Evolutionary Implications of Environmental Toxicant Exposure
Abstract
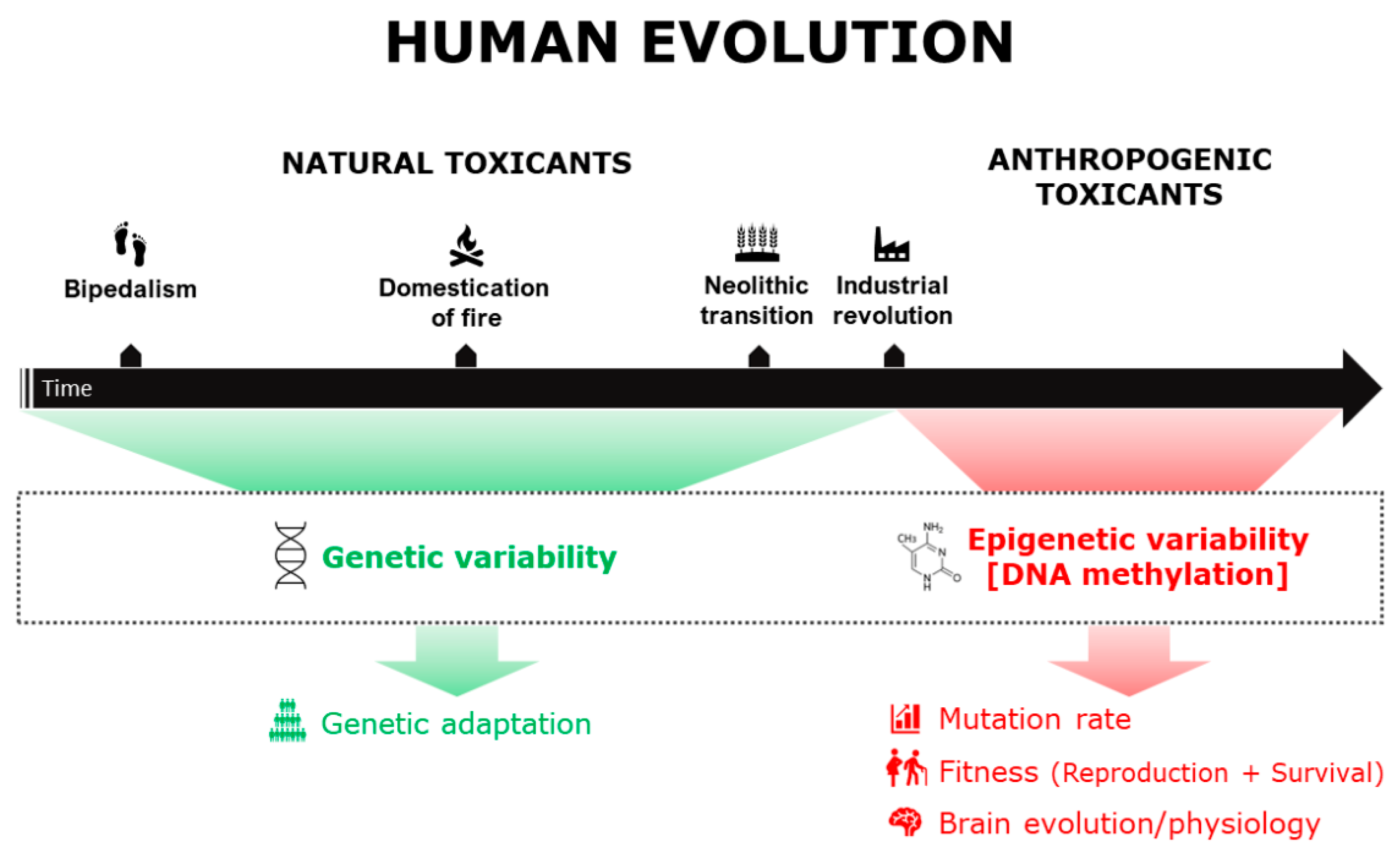
1. Introduction
2. Toxicants Exposure and Human Biodiversity
2.1. Genetic Variability
2.2. Epigenetic Variability (DNA Methylation)
3. Toxicants Exposure, DNA Methylation, and Mutation Rate
4. Link between Toxicants Exposure, DNA Methylation, and Biological Fitness
4.1. Data in Males
4.2. Data in Females
5. Link between Toxicants Exposure, DNA Methylation, and Survival
6. Link between Toxicants Exposure, DNA Methylation, and Brain Evolution
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brady, S.P.; Monosson, E.; Matson, C.W.; Bickham, J.W. Evolutionary Toxicology: Toward a Unified Understanding of Life’s Response to Toxic Chemicals. Evol. Appl. 2017, 10, 745–751. [Google Scholar] [CrossRef] [PubMed]
- Trumble, B.C.; Finch, C.E. The Exposome in Human Evolution: From Dust to Diesel. Q. Rev. Biol. 2019, 94, 333–394. [Google Scholar] [CrossRef]
- Tapia, J.; Murray, J.; Ormachea, M.; Tirado, N.; Nordstrom, D.K. Origin, Distribution, and Geochemistry of Arsenic in the Altiplano-Puna Plateau of Argentina, Bolivia, Chile, and Perú. Sci. Total Environ. 2019, 678, 309–325. [Google Scholar] [CrossRef]
- Crews, D.; Gore, A.C. Epigenetic Synthesis: A Need for a New Paradigm for Evolution in a Contaminated World. F1000 Biol. Rep. 2012, 4, 18. [Google Scholar] [CrossRef]
- Barrett, R.; Schluter, D. Adaptation from Standing Genetic Variation. Trends Ecol. Evol. 2008, 23, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Martin, E.M.; Fry, R.C. Environmental Influences on the Epigenome: Exposure- Associated DNA Methylation in Human Populations. Annu. Rev. Public Health 2018, 39, 309–333. [Google Scholar] [CrossRef]
- Russo, G.L.; Vastolo, V.; Ciccarelli, M.; Albano, L.; Macchia, P.E.; Ungaro, P. Dietary Polyphenols and Chromatin Remodeling. Crit. Rev. Food Sci. Nutr. 2017, 57, 2589–2599. [Google Scholar] [CrossRef] [PubMed]
- Nettore, I.C.; Franchini, F.; Palatucci, G.; Macchia, P.E.; Ungaro, P. Epigenetic Mechanisms of Endocrine-Disrupting Chemicals in Obesity. Biomedicines 2021, 9, 1716. [Google Scholar] [CrossRef]
- Lieb, J.D.; Beck, S.; Bulyk, M.L.; Farnham, P.; Hattori, N.; Henikoff, S.; Liu, X.S.; Okumura, K.; Shiota, K.; Ushijima, T.; et al. Applying Whole-Genome Studies of Epigenetic Regulation to Study Human Disease. Cytogenet. Genome Res. 2006, 114, 1–15. [Google Scholar] [CrossRef]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for De Novo Methylation and Mammalian Development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef]
- Rao, R.; Leibler, S. Evolutionary Dynamics, Evolutionary Forces, and Robustness: A Nonequilibrium Statistical Mechanics Perspective. Proc. Natl. Acad. Sci. USA 2022, 119, e2112083119. [Google Scholar] [CrossRef]
- Zhang, Z.; Ramstein, G.; Schuster, M.; Li, C.; Contoux, C.; Yan, Q. Aridification of the Sahara Desert Caused by Tethys Sea Shrinkage during the Late Miocene. Nature 2014, 513, 401–404. [Google Scholar] [CrossRef]
- Dupont, L. Reconstructing Pathways of Aeolian Pollen Transport to the Marine Sediments along the Coastline of SW Africa. Quat. Sci. Rev. 2003, 22, 157–174. [Google Scholar] [CrossRef]
- Thakur, S.A.; Hamilton, R.F.; Holian, A. Role of Scavenger Receptor A Family in Lung Inflammation from Exposure to Environmental Particles. J. Immunotoxicol. 2008, 5, 151–157. [Google Scholar] [CrossRef]
- Novakowski, K.E.; Yap, N.V.L.; Yin, C.; Sakamoto, K.; Heit, B.; Golding, G.B.; Bowdish, D.M.E. Human-Specific Mutations and Positively Selected Sites in MARCO Confer Functional Changes. Mol. Biol. Evol. 2018, 35, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, R.F.; Thakur, S.A.; Mayfair, J.K.; Holian, A. MARCO Mediates Silica Uptake and Toxicity in Alveolar Macrophages from C57BL/6 Mice. J. Biol. Chem. 2006, 281, 34218–34226. [Google Scholar] [CrossRef]
- Sun, W.; Hong, Y.; Li, T.; Chu, H.; Liu, J.; Feng, L. Application of Sulfur-Coated Magnetic Carbon Nanotubes for Extraction of Some Polycyclic Aromatic Hydrocarbons from Water Resources. Chemosphere 2022, 309, 136632. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, H.; Zhang, H.; Niu, Y.; Fu, Y.; Nie, J.; Yang, A.; Zhao, J.; Yang, J. Mediation Effect of AhR Expression between Polycyclic Aromatic Hydrocarbons Exposure and Oxidative DNA Damage among Chinese Occupational Workers. Environ. Pollut. 2018, 243, 972–977. [Google Scholar] [CrossRef]
- Degrelle, S.A.; Ferecatu, I.; Fournier, T. Novel Fluorescent and Secreted Transcriptional Reporters for Quantifying Activity of the Xenobiotic Sensor Aryl Hydrocarbon Receptor (AHR). Environ. Int. 2022, 169, 107545. [Google Scholar] [CrossRef]
- Hubbard, T.D.; Murray, I.A.; Bisson, W.H.; Sullivan, A.P.; Sebastian, A.; Perry, G.H.; Jablonski, N.G.; Perdew, G.H. Divergent Ah Receptor Ligand Selectivity during Hominin Evolution. Mol. Biol. Evol. 2016, 33, 2648–2658. [Google Scholar] [CrossRef]
