Abstract
Almost twenty years ago, the importance of the paracaspase MALT1 in antigen receptor-induced NF-κB activation was first described. Since then, several other immune receptors, G-protein-coupled receptors, and receptor tyrosine kinases were identified as relying on MALT1 to induce NF-κB activation. In various hematological malignancies and solid tumors, MALT1 is constitutively activated and drives chronic NF-κB target gene expression. Deregulated MALT1 activity in cancer thus promotes tumor cell survival, proliferation, and metastasis. Since the molecular function of MALT1 partially requires its protease activity, pharmacological targeting of MALT1 may represent a promising anti-cancer strategy. Here, we review the molecular features of MALT1 activation and function as well as the therapeutic potential of MALT1 inhibition in hematological malignancies and solid tumors.
1. Introduction
The transcription factor NF-κB promotes cancer development on various levels, e.g., by enhancing cell survival and proliferation, the acquisition of cancer stem cell properties, metastasis formation, metabolic reprogramming, and the suppression of anti-tumor immune responses [1,2]. Thus, targeting the activity of NF-κB has emerged as a promising anti-cancer strategy. However, a major setback of this therapeutic approach has become evident in mouse models lacking the expression of central regulators that are important for the general activation of the classical NF-κB pathway. Mice deficient in signaling components, such as the inhibitor of κB kinase (IKK) complex, are not viable and exhibit increased hepatocyte apoptosis, immune deregulation, as well as skeletal and epidermal defects, indicating an essential role of NF-κB in tissue homeostasis [3]. Accordingly, clinical trials of several IKK inhibitors have been discontinued due to unfavorable safety profiles [4]. One possibility to circumvent the complications of a general NF-κB inhibition is to focus on specific upstream regulators that can induce NF-κB activation but are not essential for the survival of the organism. One of these NF-κB regulators is the mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1) which has recently emerged as an interesting therapeutic target. Here, we will summarize what is known about the molecular function of MALT1 and how it is activated in hematological malignancies and solid cancers, and discuss the therapeutic potential of MALT1 inhibitors as well as their impact on immunotherapy.
2. MALT1 Is Activated by CARD-CC Proteins
Proteins that contain a caspase activation and recruitment domain (CARD) and a coiled-coil (CC) domain are critical for the activation of MALT1. So far, four CARD-CC proteins, which differ in their tissue expression profiles, have been described. CARD9 is mainly expressed in cells of the myeloid lineage, such as macrophages or neutrophils, and links multiple innate immune receptors, such as TREM1 or Dectin-1, to NF-κB activation [5,6,7]. Lymphocytes express the CARD-CC member CARD11 (also known as CARMA1), which transmits signals from the B-cell, T-cell, or activating Natural Killer cell receptors to MALT1 [5,8,9,10]. In contrast, the expression of CARD10 and CARD14 is mainly restricted to non-hematopoietic tissues. Whereas CARD14 (also known as CARMA2) is predominantly expressed in keratinocytes and involved in the NF-κB activation downstream of pattern recognition receptors (PRR), such as Dectin-1, CARD10 (also known as CARMA3) is expressed in several tissues (e.g., intestine, heart, and kidney) and connects some G-protein-coupled receptors (GPCRs) and receptor tyrosine kinases (RTKs) to MALT1 and NF-κB activation [5,11,12,13,14,15,16]. Even though the molecular events involved in T-cell receptor-mediated CARD11 activation have been elucidated in great detail, it is not completely understood how other immune receptors, RTKs, and GPCRs promote the activation of the different CARD-CC proteins. However, it has been proposed that, due to the homology of the CARD-CC members, major steps in their activation might be conserved [5,17]. An intramolecular interaction between the CARD, the CC domain, and a flexible linker region keeps the CARD-CC proteins in an inactive state [18,19]. This auto-inhibitory conformation is disrupted by a protein kinase C (PKC)-mediated phosphorylation of the linker region, which induces a structural constraint on this flexible domain of CARD11 [20,21]. The open conformation of phosphorylated CARD11 then allows oligomerization driven by the coiled-coil domain and the CARD-mediated recruitment of B-cell lymphoma 10 (BCL10) [5,8,22,23]. Oligomerized CARD11 serves as a platform that allows the polymerization of BCL10, leading to the formation of helical BCL10 filaments [24,25]. As BCL10 constitutively interacts with MALT1, its recruitment also translocates MALT1 to activated CARD11 oligomers and brings individual MALT1 molecules into close proximity. MALT1 possesses two distinct molecular functions promoting the activation of NF-κB. On the one hand, its protease activity acquired in the context of the CARD-BCL10-MALT1 (CBM) complex is important for the optimal activation of NF-κB (please see the following chapter). On the other hand, together with BCL10 it serves as a scaffold to recruit E3 ubiquitin ligases, such as TRAF6 and the linear ubiquitin chain assembly complex (LUBAC), resulting in K63- and M1-linked ubiquitination of BCL10 and MALT1 [26,27,28,29] (Figure 1). These polyubiquitin chains serve as docking sites for two distinct ubiquitin-binding complexes: The IKK complex is recruited via the ubiquitin-binding domain of the subunit IKKγ (also known as NEMO), whereas the transforming-growth-factor-β-activated kinase 1 (TAK1) associates with polyubiquitin chains via the adapter TAK-binding protein 2 (TAB2) [26,27,29,30]. Through ubiquitination of IKKγ and TAK1-mediated phosphorylation of IKKβ, the IKK complex becomes activated and subsequently phosphorylates the inhibitor of κB (IκB), causing its proteasomal degradation [31,32,33]. This allows the transient nuclear translocation of NF-κB members that in turn drive target gene transcription. Taken together, various receptors, such as the antigen receptors or PRRs, promote the formation of a CBM complex, and thus the activation of classical NF-κB transcription factors.
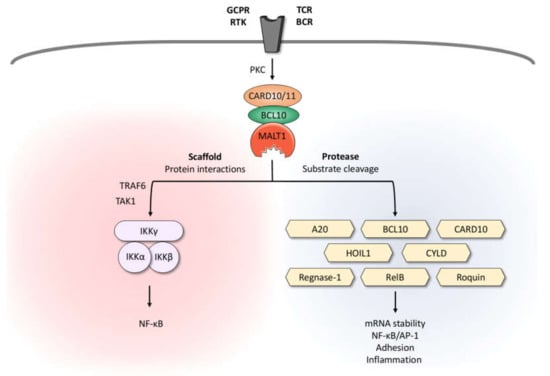
Figure 1.
Dual role of MALT1: Scaffold and protease function. Upon triggering of antigen receptors, various GPCRs and RTKs, MALT1 is activated in the context of a high-molecular-weight complex comprising BCL10 and CARD10 in non-hematopoietic or CARD11 in hematopoietic cells, respectively. By recruiting the IKK complex, MALT1 serves as a scaffold to promote optimal activation of the transcription factor NF-κB. Due to its protease activity, MALT1 cleaves various substrates involved in mRNA stability, regulation of NF-κB/AP-1 signaling, adhesion, and inflammation. BCL10, B-cell lymphoma/leukemia 10; BCR, B-cell receptor; CARD10/11, caspase recruitment domain-containing protein 10/11; GPCR, G-protein-coupled receptor; HOIL1, heme-oxidized IRP2 ubiquitin ligase 1; IKK, inhibitor of κB kinase; MALT1, mucosa-associated lymphoid tissue lymphoma translocation protein 1; PKC, protein kinase C; RTK, receptor tyrosine kinase; TAK1, transforming growth factor-β-activated kinase 1; TCR, T-cell receptor; TRAF6, TNF receptor associated factor 6.
3. MALT1 Protease Function
The central caspase-like domain of MALT1 has been bioinformatically identified due to its homology to the protease domains of caspases and it is responsible for the protease activity of MALT1 [34]. Whereas in non-activated T cells, TRAF6 and GRK2 suppress the protease activity of MALT1, monoubiquitination and dimerization activate MALT1 protease function, leading to the cleavage of selected substrates [35,36,37,38]. In contrast to caspases, which cleave their substrates after the acidic amino acid aspartate, MALT1 cleaves proteins after the positively charged arginine [39,40]. The MALT1 substrates identified so far can be divided into three groups. The first group comprises proteins that affect the activation of the transcription factors NF-κB and activator protein 1 (AP-1). MALT1-mediated inactivation of the deubiquitinating enzymes A20 and CYLD protects K63-linked polyubiquitin chains from degradation and thus sustains NF-κB and AP-1 activation [40,41]. The MALT1-dependent cleavage of the NF-κB member RelB leads to its rapid degradation by the proteasome, thus freeing the NF-κB subunits RelA and c-Rel from transcriptionally inactive complexes [42]. Interestingly, MALT1-mediated cleavage of the LUBAC member HOIL1 might represent a negative feedback loop that limits CBM-dependent NF-κB activation [43,44,45]. The second group of MALT1 substrates includes proteins that regulate the stability and the translation of selected transcripts. Regnase 1, roquin 1, and roquin 2 control a variety of inflammation-related mRNAs, such as NFKBIZ (coding for the atypical IκB member IκBζ), IL6, TNFA, ICOS, and TNFAIP3 (coding for A20). MALT1-mediated inactivation of regnase 1 and roquin 1/2 abrogates their capacity to suppress mRNA translation and thus promotes the expression of the immunoregulatory transcripts [46,47]. The third group of MALT1 substrates contains members of the CBM complex itself. MALT1 auto-proteolysis promotes NF-κB activity, even though the molecular basis of this observation is not yet fully understood [48]. In contrast, MALT1-mediated BCL10 cleavage is dispensable for NF-κB activation, but promotes T-cell receptor-induced cell adhesion to fibronectin [39].
