Alteration of Gut Immunity and Microbiome in Mixed Granulocytic Asthma
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. OVA/Poly I:C Induced Mixed Granulocytic Asthma Mouse Model
2.3. Isolation of Gut Lamina Propria Cells
2.4. Isolation of Gut mRNA and Quantitative PCR
2.5. Immunophenotyping Using Flow Cytometry
2.6. 16s rRNA Gene Amplification and Sequencing
3. Results
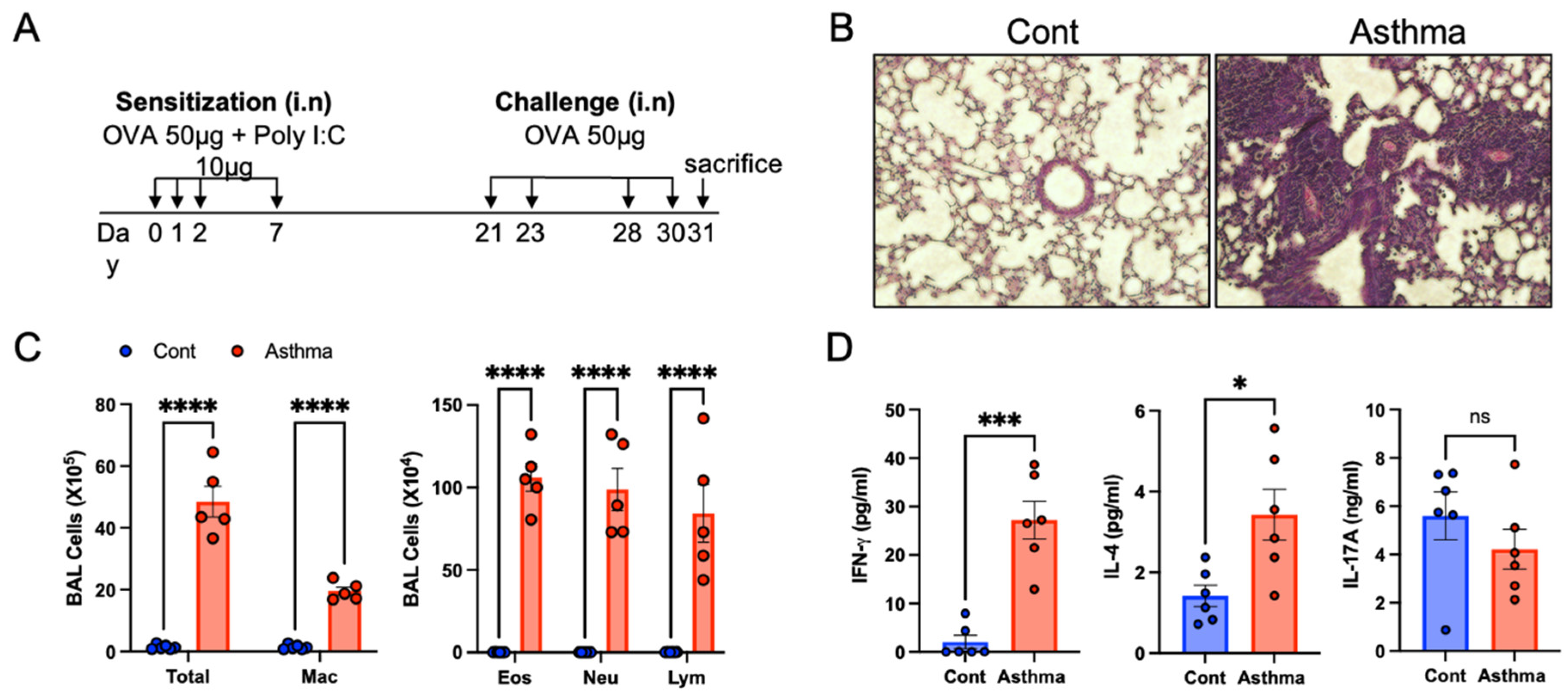
3.1. Establishment of the OVA/Poly I:C Model of Mixed Granulocytic Asthma
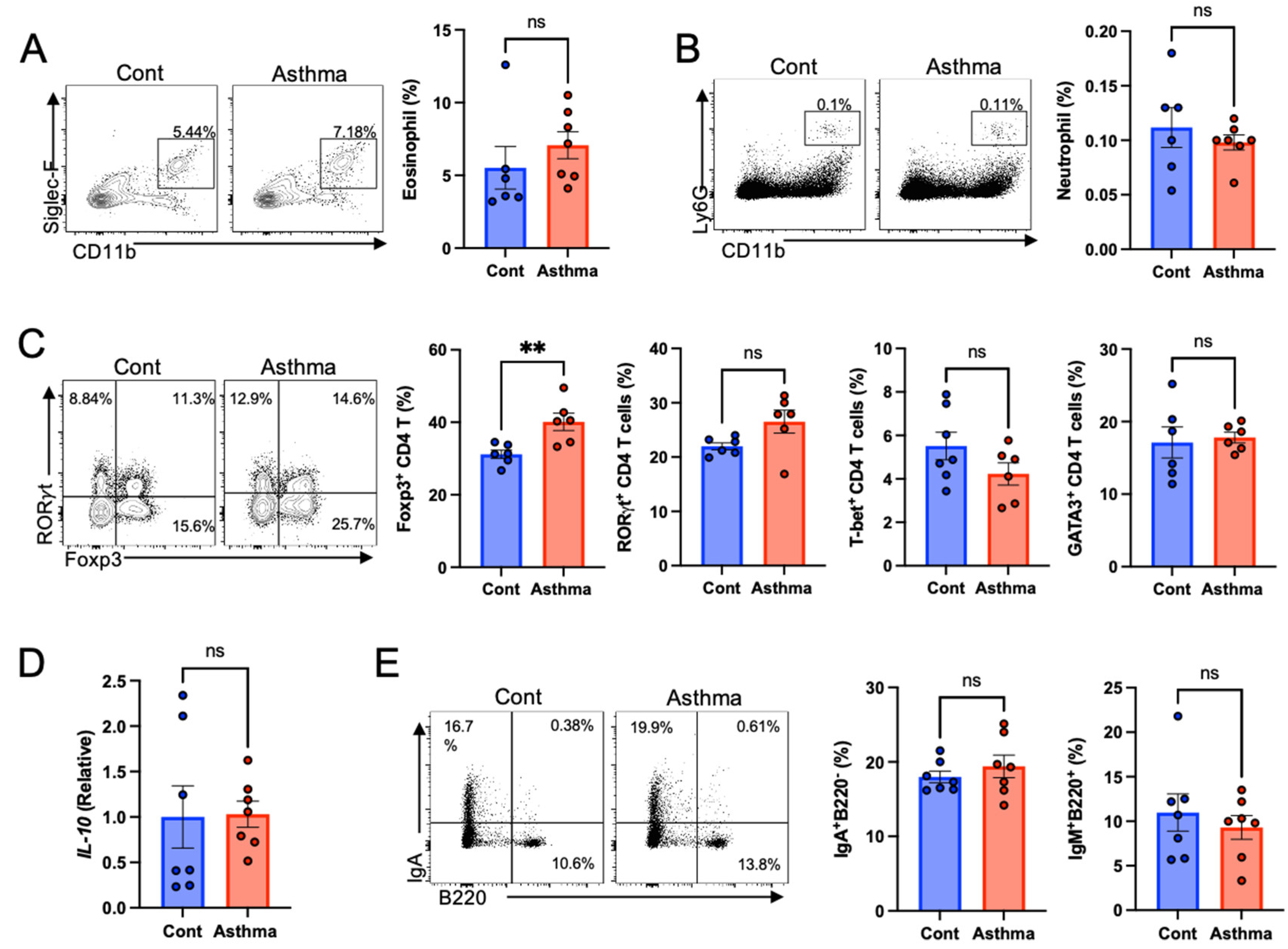
3.2. Increased Regulatory T Cells in the Gut with Mixed Granulocytic Asthma
3.3. Altered Gut Microbiome in Mixed Granulocytic Asthma
3.4. Changes of Functional Gene Abundance in Gut Microbiome of Mixed Granulocytic Asthma
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Soriano, J.B.; Abajobir, A.A.; Abate, K.H.; Abera, S.F.; Agrawal, A.; Ahmed, M.B.; Aichour, A.N.; Aichour, I.; Aichour, M.T.E.; Alam, K.; et al. Global, regional, and national deaths, prevalence, disability-adjusted life years, and years lived with disability for chronic obstructive pulmonary disease and asthma, 1990–2015, a systematic analysis for the Global Burden of Disease Study 2015. Lancet Respir. Med. 2017, 5, 691–706. [Google Scholar] [CrossRef]
- Al Heialy, S.; Ramakrishnan, R.K.; Hamid, Q. Recent advances in the immunopathogenesis of severe asthma. J. Allergy Clin. Immunol. 2022, 149, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.F.; Adcock, I.M. Precision medicine for the discovery of treatable mechanisms in severe asthma. Allergy 2019, 74, 1649–1659. [Google Scholar] [CrossRef] [PubMed]
- Fukuchi, M.; Miyabe, Y.; Furutani, C.; Saga, T.; Moritoki, Y.; Yamada, T.; Weller, P.F.; Ueki, S. How to detect eosinophil ETosis (EETosis) and extracellular traps. Allergol. Int. 2020, 70, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Stark, J.M.; Godding, V.; Sedgwick, J.B.; Busse, W.W. Respiratory syncytial virus infection enhances neutrophil and eosinophil adhesion to cultured respiratory epithelial cells. Roles of CD18 and intercellular adhesion molecule-1. J. Immunol. 1996, 156, 4774–4782. [Google Scholar]