- Pogribny, I.P.; Rusyn, I. Environmental Toxicants, Epigenetics, and Cancer. In Epigenetic Alterations in Oncogenesis; Karpf, A.R., Ed.; Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2013; Volume 754, pp. 215–232. ISBN 978-1-4419-9966-5. [Google Scholar]
- Schlebusch, C.M.; Gattepaille, L.M.; Engström, K.; Vahter, M.; Jakobsson, M.; Broberg, K. Human Adaptation to Arsenic-Rich Environments. Mol. Biol. Evol. 2015, 32, 1544–1555. [Google Scholar] [CrossRef] [PubMed]
- De Loma, J.; Vicente, M.; Tirado, N.; Ascui, F.; Vahter, M.; Gardon, J.; Schlebusch, C.M.; Broberg, K. Human Adaptation to Arsenic in Bolivians Living in the Andes. Chemosphere 2022, 301, 134764. [Google Scholar] [CrossRef]
- Schlebusch, C.M.; Lewis, C.M.; Vahter, M.; Engström, K.; Tito, R.Y.; Obregón-Tito, A.J.; Huerta, D.; Polo, S.I.; Medina, Á.C.; Brutsaert, T.D.; et al. Possible Positive Selection for an Arsenic-Protective Haplotype in Humans. Environ. Health Perspect. 2013, 121, 53–58. [Google Scholar] [CrossRef]
- Heyn, H.; Moran, S.; Hernando-Herraez, I.; Sayols, S.; Gomez, A.; Sandoval, J.; Monk, D.; Hata, K.; Marques-Bonet, T.; Wang, L.; et al. DNA Methylation Contributes to Natural Human Variation. Genome Res. 2013, 23, 1363–1372. [Google Scholar] [CrossRef] [PubMed]
- Fraga, M.F.; Ballestar, E.; Paz, M.F.; Ropero, S.; Setien, F.; Ballestar, M.L.; Heine-Suñer, D.; Cigudosa, J.C.; Urioste, M.; Benitez, J.; et al. Epigenetic Differences Arise during the Lifetime of Monozygotic Twins. Proc. Natl. Acad. Sci. USA 2005, 102, 10604–10609. [Google Scholar] [CrossRef]
- Fraser, H.B.; Lam, L.L.; Neumann, S.M.; Kobor, M.S. Population-Specificity of Human DNA Methylation. Genome Biol. 2012, 13, R8. [Google Scholar] [CrossRef]
- Galanter, J.M.; Gignoux, C.R.; Oh, S.S.; Torgerson, D.; Pino-Yanes, M.; Thakur, N.; Eng, C.; Hu, D.; Huntsman, S.; Farber, H.J.; et al. Differential Methylation between Ethnic Sub-Groups Reflects the Effect of Genetic Ancestry and Environmental Exposures. eLife 2017, 6, e20532. [Google Scholar] [CrossRef]
- McKennan, C.; Naughton, K.; Stanhope, C.; Kattan, M.; O’Connor, G.T.; Sandel, M.T.; Visness, C.M.; Wood, R.A.; Bacharier, L.B.; Beigelman, A.; et al. Longitudinal Data Reveal Strong Genetic and Weak Non-Genetic Components of Ethnicity-Dependent Blood DNA Methylation Levels. Epigenetics 2021, 16, 662–676. [Google Scholar] [CrossRef]
- Rauschert, S.; Melton, P.E.; Burdge, G.; Craig, J.M.; Godfrey, K.M.; Holbrook, J.D.; Lillycrop, K.; Mori, T.A.; Beilin, L.J.; Oddy, W.H.; et al. Maternal Smoking During Pregnancy Induces Persistent Epigenetic Changes into Adolescence, Independent of Postnatal Smoke Exposure and Is Associated with Cardiometabolic Risk. Front. Genet. 2019, 10, 770. [Google Scholar] [CrossRef]
- Giuliani, C.; Biggs, D.; Nguyen, T.T.; Marasco, E.; De Fanti, S.; Garagnani, P.; Le Phan, M.T.; Nguyen, V.N.; Luiselli, D.; Romeo, G. First Evidence of Association between Past Environmental Exposure to Dioxin and DNA Methylation of CYP1A1 and IGF2 Genes in Present Day Vietnamese Population. Environ. Pollut. 2018, 242, 976–985. [Google Scholar] [CrossRef] [PubMed]
- Alegría-Torres, J.A.; Carrizales-Yánez, L.; Díaz-Barriga, F.; Rosso-Camacho, F.; Motta, V.; Tarantini, L.; Bollati, V. DNA Methylation Changes in Mexican Children Exposed to Arsenic from Two Historic Mining Areas in San Luis Potosí: DNA Methylation Changes. Environ. Mol. Mutagen. 2016, 57, 717–723. [Google Scholar] [CrossRef]
- Gonzalez-Cortes, T.; Recio-Vega, R.; Lantz, R.C.; Chau, B.T. DNA Methylation of Extracellular Matrix Remodeling Genes in Children Exposed to Arsenic. Toxicol. Appl. Pharmacol. 2017, 329, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Reichard, J.F.; Puga, A. Effects of Arsenic Exposure on DNA Methylation and Epigenetic Gene Regulation. Epigenomics 2010, 2, 87–104. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.Q.; Young, M.R.; Diwan, B.A.; Coogan, T.P.; Waalkes, M.P. Association of Arsenic-Induced Malignant Transformation with DNA Hypomethylation and Aberrant Gene Expression. Proc. Natl. Acad. Sci. USA 1997, 94, 10907–10912. [Google Scholar] [CrossRef] [PubMed]
- Benbrahim-Tallaa, L.; Waterland, R.A.; Styblo, M.; Achanzar, W.E.; Webber, M.M.; Waalkes, M.P. Molecular Events Associated with Arsenic-Induced Malignant Transformation of Human Prostatic Epithelial Cells: Aberrant Genomic DNA Methylation and K-Ras Oncogene Activation. Toxicol. Appl. Pharmacol. 2005, 206, 288–298. [Google Scholar] [CrossRef]
- Cui, X.; Wakai, T.; Shirai, Y.; Yokoyama, N.; Hatakeyama, K.; Hirano, S. Arsenic Trioxide Inhibits DNA Methyltransferase and Restores Methylation-Silenced Genes in Human Liver Cancer Cells. Hum. Pathol. 2006, 37, 298–311. [Google Scholar] [CrossRef] [PubMed]