Since MALT1 protease activity has been shown to control glutamine uptake by ASCT2 and glutaminolysis in various cell types, MALT1 also appears to be a key regulator of glutamine metabolism, suggesting that there are so far uncharacterized MALT1 substrate(s) [49,50,51].
4. MALT1 in Hematological Malignancies
Since numerous reviews have highlighted the role of MALT1 in hematological malignancies, this review mainly focuses on the importance of MALT1 in solid tumors and only briefly summarizes its role in lymphoma and leukemia development [52,53,54]. As its name implies, MALT1 was first described in mucosa-associated lymphoid tissue (MALT) lymphoma, in which the MALT1 gene is often subject to translocations and thus was suspected to be a driver of lymphomagenesis [55]. At present, it is broadly accepted that constitutive activation of MALT1 by either genetic alterations, chronic antigen receptor signaling, overexpression, or viral proteins drives the development of several hematological malignancies.
MALT1 overexpression has been reported mainly in MALT lymphoma, but it can also be observed in other malignancies, such as B-cell acute lymphoblastic leukemia (B-ALL) and diffuse large B-cell lymphoma (DLBCL) [56,57,58,59,60]. Overexpression of MALT1 is caused either by amplification of its gene locus or by the translocation t(14;18)(q32;q21), which brings the MALT1 gene under the control of the IgG heavy chain promoter [57,59]. The oncogenic potential of MALT1 has been demonstrated in a mouse model that allows specific MALT1 expression in hematopoietic stem/progenitor cells which results in the development of MALT lymphoma [61]. Additional deletion of the tumor suppressor p53 accelerated tumor onset and resulted in an aggressive form of DLBCL, highlighting MALT1′s potential to promote B-cell survival and proliferation [61]. Similarly, the overexpression of BCL10, which induces MALT1 activation, also provokes lymphomagenesis in mice [62]. Mutations of CARD11, which can be found in an aggressive subtype of DLBCL, Sézary syndrome, angioimmunoblastic T-cell lymphoma (AITL), and adult T-cell leukemia/lymphoma (ATLL), disrupt the auto-inhibition of CARD11 and drive its receptor-independent oligomerization, thus activating MALT1 [63,64,65,66].
One third of MALT lymphomas harbor a translocation (i.e., t(11;18)(q21;q21)) which fuses the N-terminal part of cellular inhibitor of apoptosis protein 2 (cIAP2, also known as API2) to the C-terminus of MALT1 [55,67]. The cIAP2-MALT1 fusion protein is able to oligomerize independently of BCL10 and upstream signals and, through TRAF6 recruitment and MALT1 protease activation, induce NF-κB-dependent transcription [40,68,69]. Strikingly, due to cIAP2-mediated recruitment, two non-canonical MALT1 substrates are processed by the cIAP2-MALT1 fusion protein, i.e., the NF-κB-inducing kinase (NIK) and the LIM domain and actin-binding protein 1 (LIMA1). NIK cleavage results in an increased stability of its C-terminal fragment and in the activation of noncanonical NF-κB signaling, whereas LIMA1 processing converts the tumor suppressor into an oncogene [70,71].
Besides genetic alterations of components of the CBM signalosome, constitutive antigen receptor signaling is a major driver for chronic MALT1 activation and thus for lymphoma development. Chronic active B-cell receptor signaling is caused by several distinct molecular mechanisms. In DLBCL, self-antigens that are either present on the BCR itself or in apoptotic debris of the tumor cells have been proposed to promote the activation of the lymphoma cells [72]. In chronic lymphocytic leukemia (CLL), CBM activation can be driven by BCR-derived signals that are antigen-independent but rely on the recognition of an internal epitope within the BCR [73]. Moreover, antigens deriving from persistent chronic infections with viral or bacterial pathogens, such as the hepatitis C virus, can promote lymphoma development by sustained B-cell activation and proliferation [74,75].
Interestingly, some viral proteins are also able to promote the activation of NF-κB via MALT1 in a BCR-independent manner. Latent infections with Kaposi’s sarcoma-associated herpes virus (KSHV) are correlated with the development of primary effusion lymphoma (PEL). The KSHV proteins K13 and K15 promote MALT1 activity and, therefore, regulate viral latency and the growth of PEL cells [76].
In other lymphomas, such as mantle cell lymphoma (MCL), chronic activation of BCR signaling and MALT1 activity have been detected, but the molecular cause currently remains elusive [77]. Even though MALT1 exhibits two distinct molecular functions that lead either to IKK activation (scaffold function) or to the cleavage of selected substrates (protease function), the inhibition of MALT1 protease activity is sufficient to reduce tumor cell proliferation and apoptosis resistance in DLBCL, MCL, PEL, ATLL, MALT, and T-cell acute lymphoblastic leukemia (T-ALL), highlighting the therapeutic potential of MALT1 inhibitors in these hematological malignancies [70,76,77,78,79,80,81].
5. The Role of the CBM Complex in Solid Tumors
Whereas the importance of the CBM signalosome in the pathogenesis of lymphoid malignancies has been acknowledged for years, recent studies now highlight CARD10-mediated MALT1 activation in solid tumors.
Amongst the tumor types in which NF-κB is activated through the CARD10-BCL10-MALT1 (C10BM) signalosome to promote tumorigenesis, there is a subset of angiotensin II receptor 1 (AGTR1)-positive luminal breast cancers with poor prognosis [82]. A recent study has provided evidence to support the existence of an AGTR1-C10BM-NF-κB signaling axis driving breast cancer (BC) cell-intrinsic and extrinsic responses. Gene expression dataset analysis has revealed a strong upregulation of the GPCR family member AGTR1 that correlates with an NF-κB gene signature in a subset of luminal breast cancers. Even though AGTR1 overexpression alone triggers considerable NF-κB activation, as shown by RelA nuclear translocation, the presence of angiotensin II (AGTR1′s ligand) further enhances NF-κB activity more than 100-fold and leads to the development of an aggressive phenotype. Interestingly, silencing of any constituent of the C10BM complex completely abrogates the angiotensin II-induced NF-κB response and thus blocks cell-intrinsic proliferative and invasive properties, as well as cell-extrinsic effects, such as the stimulation of endothelial cells of the tumor microenvironment to promote tumor angiogenesis [82].
Similarly, in proteinase-activated receptor 1 (PAR1)-driven tumors, both inhibition and silencing of MALT1 impairs the expression of NF-κB target genes, such as IL1B, CXCL8, and MMP9, each associated with the manifestation of malignant features in breast cancer and osteosarcoma (OS) [83]. Accordingly, the number and size of metastatic tumor nodules found in the lungs of nude mice injected with MALT1-deficient MCF7-N55 breast cancer cells are greatly attenuated compared to controls. These results highlight the role of the C10BM signalosome in the acquisition of pro-tumorigenic traits in PAR-1-dependent tumor entities through activation of NF-κB. Additionally, a recent study has underlined the role of MALT1 downstream of AGTR1 and PAR1 in the induction of triple-negative breast cancer (TNBC) subtype switching [84]. Typical estrogen receptor (ER)-positive luminal breast cancers that overexpress either AGTR1 or PAR1 exhibit NF-κB-driven epithelial-mesenchymal transition (EMT)-associated alterations, such as increased expression of Snail, ZEB1, vimentin, and N-cadherin, downregulation of E-cadherin, as well as increased cell invasion and migration—all of which are abrogated upon silencing or pharmacological inhibition of MALT1 [84]. Altogether, this reveals an important role of the MALT1 protease in the induction of the EMT program in GPCR-positive TNBCs.
In the context of a GPCR-activated C10BM signalosome, a study in ovarian cancer (OC) cells has uncovered a lysophosphatidic acid (LPA) receptor-PKCα-C10BM signaling axis that induces tumor progression via activation of NF-κB and urokinase plasminogen activator (uPA) [85]. In brief, lysophosphatidic acid (LPA) binds the GPCR LPAR1, which results in Ras activation. Ras subsequently activates the PKCα isoform, thus inducing the assembly of the C10BM signalosome and MALT1 activation. This PKC-C10BM signaling axis seems to be essential for NF-κB-dependent pro-tumorigenic features, such as invasion and migration, since silencing or inhibition of MALT1 completely abrogates NF-κB activation, uPA upregulation, and the appearance of malignant traits [85].