- Bapat, S.P.; Whitty, C.; Mowery, C.T.; Liang, Y.; Yoo, A.; Jiang, Z.; Peters, M.C.; Zhang, L.-J.; Vogel, I.; Zhou, C.; et al. Obesity alters pathology and treatment response in inflammatory disease. Nature 2022, 604, 337–342. [Google Scholar] [CrossRef]
- Moore, W.C.; Hastie, A.T.; Li, X.; Li, H.; Busse, W.W.; Jarjour, N.N.; Wenzel, S.E.; Peters, S.P.; Meyers, D.A.; Bleecker, E.R. Sputum neutrophil counts are associated with more severe asthma phenotypes using cluster analysis. J. Allergy Clin. Immunol. 2013, 133, 1557–1563.e5. [Google Scholar] [CrossRef]
- Hastie, A.T.; Moore, W.C.; Meyers, D.A.; Vestal, P.L.; Li, H.; Peters, S.P.; Bleecker, E.R. Analyses of asthma severity phenotypes and inflammatory proteins in subjects stratified by sputum granulocytes. J. Allergy Clin. Immunol. 2010, 125, 1028–1036.e13. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Trompette, A.; Gollwitzer, E.S.; Pattaroni, C.; Lopez-Mejia, I.C.; Riva, E.; Pernot, J.; Ubags, N.; Fajas, L.; Nicod, L.P.; Marsland, B.J. Dietary Fiber Confers Protection against Flu by Shaping Ly6c− Patrolling Monocyte Hematopoiesis and CD8+ T Cell Metabolism. Immunity 2018, 48, 992–1005.e8. Available online: https://linkinghub.elsevier.com/retrieve/pii/S1074761318301912 (accessed on 15 May 2018). [CrossRef]
- Trompette, A.; Gollwitzer, E.S.; Yadava, K.; Sichelstiel, A.K.; Sprenger, N.; Ngom-Bru, C.; Blanchard, C.; Junt, T.; Nicod, L.P.; Harris, N.L.; et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014, 20, 159–166. Available online: http://www.nature.com/articles/nm.3444S1074761318301912 (accessed on 1 February 2014). [CrossRef]
- Kim, M.; Gu, B.; Madison, M.; Song, H.W.; Norwood, K.; Hill, A.A.; Wu, W.-J.; Corry, D.; Kheradmand, F.; Diehl, G.E. Cigarette Smoke Induces Intestinal Inflammation via a Th17 Cell-Neutrophil Axis. Front. Immunol. 2019, 10, 75. [Google Scholar] [CrossRef]
- Brassard, P.; Vutcovici, M.; Ernst, P.; Patenaude, V.; Sewitch, M.; Suissa, S.; Bitton, A. Increased incidence of inflammatory bowel disease in Québec residents with airway diseases. Eur. Respir. J. 2014, 45, 962–968. [Google Scholar] [CrossRef]