- Reichard, J.F.; Schnekenburger, M.; Puga, A. Long Term Low-Dose Arsenic Exposure Induces Loss of DNA Methylation. Biochem. Biophys. Res. Commun. 2007, 352, 188–192. [Google Scholar] [CrossRef]
- Ameer, S.S.; Engström, K.; Hossain, M.B.; Concha, G.; Vahter, M.; Broberg, K. Arsenic Exposure from Drinking Water Is Associated with Decreased Gene Expression and Increased DNA Methylation in Peripheral Blood. Toxicol. Appl. Pharmacol. 2017, 321, 57–66. [Google Scholar] [CrossRef]
- Rojas, D.; Rager, J.E.; Smeester, L.; Bailey, K.A.; Drobná, Z.; Rubio-Andrade, M.; Stýblo, M.; García-Vargas, G.; Fry, R.C. Prenatal Arsenic Exposure and the Epigenome: Identifying Sites of 5-Methylcytosine Alterations That Predict Functional Changes in Gene Expression in Newborn Cord Blood and Subsequent Birth Outcomes. Toxicol. Sci. 2015, 143, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Horsthemke, B. A Critical View on Transgenerational Epigenetic Inheritance in Humans. Nat. Commun. 2018, 9, 2973. [Google Scholar] [CrossRef]
- Chen, C.; Qi, H.; Shen, Y.; Pickrell, J.; Przeworski, M. Contrasting Determinants of Mutation Rates in Germline and Soma. Genetics 2017, 207, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; He, F.; Pu, W.; Gu, X.; Wang, J.; Su, Z. The Impact of DNA Methylation Dynamics on the Mutation Rate During Human Germline Development. G3 Genes Genomes Genet. 2020, 10, 3337–3346. [Google Scholar] [CrossRef]
- Skinner, M.K. Epigenetic Transgenerational Inheritance. Nat. Rev. Endocrinol. 2016, 12, 68–70. [Google Scholar] [CrossRef] [PubMed]
- Skinner, M.K.; Guerrero-Bosagna, C.; Haque, M.M. Environmentally Induced Epigenetic Transgenerational Inheritance of Sperm Epimutations Promote Genetic Mutations. Epigenetics 2015, 10, 762–771. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Han, L.; Zhao, Z. Investigating the Relationship of DNA Methylation with Mutation Rate and Allele Frequency in the Human Genome. BMC Genom. 2012, 13, S7. [Google Scholar] [CrossRef] [PubMed]
- Duncan, B.K.; Miller, J.H. Mutagenic Deamination of Cytosine Residues in DNA. Nature 1980, 287, 560–561. [Google Scholar] [CrossRef]
- Agarwal, I.; Przeworski, M. Mutation Saturation for Fitness Effects at Human CpG Sites. eLife 2021, 10, e71513. [Google Scholar] [CrossRef]
- Ashe, A.; Colot, V.; Oldroyd, B.P. How Does Epigenetics Influence the Course of Evolution? Philos. Trans. R. Soc. B Biol. Sci. 2021, 376, 20200111. [Google Scholar] [CrossRef]
- Zhang, A.; Pan, X.; Xia, Y.; Xiao, Q.; Huang, X. Relationship between the methylation and mutation of p53 gene and endemic arsenism caused by coal-burning. Zhonghua Yu Fang Yi Xue Za Zhi 2011, 45, 393–398. [Google Scholar]
- Neven, K.Y.; Saenen, N.D.; Tarantini, L.; Janssen, B.G.; Lefebvre, W.; Vanpoucke, C.; Bollati, V.; Nawrot, T.S. Placental Promoter Methylation of DNA Repair Genes and Prenatal Exposure to Particulate Air Pollution: An ENVIR ON AGE Cohort Study. Lancet Planet. Health 2018, 2, e174–e183. [Google Scholar] [CrossRef]
- Jobling, M.; Hollox, E.; Hurles, M.; Kivisild, T.; Tyler-Smith, C. Human Evolutionary Genetics, 2nd ed.; Garland Science: New York, NY, USA, 2014; p. 149. ISBN 978-0-8153-4148-2. [Google Scholar]
- Brüne, M.; Schiefenhöve Brüne, M.; Schiefenhövel, W. The Oxford Handbook of Evolutionary Medicine, 1st ed.; Oxford Handbooks; Oxford University Press: Oxford, UK, 2019; p. 219. ISBN 978-0-19-878966-6. [Google Scholar]
- Lidborg, L.H.; Cross, C.P.; Boothroyd, L.G. A Meta-Analysis of the Association between Male Dimorphism and Fitness Outcomes in Humans. eLife 2022, 11, e65031. [Google Scholar] [CrossRef] [PubMed]
- Schild, C.; Stern, J.; Penke, L.; Zettler, I. Voice Pitch—A Valid Indicator of One’s Unfaithfulness in Committed Relationships? Adapt. Hum. Behav. Physiol. 2021, 7, 245–260. [Google Scholar] [CrossRef]
- Krieg, S.A.; Shahine, L.K.; Lathi, R.B. Environmental Exposure to Endocrine-Disrupting Chemicals and Miscarriage. Fertil. Steril. 2016, 106, 941–947. [Google Scholar] [CrossRef] [PubMed]
- Tobi, E.W.; van den Heuvel, J.; Zwaan, B.J.; Lumey, L.H.; Heijmans, B.T.; Uller, T. Selective Survival of Embryos Can Explain DNA Methylation Signatures of Adverse Prenatal Environments. Cell Rep. 2018, 25, 2660–2667. [Google Scholar] [CrossRef] [PubMed]
- Di Nisio, A.; Sabovic, I.; Valente, U.; Tescari, S.; Rocca, M.S.; Guidolin, D.; Dall’Acqua, S.; Acquasaliente, L.; Pozzi, N.; Plebani, M.; et al. Endocrine Disruption of Androgenic Activity by Perfluoroalkyl Substances: Clinical and Experimental Evidence. J. Clin. Endocrinol. Metab. 2019, 104, 1259–1271. [Google Scholar] [CrossRef]
- Pan, Y.; Cui, Q.; Wang, J.; Sheng, N.; Jing, J.; Yao, B.; Dai, J. Profiles of Emerging and Legacy Per-/Polyfluoroalkyl Substances in Matched Serum and Semen Samples: New Implications for Human Semen Quality. Environ. Health Perspect. 2019, 127, 127005. [Google Scholar] [CrossRef]
