Metabolically Improved Stem Cell Derived Hepatocyte-Like Cells Support HBV Life Cycle and Are a Promising Tool for HBV Studies and Antiviral Drug Screenings
Abstract
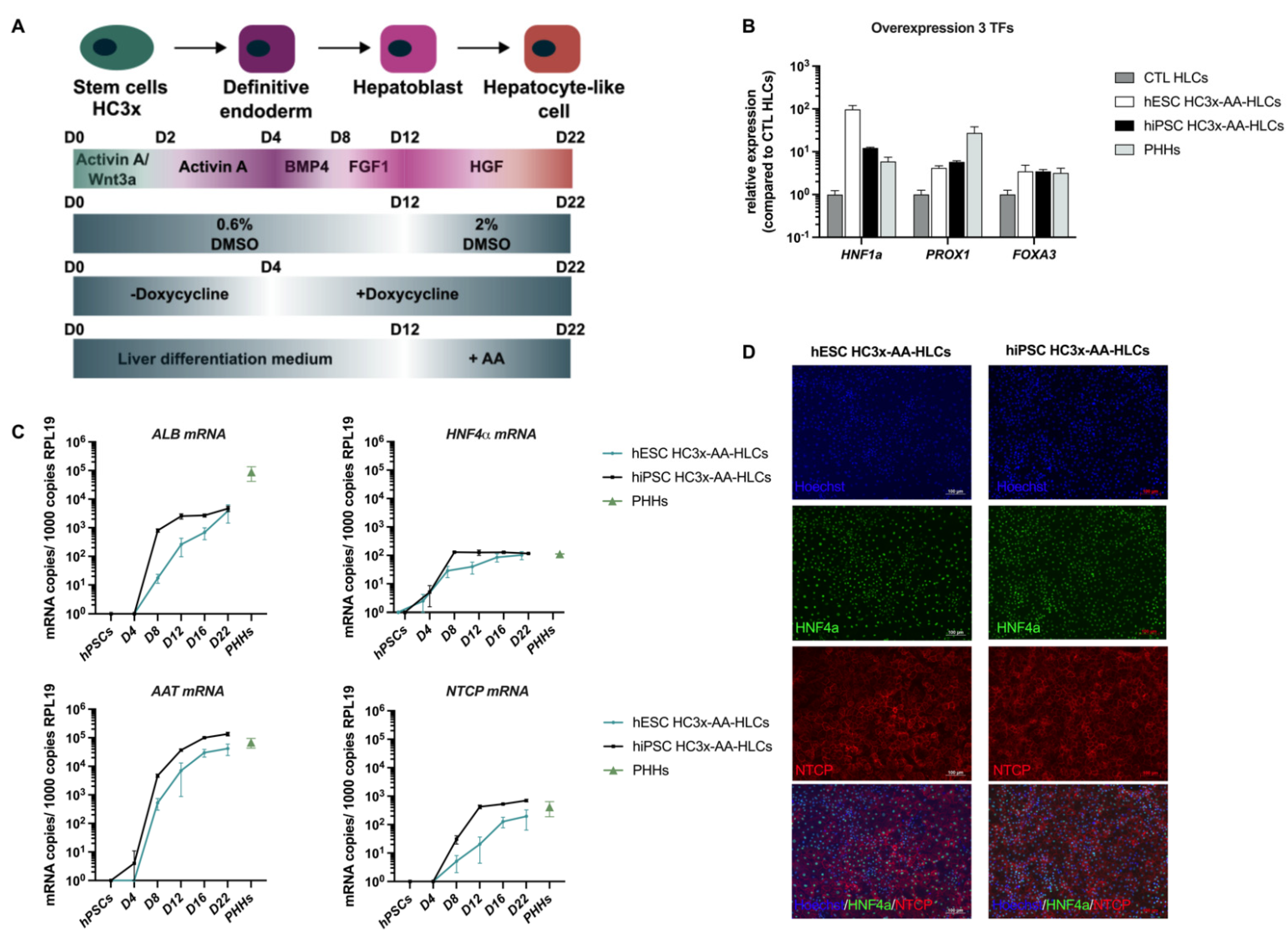
:1. Introduction
2. Materials and Methods
2.1. Cell Cultures
2.2. Primary Human Hepatocytes
2.3. Viral Inoculation
2.4. RNA Extraction and RT-qPCR
2.5. Myrcludex B Staining
2.6. Immunofluorescence Staining
2.7. HBsAg ELISA
2.8. HBeAg ELISA
2.9. Inhibition Experiments
2.10. Titrations on HepG2-NTCP Cells
2.11. Statistical Analysis
3. Results
3.1. HC3x-AA-HLCs Express the HBV Entry Receptor, NTCP
3.2. HC3x-AA-HLCs Support HBV Replication and the Production of Novel Infectious HBV Virions
3.3. Validation of HC3x-AA-HLCs with Known Anti-HBV Antivirals
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Trépo, C.; Chan, H.L.Y.; Lok, A. Hepatitis B virus infection. Lancet 2014, 384, 2053–2063. [Google Scholar] [CrossRef]
- Revill, P.A.; Penicaud, C.; Brechot, C.; Zoulim, F. Meeting the Challenge of Eliminating Chronic Hepatitis b Infection. Genes 2019, 10, 260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, J.K.; Nguyen, M.H.; Kim, W.R.; Gish, R.; Perumalswami, P.; Jacobson, I.M. Prevalence of Chronic Hepatitis B Virus Infection in the United States. Am. J. Gastroenterol. 2020, 115, 1429–1438. [Google Scholar] [CrossRef] [PubMed]
- Revill, P.; Testoni, B.; Locarnini, S.; Zoulim, F. Global strategies are required to cure and eliminate HBV infection. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 1–10. [Google Scholar] [CrossRef]
- Godoy, P.; Widera, A.; Schmidt-Heck, W.; Campos, G.; Meyer, C.; Cadenas, C.; Reif, R.; Stöber, R.; Hammad, S.; Pütter, L.; et al. Gene network activity in cultivated primary hepatocytes is highly similar to diseased mammalian liver tissue. Arch. Toxicol. 2016, 90, 2513–2529. [Google Scholar] [CrossRef] [Green Version]
- Heslop, J.A.; Rowe, C.; Walsh, J.; Sison-Young, R.; Jenkins, R.; Kamalian, L.; Kia, R.; Hay, D.; Jones, R.P.; Malik, H.Z.; et al. Mechanistic evaluation of primary human hepatocyte culture using global proteomic analysis reveals a selective dedifferentiation profile. Arch. Toxicol. 2016, 91, 439–452. [Google Scholar] [CrossRef] [Green Version]
- Shlomai, A.; Schwartz, R.E.; Ramanan, V.; Bhatta, A.; de Jong, Y.P.; Bhatia, S.N.; Rice, C.M. Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell-derived hepatocellular systems. Proc. Natl. Acad. Sci. USA 2014, 111, 12193–12198. [Google Scholar] [CrossRef] [Green Version]
- Godoy, P.; Hewitt, N.J.; Albrecht, U.; Andersen, M.E.; Ansari, N.; Bhattacharya, S.; Bode, J.G.; Bolleyn, J.; Borner, C.; Böttger, J.; et al. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch. Toxicol. 2013, 87, 1315–1530. [Google Scholar] [CrossRef] [Green Version]