As mentioned above, C10BM complex assembly can also occur downstream of RTKs. Along this line, epidermal growth factor receptor (EGFR), a member of the RTK family, is aberrantly activated in various aggressive tumors including glioblastoma multiforme (GBM) and lung cancer. In these tumor entities, deregulation of the EGFR-NF-κB signaling pathway contributes to several important malignant properties of the tumor cells. Recent studies have now identified the C10BM complex as the central signaling hub through which EGFR ligation induces NF-κB activation. In this regard, Liu et al. have reported that EGF-dependent IκBα degradation and subsequent nuclear translocation of NF-κB are dependent on the expression of MALT1 [86]. Thus, MALT1 appears to be crucial for the maintenance of the proliferative and clonogenic potential of various GBM cell lines, and its silencing or pharmacological inhibition significantly reduces the invasive and migratory potential of the tumor cells both in vitro and in vivo [86]. Consistent with these results, a study by Pan et al. further supports a role for MALT1 in EGFR-mediated NF-κB activation [87]. The authors have demonstrated that the silencing of either CARD10 or MALT1 in EGFR-overexpressing lung cancer cells correlates with reduced NF-κB activity, cancer cell proliferation, migration, survival, and tumor burden in vivo. Intriguingly, contrary to what has been observed in the GBM model, the protease activity of MALT1 is largely dispensable for EGFR-induced NF-κB activation in lung carcinoma, since the reconstitution of MALT1-silenced cells with a protease-deficient mutant MALT1 (MALT1C464A) fully restores NF-κB signaling. Furthermore, knockdown of the MALT1 interaction partner TRAF6 abolishes NF-κB activation, pinpointing the importance of MALT1 as a scaffold protein to bridge TRAF6 to the IKK complex in response to EGF in this tumor entity. Interestingly, this study has also provided genetic evidence indicating that STAT3 activation in lung carcinoma is controlled by EGFR-MALT1-dependent NF-κB activation via induction of IL-6 expression.
Another example of an RTK that activates NF-κB via the C10BM signalosome is the human epidermal growth factor receptor 2 (Her2) protein, a member of the EGF receptor subclass that is frequently overexpressed in breast cancer [88]. Silencing of either one of the CBM complex members in Her2-overexpressing cells significantly reduces Her2-induced NF-κB activation. To determine the relationship between Her2 expression and C10BM activation, Pan et al. treated SKBR3 breast cancer cells with a PKC inhibitor. Interestingly, PKC inhibition is sufficient to completely abolish NF-κB activation, suggesting PKC as the functional link between Her2 overexpression and C10BM-induced NF-κB signaling. As observed for other tumor entities, the C10BM-NF-κB axis is responsible for the induction of tumorigenic properties in SKBR3 breast cancer cells, including proliferation, clonogenic survival, invasion, and migration [88]. In this context, invasion and migration have been shown to be promoted by the NF-κB-dependent expression of the matrix-remodeling enzymes MMP1 and 13. Consistent with what has been observed in other studies, injection of CARD10-silenced SKBR3 cells into nude mice leads to the formation of significantly smaller tumors and a reduced number of lung metastatic sites [88]. Furthermore, several other studies have identified MALT1 as a central regulator of NF-κB-mediated cancer progression. Both pancreatic ductal adenocarcinoma (PDAC) and prostate cancer (PCa) cells show increased MALT1 expression that contributes to tumor malignancy and metastasis [89,90]. Silencing or pharmacological inhibition of MALT1 reduces the proliferative and invasive potential of the tumor cells both in vitro and in vivo.
In malignant melanoma (MM) and cholangiocarcinoma (CCA), MALT1 has been reported to be overexpressed and thus to contribute to survival and metastasis [91,92]. A study by Wang et al., on melanoma proposes that the elevated MALT1 levels increase the TNFα- and TRAIL-induced NF-κB and JNK/AP-1 activation, thus promoting melanoma growth and pulmonary metastasis in vivo [91]. Interestingly, using the multi-kinase inhibitor regorafenib in intrahepatic CCA, the Raf/Erk/Elk-1 pathway has been found to modulate MALT1 expression [92]. The transcription factor Elk-1 directly binds to the MALT1 promoter, thus contributing to MALT1 overexpression in the tumor cells. Accordingly, regorafenib treatment of CCA cells not only decreases MALT1 levels, but also reduces the expression of NF-κB target genes, such as IL1B and CXCL8, which correlate with CCA cell growth and survival [92].
Although the expression and activity of MALT1 and NF-κB are tightly controlled, some tumor cells can override these negative regulatory mechanisms to favor tumor progression. For instance, glioblastomas can transition from the more benign proneural (PN) subtype to the more aggressive mesenchymal (MES) subtype by downregulating the expression of the microRNA (miRNA) miR-181d [93]. It has been observed that in PN GBM, miR-181d can directly bind to the 3′-untranslated region (UTR) of MALT1 transcripts, thereby reducing its mRNA and protein expression levels. This in turn results in a decrease in NF-κB transcriptional activity, as observed by the reduction in IL-6 secretion, as well as in a suppression of glioma cell proliferation, invasion, migration, and angiogenesis. The high-grade MES GBM can downregulate miR-181d, thus increasing MALT1-dependent NF-κB activation and enhancing tumor burden and aggressiveness [93]. Likewise, TIFA (TNF receptor associated factor-interacting protein with a forkhead-associated domain), which can sensitize hepatocellular carcinoma (HCC) cells to apoptosis, competes with MALT1 for TRAF6 binding [94]. A recent study has found that TIFA is downregulated in HCC cells, allowing free access of MALT1 to TRAF6, thus driving tumorigenesis through NF-κB activation.
Whereas most studies describe a tumor-promoting role for MALT1, a recent study suggests that MALT1 can also act as a tumor suppressor. In the lung cancer cell line A549, MALT1 cleaves the C10BM complex member CARD10 after the arginine residue at position 587, thereby dampening its capacity to activate NF-κB [95]. In this manner, the cleavage of CARD10 by MALT1 acts as a negative feedback mechanism that limits the signaling potential and pro-tumorigenic role of CARD10. This is also reflected in the observation that A549 knock-in (KI) cells expressing the cleavage-deficient R587A CARD10 variant exhibit a growth advantage, increased angiogenesis, and tumorigenic potential upon i.v. injection in mice [95]. The authors identified IL-6 as the main cytokine upregulated in the A549-KI cells, suggesting that cleavage of CARD10 by MALT1 is important to limit IL-6 signaling and to suppress tumorigenesis.
Collectively, these studies highlight the role of the C10BM signalosome as a critical signaling node for the pathogenic activation of NF-κB. MALT1 activity controls the expression of several cytokines and mediators that contribute to tumorigenesis by promoting proliferation, survival, invasion, migration, angiogenesis, and metastasis (Table 1). Thus, pharmacological targeting of MALT1 could represent a potential strategy in the treatment of solid tumors.

Table 1.
Importance of MALT1 in different tumor types. AGTR1, angiotensin II receptor 1; ATL, adult T-cell lymphoma; B-ALL, B-cell acute lymphoblastic leukemia; BC, breast cancer; BCR, B-cell receptor; CARD11, caspase recruitment domain-containing protein 11; CCA, cholangiocarcinoma; CLL, chronic lymphocytic leukemia; DLBCL, diffuse large B-cell lymphoma; EGFR, epidermal growth factor receptor; GBM, glioblastoma; HER2, human epidermal growth factor receptor 2; HTLV-1; human T-cell lymphotropic virus type 1; KSHV, Karposi’s sarcoma-associated herpesvirus; LPAR, lysophosphatidic acid receptor; MALT1, mucosa-associated lymphoid tissue lymphoma translocation protein 1; MALT lymphoma; mucosa-associated lymphoid tissue lymphoma; MCL mantle cell lymphoma; MM, malignant melanoma; NSCLC, non-small cell lung cancer; OC, ovarian cancer; OS, osteosarcoma; PAR1, protease-activated receptor 1; PCa, prostate cancer; PDAC, pancreatic ductal adenocarcinoma; PEL, primary effusion lymphoma; T-ALL, T-cell acute lymphoblastic leukemia; TRAIL, tumor necrosis-factor-related apoptosis-inducing ligand.
6. MALT1 as a Potential Therapeutic Target in Solid Tumors
6.1. Effects of MALT1 Inhibition on Tumor Cells
Constitutive MALT1 activity has been found to drive proliferation, survival, and metastasis in several tumor entities, such as lymphoid malignancies and solid tumors, and it is considered a crucial player in persistent NF-κB activation. The important role of the C10BM complex in the development of malignancy features in non-hematological tumors suggests a potential therapeutic benefit by targeting one of the complex members. Since MALT1 is the only constituent of the C10BM complex that exhibits enzymatic activity and since several protease inhibitors have already been developed, as recently summarized by Hamp et al., MALT1 represents the most suitable target in solid tumors [98]. Indeed, the use of either allosteric or active site MALT1 inhibitors has revealed significant anti-tumor effects in the treatment of solid tumors both in vitro and in vivo. For instance, treatment of metastatic melanoma, cholangiocarcinoma, and glioblastoma cells with the widely used active site MALT1 inhibitor MI-2 abrogates proliferation, migration, and invasion, reduces cell viability, and diminishes tumor growth and metastasis in vivo via downregulation of the NF-κB pathway [86,91,92]. Similarly, PAR1-driven metastasis-associated gene expression in breast cancer and osteosarcoma cell lines can be impaired by treatment with MI-2 and the suicide inhibitor z-VRPR-fmk [83]. Additionally, MI-2 abrogates PAR1-driven invasion of breast cancer cells [83]. However, in the context of melanoma, MALT1 knockdown also suppresses activation of the JNK/AP-1 signaling pathway, indicating that the cellular effects observed upon MALT1 protease inhibition might not be attributable solely to the specific downregulation of NF-κB. Past studies have shown that in T cells, JNK phosphorylation depends on MALT1′s scaffolding function, indicating that the manifestation of some malignant traits might not be affected by targeting MALT1 protease function [39,99]. Allosteric MALT1 inhibitors, such as mepazine or thioridazine, have been used to treat TNBC and PDAC cells. The effects observed with these inhibitors include a reduction in the proliferative, invasive, and migratory capacities of the tumor cells, as well as an inhibition of primary tumor growth and metastatic burden in vivo. Conversely, the use of the active site inhibitor z-VRPR-fmk has no effect on lung carcinoma cells, since NF-κB activation and the development of tumorigenic features in this tumor type is regulated by MALT1′s scaffold function. In cases in which the protease function of MALT1 is dispensable, the development of inhibitors or biological entities that prevent either CARD10 oligomerization, C10BM complex formation, or TRAF6 recruitment would be inevitable [100].