- Montjean, D.; Neyroud, A.-S.; Yefimova, M.G.; Benkhalifa, M.; Cabry, R.; Ravel, C. Impact of Endocrine Disruptors upon Non-Genetic Inheritance. Int. J. Mol. Sci. 2022, 23, 3350. [Google Scholar] [CrossRef]
- Mima, M.; Greenwald, D.; Ohlander, S. Environmental Toxins and Male Fertility. Curr. Urol. Rep. 2018, 19, 50. [Google Scholar] [CrossRef]
- Luján, S.; Caroppo, E.; Niederberger, C.; Arce, J.-C.; Sadler-Riggleman, I.; Beck, D.; Nilsson, E.; Skinner, M.K. Sperm DNA Methylation Epimutation Biomarkers for Male Infertility and FSH Therapeutic Responsiveness. Sci. Rep. 2019, 9, 16786. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, S.; Li, S. Relationship between Cadmium Content in Semen and Male Infertility: A Meta-Analysis. Environ. Sci. Pollut. Res. 2019, 26, 1947–1953. [Google Scholar] [CrossRef]
- Agarwal, A.; Mulgund, A.; Hamada, A.; Chyatte, M.R. A Unique View on Male Infertility around the Globe. Reprod. Biol. Endocrinol. 2015, 13, 37. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xu, J.; Yang, T.; Chen, J.; Li, F.; Shen, B.; Fan, C. Genome-Wide Methylation Analyses of Human Sperm Unravel Novel Differentially Methylated Regions in Asthenozoospermia. Epigenomics 2022, 14, 951–964. [Google Scholar] [CrossRef] [PubMed]
- Marques, C.J.; Costa, P.; Vaz, B.; Carvalho, F.; Fernandes, S.; Barros, A.; Sousa, M. Abnormal Methylation of Imprinted Genes in Human Sperm Is Associated with Oligozoospermia. MHR Basic Sci. Reprod. Med. 2008, 14, 67–74. [Google Scholar] [CrossRef]
- Montjean, D.; Ravel, C.; Benkhalifa, M.; Cohen-Bacrie, P.; Berthaut, I.; Bashamboo, A.; McElreavey, K. Methylation Changes in Mature Sperm Deoxyribonucleic Acid from Oligozoospermic Men: Assessment of Genetic Variants and Assisted Reproductive Technology Outcome. Fertil. Steril. 2013, 100, 1241–1247. [Google Scholar] [CrossRef] [PubMed]
- Houshdaran, S.; Cortessis, V.K.; Siegmund, K.; Yang, A.; Laird, P.W.; Sokol, R.Z. Widespread Epigenetic Abnormalities Suggest a Broad DNA Methylation Erasure Defect in Abnormal Human Sperm. PLoS ONE 2007, 2, e1289. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Zhang, P.; Shen, W.; Zhao, Y.; Zhang, H. Estrogen Receptor-Related DNA and Histone Methylation May Be Involved in the Transgenerational Disruption in Spermatogenesis by Selective Toxic Chemicals. Front. Pharmacol. 2019, 10, 1012. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Cai, D.; Li, X.; Liu, B.; Chen, J.; Jiang, X.; Li, H.; Li, Z.; Teerds, K.; Sun, J.; et al. Effects of Bisphenol A on Reproductive Toxicity and Gut Microbiota Dysbiosis in Male Rats. Ecotoxicol. Environ. Saf. 2022, 239, 113623. [Google Scholar] [CrossRef]
- Aneck-Hahn, N.H.; Schulenburg, G.W.; Bornman, M.S.; Farias, P.; De Jager, C. Impaired Semen Quality Associated with Environmental DDT Exposure in Young Men Living in a Malaria Area in the Limpopo Province, South Africa. J. Androl. 2006, 28, 423–434. [Google Scholar] [CrossRef]
- Skinner, M.K.; Manikkam, M.; Tracey, R.; Guerrero-Bosagna, C.; Haque, M.; Nilsson, E.E. Ancestral Dichlorodiphenyltrichloroethane (DDT) Exposure Promotes Epigenetic Transgenerational Inheritance of Obesity. BMC Med. 2013, 11, 228. [Google Scholar] [CrossRef]
- Song, Y.; Wu, N.; Wang, S.; Gao, M.; Song, P.; Lou, J.; Tan, Y.; Liu, K. Transgenerational Impaired Male Fertility with an Igf2 Epigenetic Defect in the Rat Are Induced by the Endocrine Disruptor p,p’-DDE. Hum. Reprod. 2014, 29, 2512–2521. [Google Scholar] [CrossRef]
- Hu, J.; Yang, Y.; Lv, X.; Lao, Z.; Yu, L. Dichlorodiphenyltrichloroethane Metabolites Inhibit DNMT1 Activity Which Confers Methylation-Specific Modulation of the Sex Determination Pathway. Environ. Pollut. 2021, 279, 116828. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.S.; de Cavalcante, D.N.C.; da Araújo Santiago, M.S.; de Medeiros, P.D.C.; do Nascimento, C.C.; Fonseca, G.F.C.; Le Sueur-Maluf, L.; Perobelli, J.E. Reproductive Toxicity in Male Juvenile Rats: Antagonistic Effects between Isolated Agrochemicals and in Binary or Ternary Combinations. Ecotoxicol. Environ. Saf. 2021, 209, 111766. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, P.; Zhao, Y.; Zhang, H. Low Dose Carbendazim Disrupts Mouse Spermatogenesis Might Be through Estrogen Receptor Related Histone and DNA Methylation. Ecotoxicol. Environ. Saf. 2019, 176, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Anway, M.D.; Leathers, C.; Skinner, M.K. Endocrine Disruptor Vinclozolin Induced Epigenetic Transgenerational Adult-Onset Disease. Endocrinology 2006, 147, 5515–5523. [Google Scholar] [CrossRef] [PubMed]
- Stouder, C.; Paoloni-Giacobino, A. Transgenerational Effects of the Endocrine Disruptor Vinclozolin on the Methylation Pattern of Imprinted Genes in the Mouse Sperm. Reproduction 2010, 139, 373–379. [Google Scholar] [CrossRef]