- Ni, Y.; Lempp, F.A.; Mehrle, S.; Nkongolo, S.; Kaufman, C.; Fälth, M.; Stindt, J.; Königer, C.; Nassal, M.; Kubitz, R.; et al. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 2014, 146, 1070–1083. [Google Scholar] [CrossRef]
- Witt-Kehati, D.; Bitton Alaluf, M.; Shlomai, A. Advances and Challenges in Studying Hepatitis B Virus In Vitro. Viruses 2016, 8, 21. [Google Scholar] [CrossRef]
- Kaneko, S.; Kakinuma, S.; Asahina, Y.; Kamiya, A.; Miyoshi, M.; Tsunoda, T.; Nitta, S.; Asano, Y.; Nagata, H.; Otani, S.; et al. Human induced pluripotent stem cell-derived hepatic cell lines as a new model for host interaction with hepatitis B virus. Nat. Publ. Gr. 2016, 6, 29358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakurai, F.; Mitani, S.; Yamamoto, T.; Takayama, K.; Tachibana, M.; Watashi, K.; Wakita, T.; Iijima, S.; Tanaka, Y.; Mizuguchi, H. Human induced-pluripotent stem cell-derived hepatocyte-like cells as an in vitro model of human hepatitis B virus infection. Sci. Rep. 2017, 7, 45698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, Y.; Carpentier, A.; Cheng, X.; Block, P.D.; Zhao, Y.; Zhang, Z.; Protzer, U.; Liang, T.J. Human stem cell-derived hepatocytes as a model for hepatitis B virus infection, spreading and virus-host interactions. J. Hepatol. 2017, 66, 494–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roelandt, P.; Obeid, S.; Paeshuyse, J.; Vanhove, J.; Van Lommel, A.; Nahmias, Y.; Nevens, F.; Neyts, J.; Verfaillie, C.M. Human pluripotent stem cell-derived hepatocytes support complete replication of hepatitis C virus. J. Hepatol. 2012, 57, 246–251. [Google Scholar] [CrossRef]
- Schwartz, R.E.; Trehan, K.; Andrus, L.; Sheahan, T.P.; Ploss, A.; Duncan, S.A.; Rice, C.M.; Bhatia, S.N. Modeling hepatitis C virus infection using human induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA 2012, 109, 2544–2548. [Google Scholar] [CrossRef] [Green Version]
- Helsen, N.; Debing, Y.; Paeshuyse, J.; Dallmeier, K.; Boon, R.; Coll, M.; Sancho-Bru, P.; Claes, C.; Neyts, J.; Verfaillie, C.M. Stem cell-derived hepatocytes: A novel model for hepatitis E virus replication. J. Hepatol. 2016, 64, 565–573. [Google Scholar] [CrossRef] [Green Version]
- Lang, J.; Vera, D.; Cheng, Y.; Tang, H. Modeling Dengue Virus-Hepatic Cell Interactions Using Human Pluripotent Stem Cell-Derived Hepatocyte-like Cells. Stem Cell Rep. 2016, 7, 341–354. [Google Scholar] [CrossRef] [Green Version]
- Godoy, P.; Schmidt-Heck, W.; Natarajan, K.; Lucendo-Villarin, B.; Szkolnicka, D.; Asplund, A.; Bjorquist, P.; Widera, A.; Stoeber, R.; Campos, G.; et al. Gene networks and transcription factor motifs defining the differentiation of stem cells into hepatocyte-like cells. J. Hepatol. 2015, 63, 934–942. [Google Scholar] [CrossRef] [Green Version]