6.2. Priming of Solid Tumors via MALT1 Inhibition
A promising new line of therapeutic strategy employing MALT1 inhibitors in solid tumors is based on their effect on regulatory T cells (Tregs). Mice expressing a mutant MALT1 that lacks protease activity exhibit a reduced frequency of natural regulatory T cells and suffer from severe autoimmunity [99,101,102]. Recent studies have demonstrated that the C11BM complex is important for the conversion of resting into effector Tregs, c-Myc-mediated Treg expansion and the immunosupressive effects of Tregs [103,104]. Moreover, conditional deletion of CARD11 exclusively in Foxp3-positive cells is fatal and associated with a multi-organ inflammatory disease [105]. Excitingly, incomplete inhibition of the C11BM complex either by reducing CARD11 expression or by pharmacological inhibition of MALT1 prevents lethal autoimmune reactions, but converts tumor-infiltrating Treg cells into IFNγ-secreting effector T cells [105]. This was sufficient to reduce the growth of the melanoma cell lines D4M.3A and B16F1 in mouse xenograft models [103,105]. Since the increased IFNγ production during incomplete C11BM inhibition provokes PD-L1 expression in the melanoma cells, the combination of MALT1 inhibitors and anti-PD1 immune checkpoint therapy exhibits synergistic effects in the inhibition of tumor growth [105]. Whether it is possible to achieve a MALT1 inhibition strong enough to boost the immune response by inhibiting Treg expansion and function, while avoiding severe systemic autoimmunity at the same time, is still under debate [106,107]. Nevertheless, it is tempting to speculate that a short-term application of MALT1 inhibitors in combination with immune checkpoint inhibitors could support the T cell-mediated anti-tumor reaction and thus prove beneficial for cancer patients.
Collectively, the inhibition of the MALT1 protease appears to represent a promising treatment strategy for solid tumors, if properly balanced. Potentially, MALT1 inhibition could be used to simultaneously target both tumor cells and suppressive immune cells (Figure 2). In CBM-dependent tumor cells, MALT1 inhibition directly acts on their proliferative and invasive potential, while, at the same time, it destabilizes the Treg-mediated immunosuppression. However, it is necessary to conduct further studies to adequately adjust the dosage and timing of these inhibitors for the treatment of solid tumors to prevent unwanted side effects, as well as to further evaluate possible drug combinations that could harness an improved anti-tumor effect.
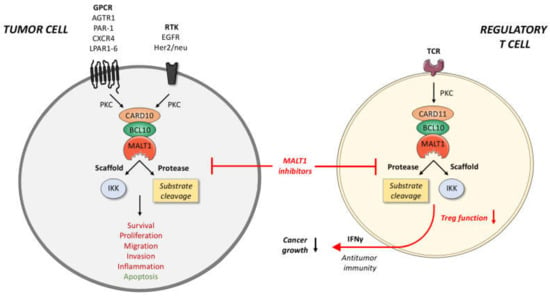
Figure 2.
Potential anti-cancer effects of MALT1 inhibitors. In tumor cells, various GPCRs and RTKs can promote the assembly of a CARD10-BCL10-MALT1 complex. Through both its scaffold and protease function, MALT1 can activate transcription factors, such as NF-κB and AP-1, that mediate key processes in cancer cells, such as cell survival, proliferation, metastasis, and inflammation. In regulatory T cells, the CARD11-BCL10-MALT1 complex has been identified as crucial factors for Treg expansion and function. Pharmacological inhibition of MALT1 protease function not only interferes with the growth and aggressiveness of the tumor, but can also dampen and reprogram Treg activity, thus provoking a T cell-based anti-tumor immune response. Tumor-suppressive functions of MALT1 are highlighted in green, oncogenic effects in red. BCL10, B-cell lymphoma/leukemia10; CARD10/11, caspase recruitment domain-containing protein 10/11; GPCR, G-protein-coupled receptor; IKK, inhibitor of κB kinase; MALT1, mucosa-associated lymphoid tissue lymphoma translocation protein 1; PKC, protein kinase C; RTK, receptor tyrosine kinase; TCR, T-cell receptor.
Author Contributions
B.G.S., A.S., K.S.-O. and S.H. wrote paragraphs of the manuscript. B.G.S. prepared the figures. All authors have accepted responsibility for the entire content of this submitted manuscript and approved its submission. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Collaborative Research Center Transregio SFB/TR 156 (to S.H.), SFB/TR 209 (to K.S.-O. and S.H.), and the Deutsche Krebshilfe (S.H.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We would like to acknowledge the Open Access Publishing Fund of the University of Tuebingen.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Taniguchi, K.; Karin, M. NF-kappaB, Inflammation, Immunity and Cancer: Coming of Age. Nat. Rev. Immunol. 2018, 18, 309–324. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Shen, S.; Verma, I.M. NF-kappaB, an Active Player in Human Cancers. Cancer Immunol. Res. 2014, 2, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Gerondakis, S.; Grumont, R.; Gugasyan, R.; Wong, L.; Isomura, I.; Ho, W.; Banerjee, A. Unravelling the Complexities of the NF-KappaB Signalling Pathway Using Mouse Knockout and Transgenic Models. Oncogene 2006, 25, 6781–6799. [Google Scholar] [CrossRef] [PubMed]
- Ramadass, V.; Vaiyapuri, T.; Tergaonkar, V. Small Molecule NF-kappaB Pathway Inhibitors in Clinic. Int. J. Mol. Sci. 2020, 21, 5164. [Google Scholar] [CrossRef]
- Ruland, J.; Hartjes, L. CARD-BCL-10-MALT1 Signalling in Protective and Pathological Immunity. Nat. Rev. Immunol. 2019, 19, 118–134. [Google Scholar] [CrossRef] [PubMed]
- Bertin, J.; Guo, Y.; Wang, L.; Srinivasula, S.M.; Jacobson, M.D.; Poyet, J.L.; Merriam, S.; Du, M.Q.; Dyer, M.J.; Robison, K.E.; et al. CARD9 Is a Novel Caspase Recruitment Domain-Containing Protein that Interacts with BCL10/CLAP and Activates NF-Kappa, B. J. Biol. Chem. 2000, 275, 41082–41086. [Google Scholar] [CrossRef] [PubMed]
- Gross, O.; Gewies, A.; Finger, K.; Schafer, M.; Sparwasser, T.; Peschel, C.; Forster, I.; Ruland, J. Card9 Controls a Non-TLR Signalling Pathway for Innate Anti-Fungal Immunity. Nature 2006, 442, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Gaide, O.; Martinon, F.; Micheau, O.; Bonnet, D.; Thome, M.; Tschopp, J. Carma1, a CARD-Containing Binding Partner of Bcl10, Induces Bcl10 Phosphorylation and NF-KappaB Activation. FEBS Lett. 2001, 496, 121–127. [Google Scholar] [CrossRef]
- Wang, D.; You, Y.; Case, S.M.; McAllister-Lucas, L.M.; Wang, L.; DiStefano, P.S.; Nunez, G.; Bertin, J.; Lin, X. A Requirement for CARMA1 in TCR-Induced NF-Kappa B Activation. Nat. Immunol. 2002, 3, 830–835. [Google Scholar] [CrossRef]
- Gaide, O.; Favier, B.; Legler, D.F.; Bonnet, D.; Brissoni, B.; Valitutti, S.; Bron, C.; Tschopp, J.; Thome, M. CARMA1 Is a Critical Lipid Raft-Associated Regulator of TCR-Induced NF-Kappa B Activation. Nat. Immunol. 2002, 3, 836–843. [Google Scholar] [CrossRef]