- Doshi, T.; Mehta, S.S.; Dighe, V.; Balasinor, N.; Vanage, G. Hypermethylation of Estrogen Receptor Promoter Region in Adult Testis of Rats Exposed Neonatally to Bisphenol A. Toxicology 2011, 289, 74–82. [Google Scholar] [CrossRef]
- El Henafy, H.M.A.; Ibrahim, M.A.; Abd El Aziz, S.A.; Gouda, E.M. Oxidative Stress and DNA Methylation in Male Rat Pups Provoked by the Transplacental and Translactational Exposure to Bisphenol A. Environ. Sci. Pollut. Res. 2020, 27, 4513–4519. [Google Scholar] [CrossRef]
- Kundakovic, M.; Gudsnuk, K.; Franks, B.; Madrid, J.; Miller, R.L.; Perera, F.P.; Champagne, F.A. Sex-Specific Epigenetic Disruption and Behavioral Changes Following Low-Dose in Utero Bisphenol A Exposure. Proc. Natl. Acad. Sci. USA 2013, 110, 9956–9961. [Google Scholar] [CrossRef]
- Li, Y.; Duan, F.; Zhou, X.; Pan, H.; Li, R. Differential Responses of GC-1 Spermatogonia Cells to High and Low Doses of Bisphenol A. Mol. Med. Rep. 2018, 18, 3034–3040. [Google Scholar] [CrossRef]
- Yin, L.; Dai, Y.; Jiang, X.; Liu, Y.; Chen, H.; Han, F.; Cao, J.; Liu, J. Role of DNA Methylation in Bisphenol A Exposed Mouse Spermatocyte. Environ. Toxicol. Pharmacol. 2016, 48, 265–271. [Google Scholar] [CrossRef]
- Miao, M.; Zhou, X.; Li, Y.; Zhang, O.; Zhou, Z.; Li, T.; Yuan, W.; Li, R.; Li, D.-K. LINE-1 Hypomethylation in Spermatozoa Is Associated with Bisphenol A Exposure. Andrology 2014, 2, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.-R.; Zhang, Y.-T.; Han, X.-Y.; Zhang, L.; Jiang, L.-G.; Ma, Y.; Meng, L.-J.; He, Q.-L.; Liu, S.-Z. Intrauterine Exposure to 2,3′,4,4′,5-Pentachlorobiphenyl Alters Spermatogenesis and Testicular DNA Methylation Levels in F1 Male Mice. Ecotoxicol. Environ. Saf. 2021, 224, 112652. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; He, X.; Liu, Q.; Wang, X.; Ding, X.; Li, H. Frequent and Mild Scrotal Heat Stress in Mice Epigenetically Alters Glucose Metabolism in the Male Offspring. Am. J. Physiol.-Endocrinol. Metab. 2020, 319, E291–E304. [Google Scholar] [CrossRef]
- Maurice, C.; Dalvai, M.; Lambrot, R.; Deschênes, A.; Scott-Boyer, M.-P.; McGraw, S.; Chan, D.; Côté, N.; Ziv-Gal, A.; Flaws, J.A.; et al. Early-Life Exposure to Environmental Contaminants Perturbs the Sperm Epigenome and Induces Negative Pregnancy Outcomes for Three Generations via the Paternal Lineage. Epigenomes 2021, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Lessard, M.; Herst, P.M.; Charest, P.L.; Navarro, P.; Joly-Beauparlant, C.; Droit, A.; Kimmins, S.; Trasler, J.; Benoit-Biancamano, M.-O.; MacFarlane, A.J.; et al. Prenatal Exposure to Environmentally-Relevant Contaminants Perturbs Male Reproductive Parameters Across Multiple Generations That Are Partially Protected by Folic Acid Supplementation. Sci. Rep. 2019, 9, 13829. [Google Scholar] [CrossRef] [PubMed]
- Leung, Y.-K.; Ouyang, B.; Niu, L.; Xie, C.; Ying, J.; Medvedovic, M.; Chen, A.; Weihe, P.; Valvi, D.; Grandjean, P.; et al. Identification of Sex-Specific DNA Methylation Changes Driven by Specific Chemicals in Cord Blood in a Faroese Birth Cohort. Epigenetics 2018, 13, 290–300. [Google Scholar] [CrossRef]
- Yauk, C.; Polyzos, A.; Rowan-Carroll, A.; Somers, C.M.; Godschalk, R.W.; Van Schooten, F.J.; Berndt, M.L.; Pogribny, I.P.; Koturbash, I.; Williams, A.; et al. Germ-Line Mutations, DNA Damage, and Global Hypermethylation in Mice Exposed to Particulate Air Pollution in an Urban/Industrial Location. Proc. Natl. Acad. Sci. USA 2008, 105, 605–610. [Google Scholar] [CrossRef]
- Schuller, A.; Bellini, C.; Jenkins, T.; Eden, M.; Matz, J.; Oakes, J.; Montrose, L. Simulated Wildfire Smoke Significantly Alters Sperm DNA Methylation Patterns in a Murine Model. Toxics 2021, 9, 199. [Google Scholar] [CrossRef]
- Cheng, Y.; Tang, Q.; Lu, Y.; Li, M.; Zhou, Y.; Wu, P.; Li, J.; Pan, F.; Han, X.; Chen, M.; et al. Semen Quality and Sperm DNA Methylation in Relation to Long-Term Exposure to Air Pollution in Fertile Men: A Cross-Sectional Study. Environ. Pollut. 2022, 300, 118994. [Google Scholar] [CrossRef]
- Vander Borght, M.; Wyns, C. Fertility and Infertility: Definition and Epidemiology. Clin. Biochem. 2018, 62, 2–10. [Google Scholar] [CrossRef]
- Zama, A.M.; Uzumcu, M. Fetal and Neonatal Exposure to the Endocrine Disruptor Methoxychlor Causes Epigenetic Alterations in Adult Ovarian Genes. Endocrinology 2009, 150, 4681–4691. [Google Scholar] [CrossRef]
- Manikkam, M.; Haque, M.M.; Guerrero-Bosagna, C.; Nilsson, E.E.; Skinner, M.K. Pesticide Methoxychlor Promotes the Epigenetic Transgenerational Inheritance of Adult-Onset Disease through the Female Germline. PLoS ONE 2014, 9, e102091. [Google Scholar] [CrossRef]
- Zama, A.M.; Uzumcu, M. Targeted Genome-Wide Methylation and Gene Expression Analyses Reveal Signaling Pathways Involved in Ovarian Dysfunction after Developmental EDC Exposure in Rats1. Biol. Reprod. 2013, 88, 1–13. [Google Scholar] [CrossRef]