- Peng, Y.-H.; Liao, W.-C.; Su, C.-H.; Chen, H.-J.; Hsia, T.-C.; Chu, C.-C.; Liu, C.-J.; Kao, C.-H. Association of inflammatory bowel disease with asthma risk: A nationwide cohort study. Allergy Asthma Proc. 2015, 36, 92–98. [Google Scholar]
- Barbiellini Amidei, C.; Zingone, F.; Zanier, L.; Canova, C. Risk of Prevalent Asthma among Children Affected by Inflammatory Bowel Disease: A Population-Based Birth Cohort Study. Int. J. Environ. Res. Public Health 2020, 17, 4255. [Google Scholar] [CrossRef]
- Benard, A.; Desreumeaux, P.; Huglo, D.; Hoorelbeke, A.; Tonnel, A.-B.; Wallaert, B. Increased intestinal permeability in bronchial asthma. J. Allergy Clin. Immunol. 1996, 97, 1173–1178. [Google Scholar] [CrossRef]
- Raj, A.A.; Birring, S.S.; Green, R.; Grant, A.; de Caestecker, J.; Pavord, I.D. Prevalence of inflammatory bowel disease in patients with airways disease. Respir. Med. 2008, 102, 780–785. [Google Scholar] [CrossRef]
- Diehl, G.E.; Longman, R.S.; Zhang, J.-X.; Breart, B.; Galan, C.; Cuesta, A.; Schwab, S.R.; Littman, D.R. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX3CR1hi cells. Nature 2013, 494, 116–120. [Google Scholar] [CrossRef]
- Sze, E.; Bhalla, A.; Nair, P. Mechanisms and therapeutic strategies for non-T2 asthma. Allergy 2019, 75, 311–325. [Google Scholar] [CrossRef]
- Krug, N.; Madden, J.; Redington, A.E.; Lackie, P.; Djukanovic, R.; Schauer, U.; Holgate, S.T.; Frew, A.J.; Howarth, P.H. T-cell cytokine profile evaluated at the single cell level in BAL and blood in allergic asthma. Am. J. Respir. Cell Mol. Biol. 1996, 14, 319–326. [Google Scholar] [CrossRef]
- Figueiredo, C.A.; Rodrigues, L.C.; Alcantara-Neves, N.M.; Cooper, P.J.; Amorim, L.D.; Silva, N.B.; Cruz, A.A.; Barreto, M.L. Does IFN-γ play a role on the pathogenesis of non-atopic asthma in Latin America children? Allergy Asthma Clin. Immunol. 2012, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Raundhal, M.; Morse, C.; Khare, A.; Oriss, T.B.; Milosevic, J.; Trudeau, J.; Huff, R.; Pilewski, J.; Holguin, F.; Kolls, J.; et al. High IFN-γ and low SLPI mark severe asthma in mice and humans. J. Clin. Investig. 2015, 125, 3037–3050. [Google Scholar] [CrossRef] [PubMed]