- McAllister-Lucas, L.M.; Inohara, N.; Lucas, P.C.; Ruland, J.; Benito, A.; Li, Q.; Chen, S.; Chen, F.F.; Yamaoka, S.; Verma, I.M.; et al. Bimp1, a MAGUK Family Member Linking Protein Kinase C Activation to Bcl10-Mediated NF-KappaB Induction. J. Biol. Chem. 2001, 276, 30589–30597. [Google Scholar] [CrossRef]
- Jordan, C.T.; Cao, L.; Roberson, E.D.; Pierson, K.C.; Yang, C.F.; Joyce, C.E.; Ryan, C.; Duan, S.; Helms, C.A.; Liu, Y.; et al. PSORS2 Is due to Mutations in CARD14. Am. J. Hum. Genet. 2012, 90, 784–795. [Google Scholar] [CrossRef]
- Schmitt, A.; Grondona, P.; Maier, T.; Brandle, M.; Schonfeld, C.; Jager, G.; Kosnopfel, C.; Eberle, F.C.; Schittek, B.; Schulze-Osthoff, K.; et al. MALT1 Protease Activity Controls the Expression of Inflammatory Genes in Keratinocytes upon Zymosan Stimulation. J. Invest. Dermatol. 2016, 136, 788–797. [Google Scholar] [CrossRef]
- McAllister-Lucas, L.M.; Ruland, J.; Siu, K.; Jin, X.; Gu, S.; Kim, D.S.; Kuffa, P.; Kohrt, D.; Mak, T.W.; Nunez, G.; et al. CARMA3/Bcl10/MALT1-Dependent NF-kappaB Activation Mediates Angiotensin II-Responsive Inflammatory Signaling in Nonimmune Cells. Proc. Natl. Acad. Sci. USA 2007, 104, 139–144. [Google Scholar] [CrossRef]
- Grabiner, B.C.; Blonska, M.; Lin, P.-C.; You, Y.; Wang, D.; Sun, J.; Darnay, B.G.; Dong, C.; Lin, X. CARMA3 Deficiency Abrogates G Protein-Coupled Receptor-Induced NF-κB Activation. Genes Dev. 2007, 21, 984–996. [Google Scholar] [CrossRef]
- Jiang, T.; Grabiner, B.; Zhu, Y.; Jiang, C.; Li, H.; You, Y.; Lang, J.; Hung, M.-C.; Lin, X. CARMA3 Is Crucial for EGFR-Induced Activation of NF-κB and Tumor Progression. Cancer Res. 2011, 71, 2183–2192. [Google Scholar] [CrossRef]
- Staal, J.; Driege, Y.; Haegman, M.; Kreike, M.; Iliaki, S.; Vanneste, D.; Lork, M.; Afonina, I.S.; Braun, H.; Beyaert, R. Defining the Combinatorial Space of PKC:CARD-CC Signal Transduction Nodes. FEBS J. 2020, 288, 1630–1647. [Google Scholar] [CrossRef]
- Holliday, M.J.; Witt, A.; Gama, A.R.; Walters, B.T.; Arthur, C.P.; Halfmann, R.; Rohou, A.; Dueber, E.C.; Fairbrother, W.J. Structures of Autoinhibited and Polymerized Forms of CARD9 Reveal Mechanisms of CARD9 and CARD11 Activation. Nat. Commun. 2019, 10, 1–17. [Google Scholar] [CrossRef]
- Rawlings, D.J.; Sommer, K.; Moreno-García, M.E. The CARMA1 Signalosome Links the Signalling Machinery of Adaptive and Innate Immunity in lymphocytes. Nat. Rev. Immunol. 2006, 6, 799–812. [Google Scholar] [CrossRef]
- Sommer, K.; Guo, B.; Pomerantz, J.L.; Bandaranayake, A.D.; Moreno-García, M.E.; Ovechkina, Y.L.; Rawlings, D.J. Phosphorylation of the CARMA1 Linker Controls NF-κB Activation. Immunity 2005, 23, 561–574. [Google Scholar] [CrossRef]
- Matsumoto, R.; Wang, D.; Blonska, M.; Li, H.; Kobayashi, M.; Pappu, B.; Chen, Y.; Wang, D.; Lin, X. Phosphorylation of CARMA1 Plays a Critical Role in T Cell Receptor-Mediated NF-κB Activation. Immunity 2005, 23, 575–585. [Google Scholar] [CrossRef]
- Tanner, M.; Hanel, W.; Gaffen, S.L.; Lin, X. CARMA1 Coiled-Coil Domain Is Involved in the Oligomerization and Subcellular Localization of CARMA1 and Is Required for T Cell Receptor-induced NF-κB Activation. J. Biol. Chem. 2007, 282, 17141–17147. [Google Scholar] [CrossRef]
- Bertin, J.; Wang, L.; Guo, Y.; Jacobson, M.D.; Poyet, J.-L.; Srinivasula, S.M.; Merriam, S.; DiStefano, P.S.; Alnemri, E.S. CARD11 and CARD14 Are Novel Caspase Recruitment Domain (CARD)/Membrane-associated Guanylate Kinase (MAGUK) Family Members that Interact with BCL10 and Activate NF-κB. J. Biol. Chem. 2001, 276, 11877–11882. [Google Scholar] [CrossRef]
- Schlauderer, F.; Seeholzer, T.; Desfosses, A.; Gehring, T.; Strauss, M.; Hopfner, K.-P.; Gutsche, I.; Krappmann, D.; Lammens, K. Molecular Architecture and Regulation of BCL10-MALT1 Filaments. Nat. Commun. 2018, 9, 4041. [Google Scholar] [CrossRef]
- David, L.; Li, Y.; Ma, J.; Garner, E.; Zhang, X.; Wu, H. Assembly Mechanism of the CARMA1–BCL10–MALT1–TRAF6 SignaloSome. Proc. Natl. Acad. Sci. USA 2018, 115, 1499–1504. [Google Scholar] [CrossRef]
- Sun, L.; Deng, L.; Ea, C.-K.; Xia, Z.-P.; Chen, Z.J. The TRAF6 Ubiquitin Ligase and TAK1 Kinase Mediate IKK Activation by BCL10 and MALT1 in T Lymphocytes. Mol. Cell 2004, 14, 289–301. [Google Scholar] [CrossRef]
- Oeckinghaus, A.; Wegener, E.; Welteke, V.; Ferch, U.; Arslan, S.; Ruland, J.; Scheidereit, C.; Krappmann, D. Malt1 Ubiquitination Triggers NF-κB Signaling upon T-Cell Activation. EMBO J. 2007, 26, 4634–4645. [Google Scholar] [CrossRef]
- Deng, L.; Wang, C.; Spencer, E.; Yang, L.; Braun, A.; You, J.; Slaughter, C.; Pickart, C.; Chen, Z.J. Activation of the IkappaB Kinase Complex by TRAF6 Requires a Dimeric Ubiquitin-Conjugating Enzyme Complex and a Unique Polyubiquitin Chain. Cell 2000, 103, 351–361. [Google Scholar] [CrossRef]
- Yang, Y.; Schmitz, R.; Mitala, J.; Whiting, A.L.; Xiao, W.; Ceribelli, M.; Wright, G.W.; Zhao, H.; Yang, Y.; Xu, W.; et al. Essential Role of the Linear Ubiquitin Chain Assembly Complex in Lymphoma Revealed by Rare Germline Polymorphisms. Cancer Discov. 2014, 4, 480–493. [Google Scholar] [CrossRef]
- Wu, C.-J.; Ashwell, J.D. NEMO Recognition of Ubiquitinated Bcl10 is Required for T Cell Receptor-Mediated NF-B Activation. Proc. Natl. Acad. Sci. USA 2008, 105, 3023–3028. [Google Scholar] [CrossRef]
- Zhou, H.; Wertz, I.; O’Rourke, K.; Ultsch, M.; Seshagiri, S.; Eby, M.; Xiao, W.; Dixit, V.M. Bcl10 Activates the NF-κB Pathway through Ubiquitination of NEMO. Nature 2003, 427, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Deng, L.; Hong, M.; Akkaraju, G.R.; Inoue, J.-I.; Chen, Z.J. TAK1 is a Ubiquitin-Dependent Kinase of MKK and IKK. Nature 2001, 412, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Emmerich, C.H.; Ordureau, A.; Strickson, S.; Arthur, J.S.C.; Pedrioli, P.G.; Komander, D.; Cohen, P. Activation of the Canonical IKK Complex by K63/M1-Linked Hybrid Ubiquitin Chains. Proc. Natl. Acad. Sci. USA 2013, 110, 15247–15252. [Google Scholar] [CrossRef] [PubMed]
- Uren, A.; O’Rourke, K.; Aravind, L.; Pisabarro, M.; Seshagiri, S.; Koonin, E.V.; Dixit, V.M. Identification of Paracaspases and Metacaspases: Two Ancient Families of Caspase-like Proteins, One of which Plays a Key Role in MALT Lymphoma. Mol. Cell 2000, 6, 961–967. [Google Scholar] [CrossRef]
- Cheng, J.; Klei, L.R.; Hubel, N.E.; Zhang, M.; Schairer, R.; Maurer, L.M.; Klei, H.B.; Kang, H.; Concel, V.J.; Delekta, P.C.; et al. GRK2 Suppresses Lymphomagenesis by Inhibiting the MALT1 Proto-Oncoprotein. J. Clin. Investig. 2019, 130, 1036–1051. [Google Scholar] [CrossRef]