- Gao, Y.; Zheng, H.; Xia, Y.; Cai, M. Global Scale Distribution, Seasonal Changes and Long-Range Transport Potentiality of Endosulfan in the Surface Seawater and Air. Chemosphere 2020, 260, 127634. [Google Scholar] [CrossRef]
- Milesi, M.M.; Lorenz, V.; Varayoud, J. Aberrant Hoxa10 Gene Methylation as a Mechanism for Endosulfan-Induced Implantation Failures in Rats. Mol. Cell. Endocrinol. 2022, 547, 111576. [Google Scholar] [CrossRef]
- Milesi, M.M.; Varayoud, J.; Ramos, J.G.; Luque, E.H. Uterine ERα Epigenetic Modifications Are Induced by the Endocrine Disruptor Endosulfan in Female Rats with Impaired Fertility. Mol. Cell. Endocrinol. 2017, 454, 1–11. [Google Scholar] [CrossRef]
- Qu, X.-L.; Zhang, M.; Fang, Y.; Wang, H.; Zhang, Y.-Z. Effect of 2,3′,4,4′,5-Pentachlorobiphenyl Exposure on Endometrial Receptivity and the Methylation of HOXA10. Reprod. Sci. 2018, 25, 256–268. [Google Scholar] [CrossRef]
- Bromer, J.G.; Zhou, Y.; Taylor, M.B.; Doherty, L.; Taylor, H.S. Bisphenol-A Exposure in Utero Leads to Epigenetic Alterations in the Developmental Programming of Uterine Estrogen Response. FASEB J. 2010, 24, 2273–2280. [Google Scholar] [CrossRef]
- Cohn, B.A.; Cirillo, P.M.; Wolff, M.S.; Schwingl, P.J.; Cohen, R.D.; Sholtz, R.I.; Ferrara, A.; Christianson, R.E.; van den Berg, B.J.; Siiteri, P.K. DDT and DDE Exposure in Mothers and Time to Pregnancy in Daughters. Lancet 2003, 361, 2205–2206. [Google Scholar] [CrossRef]
- Franceschi, C.; Garagnani, P.; Vitale, G.; Capri, M.; Salvioli, S. Inflammaging and ‘Garb-Aging’. Trends Endocrinol. Metab. 2017, 28, 199–212. [Google Scholar] [CrossRef]
- Derevyanko, A.; Skowronska, A.; Skowronski, M.T.; Kordowitzki, P. The Interplay between Telomeres, Mitochondria, and Chronic Stress Exposure in the Aging Egg. Cells 2022, 11, 2612. [Google Scholar] [CrossRef]
- Zhang, F.-L.; Li, W.-D.; Chu, H.T.; Luk, A.C.S.; Tsang, S.W.; Lee, W.K.; Tang, P.M.-K.; Chan, W.-Y.; Chow, K.L.; Chan, D.Y.L.; et al. Parallel Bimodal Single-Cell Sequencing of Transcriptome and Methylome Provides Molecular and Translational Insights on Oocyte Maturation and Maternal Aging. Genomics 2022, 114, 110379. [Google Scholar] [CrossRef]
- Marioni, R.E.; Shah, S.; McRae, A.F.; Chen, B.H.; Colicino, E.; Harris, S.E.; Gibson, J.; Henders, A.K.; Redmond, P.; Cox, S.R.; et al. DNA Methylation Age of Blood Predicts All-Cause Mortality in Later Life. Genome Biol. 2015, 16, 25. [Google Scholar] [CrossRef]
- Horvath, S. DNA Methylation Age of Human Tissues and Cell Types. Genome Biol. 2013, 14, R115. [Google Scholar] [CrossRef]
- Ryan, C.P. “Epigenetic Clocks”: Theory and Applications in Human Biology. Am. J. Hum. Biol. 2021, 33, e23488. [Google Scholar] [CrossRef]
- Horvath, S.; Oshima, J.; Martin, G.M.; Lu, A.T.; Quach, A.; Cohen, H.; Felton, S.; Matsuyama, M.; Lowe, D.; Kabacik, S.; et al. Epigenetic Clock for Skin and Blood Cells Applied to Hutchinson Gilford Progeria Syndrome and Ex Vivo Studies. Aging 2018, 10, 1758–1775. [Google Scholar] [CrossRef]
- Levine, M.E.; Lu, A.T.; Quach, A.; Chen, B.H.; Assimes, T.L.; Bandinelli, S.; Hou, L.; Baccarelli, A.A.; Stewart, J.D.; Li, Y.; et al. An Epigenetic Biomarker of Aging for Lifespan and Healthspan. Aging 2018, 10, 573–591. [Google Scholar] [CrossRef]
- Lu, A.T.; Quach, A.; Wilson, J.G.; Reiner, A.P.; Aviv, A.; Raj, K.; Hou, L.; Baccarelli, A.A.; Li, Y.; Stewart, J.D.; et al. DNA Methylation GrimAge Strongly Predicts Lifespan and Healthspan. Aging 2019, 11, 303–327. [Google Scholar] [CrossRef]
- Ross, K.M.; Carroll, J.E.; Horvath, S.; Hobel, C.J.; Coussons-Read, M.E.; Dunkel Schetter, C. Epigenetic Age and Pregnancy Outcomes: GrimAge Acceleration Is Associated with Shorter Gestational Length and Lower Birthweight. Clin. Epigenetics 2020, 12, 120. [Google Scholar] [CrossRef]
- van der Laan, L.; Cardenas, A.; Vermeulen, R.; Fadadu, R.P.; Hubbard, A.E.; Phillips, R.V.; Zhang, L.; Breeze, C.; Hu, W.; Wen, C.; et al. Epigenetic Aging Biomarkers and Occupational Exposure to Benzene, Trichloroethylene and Formaldehyde. Environ. Int. 2022, 158, 106871. [Google Scholar] [CrossRef]
- de Prado-Bert, P.; Ruiz-Arenas, C.; Vives-Usano, M.; Andrusaityte, S.; Cadiou, S.; Carracedo, Á.; Casas, M.; Chatzi, L.; Dadvand, P.; González, J.R.; et al. The Early-Life Exposome and Epigenetic Age Acceleration in Children. Environ. Int. 2021, 155, 106683. [Google Scholar] [CrossRef]
- White, A.J.; Kresovich, J.K.; Keller, J.P.; Xu, Z.; Kaufman, J.D.; Weinberg, C.R.; Taylor, J.A.; Sandler, D.P. Air Pollution, Particulate Matter Composition and Methylation-Based Biologic Age. Environ. Int. 2019, 132, 105071. [Google Scholar] [CrossRef]