- Naumov, D.; Gassan, D.; Kotova, O.; Sheludko, E.; Afanaseva, E.; Perelman, J.; Shvetcova, Y. Role of interferon-gamma as a marker of asthma severity and control. Eur. Respir. J. Suppl. 2019, 54, 4378. [Google Scholar]
- Fujimura, K.E.; Sitarik, A.R.; Havstad, S.; Lin, D.L.; LeVan, S.; Fadrosh, D.; Panzer, A.R.; LaMere, B.; Rackaityte, E.; Lukacs, N.W.; et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 2016, 22, 1187–1191. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, M.-C.; Arévalo, A.; Stiemsma, L.; Dimitriu, P.; Chico, M.E.; Loor, S.; Vaca, M.; Boutin, R.C.; Morien, E.; Jin, M.; et al. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J. Allergy Clin. Immunol. 2018, 142, 424–434.e10. [Google Scholar] [CrossRef]
- McLoughlin, R.; Berthon, B.S.; Rogers, G.B.; Baines, K.J.; Leong, L.E.; Gibson, P.G.; Williams, E.J.; Wood, L.G. Soluble fibre supplementation with and without a probiotic in adults with asthma: A 7-day randomised, double blind, three way cross-over trial. eBioMedicine 2019, 46, 473–485. [Google Scholar] [CrossRef]
- Sefik, E.; Geva-Zatorsky, N.; Oh, S.; Konnikova, L.; Zemmour, D.; McGuire, A.M.; Burzyn, D.; Ortiz-Lopez, A.; Lobera, M.; Yang, J.; et al. Individual intestinal symbionts induce a distinct population of RORγ + regulatory T cells. Science 2015, 349, 993–997. [Google Scholar] [CrossRef]
- Yang, B.-H.; Hagemann, S.; Mamareli, P.; Lauer, U.M.; Hoffmann, U.; Beckstette, M.; Fohse, L.; Prinz, I.; Pezoldt, J.; Suerbaum, S.; et al. Foxp3+ T cells expressing RORγt represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunol. 2015, 9, 444–457. [Google Scholar] [CrossRef]
- Lochner, M.; Peduto, L.; Cherrier, M.; Sawa, S.; Langa, F.; Varona, R.; Riethmacher, D.; Si-Tahar, M.; Di Santo, J.; Eberl, G. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORγt+ T cells. J. Exp. Med. 2008, 205, 1381–1393. [Google Scholar] [CrossRef]
- Zhao, Y.; Wu, J.; Li, J.V.; Zhou, N.-Y.; Tang, H.; Wang, Y. Gut Microbiota Composition Modifies Fecal Metabolic Profiles in Mice. J. Proteome Res. 2013, 12, 2987–2999. [Google Scholar] [CrossRef]