- O’Neill, T.J.; Seeholzer, T.; Gewies, A.; Gehring, T.; Giesert, F.; Hamp, I.; Graß, C.; Schmidt, H.; Kriegsmann, K.; Tofaute, M.J.; et al. TRAF6 Prevents Fatal Inflammation by Homeostatic Suppression of MALT1 Protease. Sci. Immunol. 2021, 6, eabh2095. [Google Scholar] [CrossRef]
- Pelzer, C.; Cabalzar, K.; Wolf, A.; Gonzalez, M.; Lenz, G.; Thome, M. The Protease Activity of the Paracaspase MALT1 Is Controlled by Monoubiquitination. Nat. Immunol. 2013, 14, 337–345. [Google Scholar] [CrossRef]
- Yu, J.W.; Jeffrey, P.D.; Ha, J.Y.; Yang, X.; Shi, Y. Crystal Structure of the Mucosa-Associated Lymphoid Tissue Lymphoma Translocation 1 (MALT1) Paracaspase Region. Proc. Natl. Acad. Sci. USA 2011, 108, 21004–21009. [Google Scholar] [CrossRef]
- Rebeaud, F.; Hailfinger, S.; Posevitz-Fejfar, A.; Tapernoux, M.; Moser, R.; Rueda, D.; Gaide, O.; Guzzardi, M.; Iancu, E.M.; Rufer, N.; et al. The Proteolytic Activity of the Paracaspase MALT1 is Key in T Cell Activation. Nat. Immunol. 2008, 9, 272–281. [Google Scholar] [CrossRef]
- Coornaert, B.; Baens, M.; Heyninck, K.; Bekaert, T.; Haegman, M.; Staal, J.; Sun, L.; Chen, Z.J.; Marynen, P.; Beyaert, R. T Cell Antigen Receptor Stimulation Induces MALT1 Paracaspase–Mediated Cleavage of the NF-κB inhibitor A20. Nat. Immunol. 2008, 9, 263–271. [Google Scholar] [CrossRef]
- Staal, J.; Driege, Y.; Bekaert, T.; Demeyer, A.; Muyllaert, D.; Van Damme, P.; Gevaert, K.; Beyaert, R. T-Cell Receptor-Induced JNK Activation Requires Proteolytic Inactivation of CYLD by MALT1. EMBO J. 2011, 30, 1742–1752. [Google Scholar] [CrossRef]
- Hailfinger, S.; Nogai, H.; Pelzer, C.; Jaworski, M.; Cabalzar, K.; Charton, J.-E.; Guzzardi, M.; Décaillet, C.; Grau, M.; Dörken, B.; et al. Malt1-Dependent RelB Cleavage Promotes Canonical NF-B Activation in Lymphocytes and Lymphoma Cell Lines. Proc. Natl. Acad. Sci. USA 2011, 108, 14596–14601. [Google Scholar] [CrossRef]
- Douanne, T.; Gavard, J.; Bidère, N. The Paracaspase MALT1 Cleaves the LUBAC Subunit HOIL1 during Antigen Receptor Signaling. J. Cell Sci. 2016, 129, 1775–1780. [Google Scholar] [CrossRef]
- Elton, L.; Carpentier, I.; Staal, J.; Driege, Y.; Haegman, M.; Beyaert, R. MALT1 Cleaves the E3 Ubiquitin Ligase HOIL-1 in Activated T Cells, Generating a Dominant Negative Inhibitor of LUBAC-Induced NF-κB Signaling. FEBS J. 2015, 283, 403–412. [Google Scholar] [CrossRef]
- Klein, T.; Fung, S.-Y.; Renner, F.; Blank, M.A.; Dufour, A.; Kang, S.; Bolger-Munro, M.; Scurll, J.M.; Priatel, J.; Schweigler, P.; et al. The Paracaspase MALT1 Cleaves HOIL1 Reducing Linear Ubiquitination by LUBAC to Dampen Lymphocyte NF-κB Signalling. Nat. Commun. 2015, 6, 8777. [Google Scholar] [CrossRef]
- Uehata, T.; Iwasaki, H.; Vandenbon, A.; Matsushita, K.; Hernandez-Cuellar, E.; Kuniyoshi, K.; Satoh, T.; Mino, T.; Suzuki, Y.; Standley, D.M.; et al. Malt1-Induced Cleavage of Regnase-1 in CD4+ Helper T Cells Regulates Immune Activation. Cell 2013, 153, 1036–1049. [Google Scholar] [CrossRef]
- Jeltsch, K.M.; Hu, D.; Brenner, S.; Zöller, J.; Heinz, G.A.; Nagel, D.; Vogel, K.U.; Rehage, N.; Warth, S.C.; Edelmann, S.L.; et al. Cleavage of Roquin and Regnase-1 by the Paracaspase MALT1 Releases their Cooperatively Repressed Targets to Promote TH17 Differentiation. Nat. Immunol. 2014, 15, 1079–1089. [Google Scholar] [CrossRef]
- Baens, M.; Bonsignore, L.; Somers, R.; Vanderheydt, C.; Weeks, S.; Gunnarsson, J.; Nilsson, E.; Roth, R.G.; Thome, M.; Marynen, P. MALT1 Auto-Proteolysis Is Essential for NF-κB-Dependent Gene Transcription in Activated Lymphocytes. PLoS ONE 2014, 9, e103774. [Google Scholar] [CrossRef]
- Xia, X.; Cao, G.; Sun, G.; Zhu, L.; Tian, Y.; Song, Y.; Guo, C.; Wang, X.; Zhong, J.; Zhou, W.; et al. GLS1-Mediated Glutaminolysis Unbridled by MALT1 Protease Promotes Psoriasis Pathogenesis. J. Clin. Investig. 2020, 130, 5180–5196. [Google Scholar] [CrossRef]
- Nakaya, M.; Xiao, Y.; Zhou, X.; Chang, J.-H.; Chang, M.; Cheng, X.; Blonska, M.; Lin, X.; Sun, S.-C. Inflammatory T Cell Responses Rely on Amino Acid Transporter ASCT2 Facilitation of Glutamine Uptake and mTORC1 Kinase Activation. Immunity 2014, 40, 692–705. [Google Scholar] [CrossRef]
- Xia, X.; Zhou, W.; Guo, C.; Fu, Z.; Zhu, L.; Li, P.; Xu, Y.; Zheng, L.; Zhang, H.; Shan, C.; et al. Glutaminolysis Mediated by MALT1 Protease Activity Facilitates PD-L1 Expression on ABC-DLBCL Cells and Contributes to Their Immune Evasion. Front. Oncol. 2018, 8, 632. [Google Scholar] [CrossRef]
- Juilland, M.; Thome, M. Role of the CARMA1/BCL10/MALT1 Complex in Lymphoid Malignancies. Curr. Opin. Hematol. 2016, 23, 402–409. [Google Scholar] [CrossRef]
- Hailfinger, S.; Lenz, G.; Thome, M. Targeting B-Cell Lymphomas with Inhibitors of the MALT1 Paracaspase. Curr. Opin. Chem. Biol. 2014, 23, 47–55. [Google Scholar] [CrossRef]
- Grondona, P.; Bucher, P.; Schulze-Osthoff, K.; Hailfinger, S.; Schmitt, A. NF-κB Activation in Lymphoid Malignancies: Genetics, Signaling, and Targeted Therapy. Biomedicines 2018, 6, 38. [Google Scholar] [CrossRef] [PubMed]
- Akagi, T.; Motegi, M.; Tamura, A.; Suzuki, R.; Hosokawa, Y.; Suzuki, H.; Ota, H.; Nakamura, S.; Morishima, Y.; Taniwaki, M.; et al. A Novel Gene, MALT1 at 18q21, is Involved in t(11;18) (q21;q21) Found in Low-Grade B-Cell Lymphoma of Mucosa-Associated Lymphoid Tissue. Oncogene 1999, 18, 5785–5794. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Hu, J.; Wang, X.; Xuan, L.; Lai, J.; Xu, L.; Chen, S.; Yang, L.; Luo, G.; Zhu, K.; et al. Overexpression of MALT1-A20-NF-κB in Adult B-Cell Acute Lymphoblastic Leukemia. Cancer Cell Int. 2015, 15, 73. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Izquierdo, D.; Buchonnet, G.; Siebert, R.; Gascoyne, R.D.; Climent, J.; Karran, L.; Marin, M.; Blesa, D.; Horsman, D.; Rosenwald, A.; et al. MALT1 is Deregulated by Both Chromosomal Translocation and Amplification in B-Cell Non-Hodgkin Lymphoma. Blood 2003, 101, 4539–4546. [Google Scholar] [CrossRef]
- Kuper-Hommel, M.J.J.; Schreuder, M.I.; Gemmink, A.H.; Van Krieken, J.H.J.M. T(14;18)(q32;q21) Involving MALT1 and IGH Genes Occurs in Extranodal Diffuse Large B-Cell Lymphomas of the Breast and Testis. Mod. Pathol. 2012, 26, 421–427. [Google Scholar] [CrossRef][Green Version]
- Streubel, B.; Lamprecht, A.; Dierlamm, J.; Cerroni, L.; Stolte, M.; Ott, G.; Raderer, M.; Chott, A. T(14;18)(q32;q21) Involving IGH and MALT1 Is a Frequent Chromosomal Aberration in MALT Lymphoma. Blood 2003, 101, 2335–2339. [Google Scholar] [CrossRef]
- Cook, J.R.; Sherer, M.; Craig, F.E.; Shekhter-Levin, S.; Swerdlow, S.H. T(14;18)(q32;q21) Involving MALT1 and IGH Genes in an Extranodal Diffuse Large B-Cell Lymphoma. Hum. Pathol. 2003, 34, 1212–1215. [Google Scholar] [CrossRef]