- Nwanaji-Enwerem, J.C.; Dai, L.; Colicino, E.; Oulhote, Y.; Di, Q.; Kloog, I.; Just, A.C.; Hou, L.; Vokonas, P.; Baccarelli, A.A.; et al. Associations between Long-Term Exposure to PM2.5 Component Species and Blood DNA Methylation Age in the Elderly: The VA Normative Aging Study. Environ. Int. 2017, 102, 57–65. [Google Scholar] [CrossRef]
- Cardenas, A.; Ecker, S.; Fadadu, R.P.; Huen, K.; Orozco, A.; McEwen, L.M.; Engelbrecht, H.-R.; Gladish, N.; Kobor, M.S.; Rosero-Bixby, L.; et al. Epigenome-Wide Association Study and Epigenetic Age Acceleration Associated with Cigarette Smoking among Costa Rican Adults. Sci. Rep. 2022, 12, 4277. [Google Scholar] [CrossRef]
- Bozack, A.K.; Boileau, P.; Hubbard, A.E.; Sillé, F.C.M.; Ferreccio, C.; Steinmaus, C.M.; Smith, M.T.; Cardenas, A. The Impact of Prenatal and Early-Life Arsenic Exposure on Epigenetic Age Acceleration among Adults in Northern Chile. Environ. Epigenetics 2022, 8, dvac014. [Google Scholar] [CrossRef]
- Herrera-Moreno, J.F.; Estrada-Gutierrez, G.; Wu, H.; Bloomquist, T.R.; Rosa, M.J.; Just, A.C.; Lamadrid-Figueroa, H.; Téllez-Rojo, M.M.; Wright, R.O.; Baccarelli, A.A. Prenatal Lead Exposure, Telomere Length in Cord Blood, and DNA Methylation Age in the PROGRESS Prenatal Cohort. Environ. Res. 2022, 205, 112577. [Google Scholar] [CrossRef]
- Xu, Y.; Jurkovic-Mlakar, S.; Lindh, C.H.; Scott, K.; Fletcher, T.; Jakobsson, K.; Engström, K. Associations between Serum Concentrations of Perfluoroalkyl Substances and DNA Methylation in Women Exposed through Drinking Water: A Pilot Study in Ronneby, Sweden. Environ. Int. 2020, 145, 106148. [Google Scholar] [CrossRef]
- Lister, R.; Mukamel, E.A.; Nery, J.R.; Urich, M.; Puddifoot, C.A.; Johnson, N.D.; Lucero, J.; Huang, Y.; Dwork, A.J.; Schultz, M.D.; et al. Global Epigenomic Reconfiguration During Mammalian Brain Development. Science 2013, 341, 1237905. [Google Scholar] [CrossRef]
- Feng, J.; Fouse, S.; Fan, G. Epigenetic Regulation of Neural Gene Expression and Neuronal Function. Pediatr. Res. 2007, 61, 58R–63R. [Google Scholar] [CrossRef]
- Jeong, H.; Mendizabal, I.; Berto, S.; Chatterjee, P.; Layman, T.; Usui, N.; Toriumi, K.; Douglas, C.; Singh, D.; Huh, I.; et al. Evolution of DNA Methylation in the Human Brain. Nat. Commun. 2021, 12, 2021. [Google Scholar] [CrossRef]
- Keil, K.P.; Lein, P.J. DNA Methylation: A Mechanism Linking Environmental Chemical Exposures to Risk of Autism Spectrum Disorders? Environ. Epigenetics 2016, 2, dvv012. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Martín, F.J.; Lindquist, D.M.; Landero-Figueroa, J.; Zhang, X.; Chen, J.; Cecil, K.M.; Medvedovic, M.; Puga, A. Sex- and Tissue-Specific Methylome Changes in Brains of Mice Perinatally Exposed to Lead. Neurotoxicology 2015, 46, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Senut, M.-C.; Cingolani, P.; Sen, A.; Kruger, A.; Shaik, A.; Hirsch, H.; Suhr, S.T.; Ruden, D. Epigenetics of Early-Life Lead Exposure and Effects on Brain Development. Epigenomics 2012, 4, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Kochmanski, J.; VanOeveren, S.E.; Patterson, J.R.; Bernstein, A.I. Developmental Dieldrin Exposure Alters DNA Methylation at Genes Related to Dopaminergic Neuron Development and Parkinson’s Disease in Mouse Midbrain. Toxicol. Sci. 2019, 169, 593–607. [Google Scholar] [CrossRef] [PubMed]
- Paul, K.C.; Horvath, S.; Del Rosario, I.; Bronstein, J.M.; Ritz, B. DNA Methylation Biomarker for Cumulative Lead Exposure Is Associated with Parkinson’s Disease. Clin. Epigenetics 2021, 13, 59. [Google Scholar] [CrossRef]
- Bihaqi, S.W.; Bahmani, A.; Subaiea, G.M.; Zawia, N.H. Infantile Exposure to Lead and Late-age Cognitive Decline: Relevance to AD. Alzheimers Dement. 2014, 10, 187–195. [Google Scholar] [CrossRef]
- Brown, E.E.; Shah, P.; Pollock, B.G.; Gerretsen, P.; Graff-Guerrero, A. Lead (Pb) in Alzheimer’s Dementia: A Systematic Review of Human Case- Control Studies. Curr. Alzheimer Res. 2019, 16, 353–361. [Google Scholar] [CrossRef]
- Dosunmu, R.; Alashwal, H.; Zawia, N.H. Genome-Wide Expression and Methylation Profiling in the Aged Rodent Brain Due to Early-Life Pb Exposure and Its Relevance to Aging. Mech. Ageing Dev. 2012, 133, 435–443. [Google Scholar] [CrossRef]
- Nazeer, A.; Ghaziuddin, M. Autism Spectrum Disorders: Clinical Features and Diagnosis. Pediatr. Clin. N. Am. 2012, 59, 19–25. [Google Scholar] [CrossRef]
- Sherman, H.T.; Liu, K.; Kwong, K.; Chan, S.-T.; Li, A.C.; Kong, X.-J. Carbon Monoxide (CO) Correlates with Symptom Severity, Autoimmunity, and Responses to Probiotics Treatment in a Cohort of Children with Autism Spectrum Disorder (ASD): A Post-Hoc Analysis of a Randomized Controlled Trial. BMC Psychiatry 2022, 22, 536. [Google Scholar] [CrossRef]
- Senut, M.-C.; Sen, A.; Cingolani, P.; Shaik, A.; Land, S.J.; Ruden, D.M. Lead Exposure Disrupts Global DNA Methylation in Human Embryonic Stem Cells and Alters Their Neuronal Differentiation. Toxicol. Sci. Off. J. Soc. Toxicol. 2014, 139, 142–161. [Google Scholar] [CrossRef]
