphocyte ratio and lymphocyte-to-C-reactive protein ratio in patients with severe coronavirus disease 2019 (COVID-19): A meta-analysis. J. Med. Virol. 2020, 92, 1733–1734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, S.; Cai, X.; Wang, H.; He, G.; Lin, Y.; Lu, B.; Chen, C.; Pan, Y.; Hu, X. Abnormalities of peripheral blood system in patients with COVID-19 in Wenzhou, China. Clin. Chim. Acta 2020, 507, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Barnes, B.J.; Adrover, J.M.; Baxter-Stoltzfus, A.; Borczuk, A.; Cools-Lartigue, J.; Crawford, J.M.; Daßler-Plenker, J.; Guerci, P.; Huynh, C.; Knight, J.S.; et al. Targeting potential drivers of COVID-19: Neutrophil extracellular traps. J. Exp. Med. 2020, 217, e20200652. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muralidharan, A.; Wyatt, T.A.; Reid, S.P. SARS-CoV-2 Dysregulates Neutrophil Degranulation and Reduces Lymphocyte Counts. Biomedicines 2022, 10, 382. https://doi.org/10.3390/biomedicines10020382
Muralidharan A, Wyatt TA, Reid SP. SARS-CoV-2 Dysregulates Neutrophil Degranulation and Reduces Lymphocyte Counts. Biomedicines. 2022; 10(2):382. https://doi.org/10.3390/biomedicines10020382
Chicago/Turabian StyleMuralidharan, Abenaya, Todd A. Wyatt, and St Patrick Reid. 2022. "SARS-CoV-2 Dysregulates Neutrophil Degranulation and Reduces Lymphocyte Counts" Biomedicines 10, no. 2: 382. https://doi.org/10.3390/biomedicines10020382
APA StyleMuralidharan, A., Wyatt, T. A., & Reid, S. P. (2022). SARS-CoV-2 Dysregulates Neutrophil Degranulation and Reduces Lymphocyte Counts. Biomedicines, 10(2), 382. https://doi.org/10.3390/biomedicines10020382