- Vicente-Dueñas, C.; Fontan, L.; Gonzalez-Herrero, I.; Romero-Camarero, I.; Segura, V.; Aznar, M.A.; Alonso-Escudero, E.; Campos-Sanchez, E.; Ruiz, L.; Barajas-Diego, M.; et al. Expression of MALT1 Oncogene in Hematopoietic Stem/Progenitor Cells RecapituLates the Pathogenesis of Human Lymphoma in Mice. Proc. Natl. Acad. Sci. USA 2012, 109, 10534–10539. [Google Scholar] [CrossRef]
- Li, Z.; Wang, H.; Xue, L.; Shin, N.-M.; Roopenian, D.; Xu, W.; Qi, C.-F.; Sangster, M.; Orihuela, C.J.; Tuomanen, E.; et al. Eμ-BCL10 Mice Exhibit Constitutive Activation of Both Canonical and Noncanonical NF-κB Pathways Generating Marginal Zone (MZ) B-Cell Expansion as a Precursor to Splenic MZ Lymphoma. Blood 2009, 114, 4158–4168. [Google Scholar] [CrossRef]
- Lenz, G.; Davis, R.E.; Ngo, V.N.; Lam, L.; George, T.C.; Wright, G.W.; Dave, S.S.; Zhao, H.; Xu, W.; Rosenwald, A.; et al. Oncogenic CARD11 Mutations in Human Diffuse Large B Cell Lymphoma. Science 2008, 319, 1676–1679. [Google Scholar] [CrossRef]
- Almeida, A.C.D.S.; Abate, F.; Khiabanian, H.; Martinez-Escala, E.; Guitart, J.; Tensen, C.; Vermeer, M.; Rabadan, R.; Ferrando, A.; Palomero, T. The Mutational Landscape of Cutaneous T Cell Lymphoma and Sézary Syndrome. Nat. Genet. 2015, 47, 1465–1470. [Google Scholar] [CrossRef]
- Wang, L.; Ni, X.; Covington, K.R.; Yang, B.Y.; Shiu, J.; Zhang, X.; Xi, L.; Meng, Q.; Langridge, T.; Drummond, J.; et al. Genomic Profiling of Sézary Syndrome Identifies Alterations of Key T Cell Signaling and Differentiation Genes. Nat. Genet. 2015, 47, 1426–1434. [Google Scholar] [CrossRef]
- Vallois, D.; Dobay, M.P.D.; Morin, R.D.; Lemonnier, F.; Missiaglia, E.; Juilland, M.; Iwaszkiewicz, J.; Fataccioli, V.; Bisig, B.; Roberti, A.; et al. Activating Mutations in Genes Related to TCR Signaling in Angioimmunoblastic and Other Follicular Helper T-Cell–Derived Lymphomas. Blood 2016, 128, 1490–1502. [Google Scholar] [CrossRef]
- Dierlamm, J.; Baens, M.; Wlodarska, I.; Stefanova-Ouzounova, M.; Hernandez, J.M.; Hossfeld, D.K.; De Wolf-Peeters, C.; Hagemeijer, A.; Van den Berghe, H.; Marynen, P. The Apoptosis Inhibitor Gene API2 and a Novel 18q Gene, MLT, are Recurrently Rearranged in the t(11;18)(q21;q21) Associated with Mucosa-Associated Lymphoid Tissue Lymphomas. Blood 1999, 93, 3601–3609. [Google Scholar] [CrossRef]
- Lucas, P.; Kuffa, P.; Gu, S.; Kohrt, D.; Kim, D.S.L.; Siu, K.; Jin, X.; Swenson, J.; McAllister-Lucas, L.M. A Dual Role for the API2 Moiety in API2-MALT1-Dependent NF-κB Activation: Heterotypic Oligomerization and TRAF2 Recruitment. Oncogene 2007, 26, 5643–5654. [Google Scholar] [CrossRef]
- Noels, H.; van Loo, G.; Hagens, S.; Broeckx, V.; Beyaert, R.; Marynen, P.; Baens, M. A Novel TRAF6 Binding Site in MALT1 Defines Distinct Mechanisms of NF-κB Activation by API2·MALT1 Fusions. J. Biol. Chem. 2007, 282, 10180–10189. [Google Scholar] [CrossRef]
- Rosebeck, S.; Madden, L.; Jin, X.; Gu, S.; Apel, I.J.; Appert, A.; Hamoudi, R.A.; Noels, H.; Sagaert, X.; Van Loo, P.; et al. Cleavage of NIK by the API2-MALT1 Fusion Oncoprotein Leads to Noncanonical NF-κB Activation. Science 2011, 331, 468–472. [Google Scholar] [CrossRef]
- Nie, Z.; Du, M.-Q.; McAllister-Lucas, L.M.; Lucas, P.; Bailey, N.; Hogaboam, C.M.; Lim, M.; Elenitoba-Johnson, K. Conversion of the LIMA1 Tumour Suppressor into an Oncogenic LMO-Like Protein by API2–MALT1 in MALT Lymphoma. Nat. Commun. 2015, 6, 5908. [Google Scholar] [CrossRef]
- Young, R.M.; Wu, T.; Schmitz, R.; Dawood, M.; Xiao, W.; Phelan, J.D.; Xu, W.; Menard, L.; Meffre, E.; Chan, W.-C.C.; et al. Survival of Human Lymphoma Cells Requires B-Cell Receptor Engagement by Self-Antigens. Proc. Natl. Acad. Sci. USA 2015, 112, 13447–13454. [Google Scholar] [CrossRef] [PubMed]
- Minden, M.D.-V.; Übelhart, R.; Schneider, D.; Wossning, T.; Bach, M.P.; Buchner, M.; Hofmann, D.; Surova, E.; Follo, M.; Köhler, F.; et al. Chronic Lymphocytic Leukaemia is Driven by Antigen-Independent Cell-Autonomous Signalling. Nature 2012, 489, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Quinn, E.R.; Chan, C.H.; Hadlock, K.G.; Foung, S.K.H.; Flint, M.; Levy, S. The B-Cell Receptor of a Hepatitis C Virus (HCV)–Associated Non-Hodgkin Lymphoma Binds the Viral E2 Envelope Protein, Implicating HCV in Lymphomagenesis. Blood 2001, 98, 3745–3749. [Google Scholar] [CrossRef] [PubMed]
- Suarez, F.; Lortholary, O.; Hermine, O.; Lecuit, M. Infection-Associated Lymphomas Derived from Marginal Zone B Cells: A Model of Antigen-Driven Lymphoproliferation. Blood 2006, 107, 3034–3044. [Google Scholar] [CrossRef]
- Bonsignore, L.; Passelli, K.; Pelzer, C.; Perroud, M.; Konrad, A.; Thurau, M.; Stürzl, M.; Dai, L.; Trillo-Tinoco, J.; Del Valle, L.; et al. A Role for MALT1 Activity in Kaposi’s Sarcoma-Associated Herpes Virus Latency and Growth of Primary Effusion Lymphoma. Leukemia 2016, 31, 614–624. [Google Scholar] [CrossRef] [PubMed]
- Dai, B.; Grau, M.; Juilland, M.; Klener, P.; Höring, E.; Molinsky, J.; Schimmack, G.; Aukema, S.M.; Hoster, E.; Vogt, N.; et al. B-Cell Receptor–Driven MALT1 Activity Regulates MYC Signaling in Mantle Cell Lymphoma. Blood 2017, 129, 333–346. [Google Scholar] [CrossRef]
- Hailfinger, S.; Lenz, G.; Ngo, V.; Posvitz-Fejfar, A.; Rebeaud, F.; Guzzardi, M.; Penas, E.-M.M.; Dierlamm, J.; Chan, W.C.; Staudt, L.M.; et al. Essential Role of MALT1 Protease Activity in Activated B Cell-Like Diffuse Large B-Cell Lymphoma. Proc. Natl. Acad. Sci. USA 2009, 106, 19946–19951. [Google Scholar] [CrossRef]
- Ferch, U.; Kloo, B.; Gewies, A.; Pfänder, V.; Düwel, M.; Peschel, C.; Krappmann, D.; Ruland, J. Inhibition of MALT1 Protease Activity is Selectively Toxic for Activated B Cell–Like Diffuse Large B Cell Lymphoma Cells. J. Exp. Med. 2009, 206, 2313–2320. [Google Scholar] [CrossRef]
- Saba, N.S.; Wong, D.; Tanios, G.; Iyer, J.R.; Lobelle-Rich, P.; Dadashian, E.L.; Liu, D.; Fontan, L.; Flemington, E.K.; Nichols, C.M.; et al. MALT1 Inhibition Is Efficacious in Both Naïve and Ibrutinib-Resistant Chronic Lymphocytic Leukemia. Cancer Res. 2017, 77, 7038–7048. [Google Scholar] [CrossRef]
- Ishikawa, C.; Mori, N. MALT-1 as a Novel Therapeutic Target for Adult T-Cell Leukemia. Eur. J. Haematol. 2020, 105, 460–467. [Google Scholar] [CrossRef]
- Ekambaram, P.; Lee, J.-Y.; Hubel, N.E.; Hu, N.; Yerneni, S.; Campbell, P.G.; Pollock, N.; Klei, L.R.; Concel, V.J.; Delekta, P.C.; et al. The CARMA3-Bcl10-MALT1 Signalosome Drives NFκB Activation and Promotes Aggressiveness in Angiotensin II Receptor-Positive Breast Cancer. Cancer Res. 2017, 78, 1225–1240. [Google Scholar] [CrossRef]