- Stamou, M.; Streifel, K.M.; Goines, P.E.; Lein, P.J. Neuronal Connectivity as a Convergent Target of Gene × Environment Interactions That Confer Risk for Autism Spectrum Disorders. Neurotoxicol. Teratol. 2013, 36, 3–16. [Google Scholar] [CrossRef]
- Penzes, P.; Cahill, M.E.; Jones, K.A.; VanLeeuwen, J.-E.; Woolfrey, K.M. Dendritic Spine Pathology in Neuropsychiatric Disorders. Nat. Neurosci. 2011, 14, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, O.F.; Lee, J.; Yu King Hing, N.; Kim, S.-E.; Freeman, J.L.; Yuan, C. Lead (Pb) Exposure Reduces Global DNA Methylation Level by Non-Competitive Inhibition and Alteration of Dnmt Expression. Metallomics 2017, 9, 149–160. [Google Scholar] [CrossRef]
- Wolstenholme, J.T.; Taylor, J.A.; Shetty, S.R.J.; Edwards, M.; Connelly, J.J.; Rissman, E.F. Gestational Exposure to Low Dose Bisphenol A Alters Social Behavior in Juvenile Mice. PLoS ONE 2011, 6, e25448. [Google Scholar] [CrossRef]
- Kundakovic, M.; Gudsnuk, K.; Herbstman, J.B.; Tang, D.; Perera, F.P.; Champagne, F.A. DNA Methylation of BDNF as a Biomarker of Early-Life Adversity. Proc. Natl. Acad. Sci. USA 2015, 112, 6807–6813. [Google Scholar] [CrossRef]
- Honkova, K.; Rossnerova, A.; Chvojkova, I.; Milcova, A.; Margaryan, H.; Pastorkova, A.; Ambroz, A.; Rossner, P.; Jirik, V.; Rubes, J.; et al. Genome-Wide DNA Methylation in Policemen Working in Cities Differing by Major Sources of Air Pollution. Int. J. Mol. Sci. 2022, 23, 1666. [Google Scholar] [CrossRef]


| Toxicant | Species | Tissue | Genes | Technology | Reference |
|---|---|---|---|---|---|
| DDT | rat | sperm | DMRs | MeDIP-chip | [72] |
| DDE | rat | sperm | IGF2 | bisulfite sequencing | [73] |
| DDA DDMU | human | embryonic stem cells | DNMT1, Sox9, Oct4 | HPLC-MS-MS | [74] |
| CBZ | mouse | leydig cells | 5 mC | immunofluorescence staining | [76] |
| VCZ | mouse | sperm | H19, Gtl2, Peg1, Snrpn, Peg3 | bisulfite-pyrosequencing | [77,78] |
| BPA | rat | testis | Er(α,β) DNMT3A | bisulfite sequencing methylation-specific PCR | [79] [80] |
| mouse | sperm | whole genome | dot-blot assays | [82] | |
| DNMT1 | RT-qPCR and western blot | ||||
| mybph and prkcd | Affymetrix Mouse Promoter 1.0R Array | [83] | |||
| human | sperm | LINE-1 | methylation-specific real-time PCR | [84] | |
| PCB118 | mouse | testis | whole genome DNMT1, PCNA, STRA8 | 5 mC immunohistochemistry Western blot and qPCR | [85] |
| Heat stress | mouse | sperm | DMRs, Pik3cg, Nr4a1 | bisulfite sequencing | [86] |
| POPs | rat | sperm | DNMT3L, DNMT3(A,B), Tbx2, Rapsn | RRBS and pyrosequencing | [87] |
| Mixture of MeHg, PCBs, HCB, DDE, DDT, PFAS | human | cord blood | whole genome | Infinium HumanMethylation450 BeadChip (Illumina) | [89] |
| Air pollution | mouse | sperm | whole genome | cytosine extension and methyl-acceptance assays | [90] |
| human | sperm | whole genome | MethylFlash™ Global DNA Methylation (5 mC) ELISA Easy Kit (Epigentek) | [92] | |
| Fire smoke | mouse | sperm | DMRs | RRBS | [91] |
| Toxicant | Species | Tissue | Genes | Technology | Reference |
|---|---|---|---|---|---|
| MXC | rat | ovaries | Erβ | BSPCR and MSPCR | [94,95] |
| whole genome | methylation-sensitive AP-PCR | ||||
| Dnmt3b | semiquantitative RT-PCR | ||||
| PTEN, IGF-1, Rapid estrogen signaling | Nimblegen 3x720K CpG Island Plus RefSeq Promoter Arrays | [96] | |||
| Endosulfan | rat | utero | Erα, Hoxa10 Dnmt3(a,b) Erα | MSRE-PCR qRT-PCR immunohistochemistry and real-time RT-PCR | [98] [99] |
| PCB118 | mouse | endometrium | Hoxa10, integrin subunit β, ER1 | bisulfite genomic sequencing | [100] |
| BPA | mouse | uterus | Hoxa10 | bisulfite sequencing | [101] |
| DDT DDE | rat | ovary, uterus, blood | - | - | [72,102] |
| oxidative stress (childbirth delay) | mouse | oocytes | whole genome, Dnmt3a, Dnmt3l | scBS-seq | [104,105] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bolognesi, G.; Bacalini, M.G.; Pirazzini, C.; Garagnani, P.; Giuliani, C. Evolutionary Implications of Environmental Toxicant Exposure. Biomedicines 2022, 10, 3090. https://doi.org/10.3390/biomedicines10123090
Bolognesi G, Bacalini MG, Pirazzini C, Garagnani P, Giuliani C. Evolutionary Implications of Environmental Toxicant Exposure. Biomedicines. 2022; 10(12):3090. https://doi.org/10.3390/biomedicines10123090
Chicago/Turabian StyleBolognesi, Giorgia, Maria Giulia Bacalini, Chiara Pirazzini, Paolo Garagnani, and Cristina Giuliani. 2022. "Evolutionary Implications of Environmental Toxicant Exposure" Biomedicines 10, no. 12: 3090. https://doi.org/10.3390/biomedicines10123090
APA StyleBolognesi, G., Bacalini, M. G., Pirazzini, C., Garagnani, P., & Giuliani, C. (2022). Evolutionary Implications of Environmental Toxicant Exposure. Biomedicines, 10(12), 3090. https://doi.org/10.3390/biomedicines10123090