- Baxter, M.; Withey, S.; Harrison, S.; Segeritz, C.-P.; Zhang, F.; Atkinson-Dell, R.; Rowe, C.; Gerrard, D.T.; Sison-Young, R.; Jenkins, R.; et al. Phenotypic and functional analyses show stem cell-derived hepatocyte-like cells better mimic fetal rather than adult hepatocytes. J. Hepatol. 2015, 62, 581–589. [Google Scholar] [CrossRef]
- Ulvestad, M.; Nordell, P.; Asplund, A.; Rehnström, M.; Jacobsson, S.; Holmgren, G.; Davidson, L.; Brolén, G.; Edsbagge, J.; Björquist, P.; et al. Drug metabolizing enzyme and transporter protein profiles of hepatocytes derived from human embryonic and induced pluripotent stem cells. Biochem. Pharmacol. 2013, 86, 691–702. [Google Scholar] [CrossRef]
- Ordovas, L.; Boon, R.; Pistoni, M.; Chen, Y.; Wolfs, E.; Guo, W.; Sambathkumar, R.; Bobis-wozowicz, S.; Helsen, N.; Vanhove, J.; et al. Efficient Recombinase-Mediated Cassette Exchange in hPSCs to study Hepatocyte Lineage Reveals AAVS 1 Locus-Mediated Transgene Inhibition. Stem Cell Rep. 2015, 5, 918–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boon, R.; Kumar, M.; Tricot, T.; Elia, I.; Ordovas, L.; Jacobs, F.; One, J.; De Smedt, J.; Eelen, G.; Bird, M.; et al. Amino acid levels determine metabolism and CYP450 function of stem cell derived hepatocytes and hepatoma cell lines. Nat. Commun. 2020, 11, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sison-Young, R.L.; Lauschke, V.M.; Johann, E.; Alexandre, E.; Antherieu, S.; Aerts, H.; Gerets, H.H.J.; Labbe, G.; Hoët, D.; Dorau, M.; et al. A multicenter assessment of single-cell models aligned to standard measures of cell health for prediction of acute hepatotoxicity. Arch. Toxicol. 2017, 91, 1385–1400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanhove, J.; Pistoni, M.; Welters, M.; Eggermont, K.; Vanslembrouck, V.; Helsen, N.; Boon, R.; Najimi, M.; Sokal, E.; Collas, P.; et al. H3K27me3 Does Not Orchestrate the Expression of Lineage-Specific Markers in hESC-Derived Hepatocytes In Vitro. Stem Cell Rep. 2016, 7, 192–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gripon, P.; Diot, C.; Thézé, N.; Fourel, I.; Loreal, O.; Brechot, C.; Guguen-Guillouzo, C. Hepatitis B virus infection of adult human hepatocytes cultured in the presence of dimethyl sulfoxide. J. Virol. 1988, 62, 4136–4143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Zong, L.; Sureau, C.; Barker, L.; Wands, J.R.; Tong, S. Unusual Features of Sodium Taurocholate Cotransporting Polypeptide as a Hepatitis B Virus Receptor. J. Virol. 2016, 90, 8302–8313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konig, A.; Doring, B.; Mohr, C.; Geipel, A.; Geyer, J.; Glebe, D. Kinetics of the bile acid transporter and hepatitis B virus receptor Na+/taurocholate cotransporting polypeptide (NTCP) in hepatocytes. J. Hepatol. 2014, 61, 867–875. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; Allweiss, L.; Yang, D.; Kang, J.; Wang, J.; Qian, X.; Zhang, T.; Liu, H.; Wang, L.; Liu, S.; et al. Down-regulation of cell membrane localized NTCP expression in proliferating hepatocytes prevents hepatitis B virus infection. Emerg. Microbes Infect. 2019, 8, 879–894. [Google Scholar] [CrossRef]