- McAuley, J.R.; Bailey, K.M.; Ekambaram, P.; Klei, L.R.; Kang, H.; Hu, D.; Freeman, T.J.; Concel, V.J.; Hubel, N.E.; Lee, J.-Y.; et al. MALT1 Is a Critical Mediator of PAR1-Driven NF-κB Activation and Metastasis in Multiple Tumor Types. Oncogene 2019, 38, 7384–7398. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Ekambaram, P.; Carleton, N.M.; Hu, D.; Klei, L.R.; Cai, Z.; Myers, M.I.; Hubel, N.E.; Covic, L.; Agnihotri, S.; et al. MALT1 Is a Targetable Driver of Epithelial-to-Mesenchymal Transition in Claudin-Low, Triple-Negative Breast Cancer. Mol. Cancer Res. 2021. [Google Scholar] [CrossRef]
- Mahanivong, C.; Chen, H.M.; Yee, S.W.; Pan, Z.K.; Dong, Z.; Huang, S. Protein Kinase Cα-CARMA3 Signaling Axis Links Ras to NF-κB for Lysophosphatidic Acid-Induced Urokinase Plasminogen Activator Expression in Ovarian Cancer Cells. Oncogene 2007, 27, 1273–1280. [Google Scholar] [CrossRef]
- Liu, X.; Yue, C.; Shi, L.; Liu, G.; Cao, Q.; Shan, Q.; Wang, Y.; Chen, X.; Li, H.; Wang, J.; et al. MALT1 Is a Potential Therapeutic Target in Glioblastoma and Plays a Crucial Role in EGFR-Induced NF-κB Activation. J. Cell Mol. Med. 2020, 24, 7550–7562. [Google Scholar] [CrossRef]
- Pan, D.; Jiang, C.; Ma, Z.-L.; Blonska, M.; You, M.J.; Lin, X. MALT1 Is Required for EGFR-Induced NF-κB Activation and Contributes to EGFR-Driven Lung Cancer Progression. Oncogene 2015, 35, 919–928. [Google Scholar] [CrossRef]
- Pan, D.; Zhu, Y.; Zhou, Z.; Wang, T.; You, H.; Jiang, C.; Lin, X. The CBM Complex Underwrites NF-κB Activation to Promote HER2-Associated Tumor Malignancy. Mol. Cancer Res. 2015, 14, 93–102. [Google Scholar] [CrossRef]
- Konczalla, L.; Perez, D.R.; Wenzel, N.; Wolters-Eisfeld, G.; Klemp, C.; Lüddeke, J.; Wolski, A.; Landschulze, D.; Meier, C.; Buchholz, A.; et al. Biperiden and Mepazine Effectively Inhibit MALT1 Activity and Tumor Growth in Pancreatic Cancer. Int. J. Cancer 2019, 146, 1618–1630. [Google Scholar] [CrossRef]
- Tsui, K.-H.; Chang, K.-S.; Sung, H.-C.; Hsu, S.-Y.; Lin, Y.-H.; Hou, C.-P.; Yang, P.-S.; Chen, C.-L.; Feng, T.-H.; Juang, H.-H. Mucosa-Associated Lymphoid Tissue 1 Is an Oncogene Inducing Cell Proliferation, Invasion, and Tumor Growth via the Upregulation of NF-κB Activity in Human Prostate Carcinoma Cells. Biomedicines 2021, 9, 250. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, G.; Jin, J.; Degan, S.; Tameze, Y.; Zhang, J.Y. MALT1 Promotes Melanoma Progression through JNK/c-Jun Signaling. Oncogenesis 2017, 6, e365. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.-N.; Chang, Y.-C.; Su, Y.; Hsu, D.S.-S.; Cheng, C.-T.; Wu, R.-C.; Chung, Y.-H.; Chiang, K.-C.; Yeh, T.-S.; Lu, M.-L.; et al. Identification of MALT1 as Both a Prognostic Factor and a Potential Therapeutic Target of Regorafenib in Cholangiocarcinoma Patients. Oncotarget 2017, 8, 113444–113459. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Liu, X.; Liu, Y.; Liu, Y.; Zhang, C.; Wang, Z.; Jiang, T.; Wang, Y. miR-181d/MALT1 Regulatory Axis Attenuates Mesenchymal Phenotype through NF-κB Pathways in Glioblastoma. Cancer Lett. 2017, 396, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Du, R.; Li, J.; Luo, X.; Zhao, S.; Chang, A.; Zhou, W.; Gao, R.; Luo, D.; Wang, J.; et al. TIFA Suppresses Hepatocellular Carcinoma Progression via MALT1-Dependent and-Independent Signaling Pathways. Signal Transduct. Target. Ther. 2016, 1, 16013. [Google Scholar] [CrossRef]
- Israël, L.; Glück, A.; Berger, M.; Coral, M.; Ceci, M.; Unterreiner, A.; Rubert, J.; Bardet, M.; Ginster, S.; Golding-Ochsenbein, A.M.; et al. CARD10 Cleavage by MALT1 Restricts Lung Carcinoma Growth in vivo. Oncogenesis 2021, 10, 32. [Google Scholar] [CrossRef]
- Rosebeck, S.; Rehman, A.O.; Lucas, P.C.; McAllister-Lucas, L.M. From MALT Lymphoma to the CBM Signalosome: Three Decades of Discovery. Cell Cycle 2011, 10, 2485–2496. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, H.; Xu, J.; Zhang, N.; Pan, T.; Zhong, X.; Zhang, H.; Yin, L.; Yao, Y.; Wu, Q.; et al. MALT1 Inhibition as a Therapeutic Strategy in T-Cell Acute Lymphoblastic Leukemia by Blocking Notch1-Induced NF-κB Activation. Front. Oncol. 2020, 10, 558339. [Google Scholar] [CrossRef]
- Hamp, I.; O’Neill, T.J.; Plettenburg, O.; Krappmann, D. A Patent Review of MALT1 inhibitors (2013-present). Expert Opin. Ther. Pat. 2021, 31, 1079–1096. [Google Scholar] [CrossRef]
- Jaworski, M.; Marsland, B.J.; Gehrig, J.; Held, W.; Favre, S.; Luther, S.; Perroud, M.; Golshayan, D.; Gaide, O.; Thome, M. Malt1 Protease Inactivation Efficiently Dampens Immune Responses but Causes Spontaneous Autoimmunity. EMBO J. 2014, 33, 2765–2781. [Google Scholar] [CrossRef]
- Demeyer, A.; Staal, J.; Beyaert, R. Targeting MALT1 Proteolytic Activity in Immunity, Inflammation and Disease: Good or Bad? Trends Mol. Med. 2016, 22, 135–150. [Google Scholar] [CrossRef]
- Gewies, A.; Gorka, O.; Bergmann, H.; Pechloff, K.; Petermann, F.; Jeltsch, K.M.; Rudelius, M.; Kriegsmann, M.; Weichert, W.; Horsch, M.; et al. Uncoupling Malt1 Threshold Function from Paracaspase Activity Results in Destructive Autoimmune Inflammation. Cell Rep. 2014, 9, 1292–1305. [Google Scholar] [CrossRef]
- Brustle, A.; Brenner, D.; Knobbethomsen, C.B.; Cox, M.; Lang, P.A.; Lang, K.; Mak, T.W. MALT1 Is an Intrinsic Regulator of Regulatory T Cells. Cell Death Differ. 2015, 24, 1214–1223. [Google Scholar] [CrossRef]
- Rosenbaum, M.; Gewies, A.; Pechloff, K.; Heuser, C.; Engleitner, T.; Gehring, T.; Hartjes, L.; Krebs, S.; Krappmann, D.; Kriegsmann, M.; et al. Bcl10-Controlled Malt1 Paracaspase Activity Is Key for the Immune Suppressive Function of Regulatory T Cells. Nat. Commun. 2019, 10, 2352. [Google Scholar] [CrossRef]
- Rosenbaum, M.; Schnalzger, T.; Engleitner, T.; Weiß, C.; Mishra, R.; Mibus, C.; Mitterer, T.; Rad, R.; Ruland, J. MALT1 Protease Function in Regulatory T Cells Induces MYC Activity to Promote Mitochondrial Function and Cellular Expansion. Eur. J. Immunol. 2021, 52, 85–95. [Google Scholar] [CrossRef]
- Di Pilato, M.; Kim, E.Y.; Cadilha, B.; Prüßmann, J.N.; Nasrallah, M.N.; Seruggia, D.; Usmani, S.; Misale, S.; Zappulli, V.; Carrizosa, E.; et al. Targeting the CBM Complex Causes Treg Cells to Prime Tumours for Immune Checkpoint Therapy. Nature 2019, 570, 112–116. [Google Scholar] [CrossRef]
- Demeyer, A.; Driege, Y.; Skordos, I.; Coudenys, J.; Lemeire, K.; Elewaut, D.; Staal, J.; Beyaert, R. Long-Term MALT1 Inhibition in Adult Mice Without Severe Systemic Autoimmunity. iScience 2020, 23, 101557. [Google Scholar] [CrossRef]
- Martin, K.; Junker, U.; Tritto, E.; Sutter, E.; Rubic-Schneider, T.; Morgan, H.; Niwa, S.; Li, J.; Schlapbach, A.; Walker, D.; et al. Pharmacological Inhibition of MALT1 Protease Leads to a Progressive IPEX-Like Pathology. Front. Immunol. 2020, 11, 745. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).