- Volz, T.; Allweiss, L.; ḾBarek, M.B.; Warlich, M.; Lohse, A.W.; Pollok, J.M.; Alexandrov, A.; Urban, S.; Petersen, J.; Lütgehetmann, M.; et al. The entry inhibitor Myrcludex-B efficiently blocks intrahepatic virus spreading in humanized mice previously infected with hepatitis B virus. J. Hepatol. 2013, 58, 861–867. [Google Scholar] [CrossRef]
- Li, W.; Urban, S. Entry of hepatitis B and hepatitis D virus into hepatocytes: Basic insights and clinical implications. J. Hepatol. 2016, 64, S32–S40. [Google Scholar] [CrossRef]
- Tipples, G.A.; Ma, M.M.; Fischer, K.P.; Bain, V.G.; Kneteman, N.M.; Tyrrell, D.L. Mutation in HBV RNA-dependent DNA polymerase confers resistance to lamivudine in vivo. Hepatology 1996, 24, 714–717. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.S.Y.; Covert, E.; Wilson, E.; Kottilil, S. Chronic Hepatitis B infection a review. JAMA J. Am. Med. Assoc. 2018, 319, 1802–1813. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tricot, T.; Thibaut, H.J.; Abbasi, K.; Boon, R.; Helsen, N.; Kumar, M.; Neyts, J.; Verfaillie, C. Metabolically Improved Stem Cell Derived Hepatocyte-Like Cells Support HBV Life Cycle and Are a Promising Tool for HBV Studies and Antiviral Drug Screenings. Biomedicines 2022, 10, 268. https://doi.org/10.3390/biomedicines10020268
Tricot T, Thibaut HJ, Abbasi K, Boon R, Helsen N, Kumar M, Neyts J, Verfaillie C. Metabolically Improved Stem Cell Derived Hepatocyte-Like Cells Support HBV Life Cycle and Are a Promising Tool for HBV Studies and Antiviral Drug Screenings. Biomedicines. 2022; 10(2):268. https://doi.org/10.3390/biomedicines10020268
Chicago/Turabian StyleTricot, Tine, Hendrik Jan Thibaut, Kayvan Abbasi, Ruben Boon, Nicky Helsen, Manoj Kumar, Johan Neyts, and Catherine Verfaillie. 2022. "Metabolically Improved Stem Cell Derived Hepatocyte-Like Cells Support HBV Life Cycle and Are a Promising Tool for HBV Studies and Antiviral Drug Screenings" Biomedicines 10, no. 2: 268. https://doi.org/10.3390/biomedicines10020268
APA StyleTricot, T., Thibaut, H. J., Abbasi, K., Boon, R., Helsen, N., Kumar, M., Neyts, J., & Verfaillie, C. (2022). Metabolically Improved Stem Cell Derived Hepatocyte-Like Cells Support HBV Life Cycle and Are a Promising Tool for HBV Studies and Antiviral Drug Screenings. Biomedicines, 10(2), 268. https://doi.org/10.3390/biomedicines10020268