- Palm, N.W.; De Zoete, M.R.; Cullen, T.W.; Barry, N.A.; Stefanowski, J.; Hao, L.; Degnan, P.H.; Hu, J.; Peter, I.; Zhang, W.; et al. Immunoglobulin A Coating Identifies Colitogenic Bacteria in Inflammatory Bowel Disease. Cell 2014, 158, 1000–1010. [Google Scholar] [CrossRef]
- Dinh, D.M.; Volpe, G.E.; Duffalo, C.; Bhalchandra, S.; Tai, A.K.; Kane, A.V.; Wanke, C.A.; Ward, H.D. Intestinal Microbiota, Microbial Translocation, and Systemic Inflammation in Chronic HIV Infection. J. Infect. Dis. 2014, 211, 19–27. [Google Scholar] [CrossRef]
- Cao, Y.G.; Bae, S.; Villarreal, J.; Moy, M.; Chun, E.; Michaud, M.; Lang, J.K.; Glickman, J.N.; Lobel, L.; Garrett, W.S. Faecalibaculum rodentium remodels retinoic acid signaling to govern eosinophil-dependent intestinal epithelial homeostasis. Cell Host Microbe 2022, 30, 1295–1310.e8. [Google Scholar] [CrossRef]
- Fleissner, C.K.; Huebel, N.; El-Bary, M.M.A.; Loh, G.; Klaus, S.; Blaut, M. Absence of intestinal microbiota does not protect mice from diet-induced obesity. Br. J. Nutr. 2010, 104, 919–929. [Google Scholar] [CrossRef]
- Zhang, H.; DiBaise, J.K.; Zuccolo, A.; Kudrna, D.; Braidotti, M.; Yu, Y.; Parameswaran, P.; Crowell, M.D.; Wing, R.; Rittmann, B.E.; et al. Human Gut Microbiota in Obesity and after Gastric Bypass. Proc. Natl. Acad. Sci. USA 2009, 106, 2365–2370. [Google Scholar] [CrossRef]
- Spencer, M.; Hamp, T.J.; Reid, R.; Fischer, L.M.; Zeisel, S.H.; Fodor, A.A. Association Between Composition of the Human Gastrointestinal Microbiome and Development of Fatty Liver With Choline Deficiency. Gastroenterology 2011, 140, 976–986. [Google Scholar] [CrossRef]
- Martínez, I.; Wallace, G.; Zhang, C.; Legge, R.; Benson, A.K.; Carr, T.P.; Moriyama, E.N.; Walter, J. Diet-Induced Metabolic Improvements in a Hamster Model of Hypercholesterolemia Are Strongly Linked to Alterations of the Gut Microbiota. Appl. Environ. Microbiol. 2009, 75, 4175–4184. [Google Scholar] [CrossRef]
- Martínez, I.; Perdicaro, D.J.; Brown, A.W.; Hammons, S.; Carden, T.J.; Carr, T.P.; Eskridge, K.M.; Walter, J. Diet-Induced Alterations of Host Cholesterol Metabolism Are Likely To Affect the Gut Microbiota Composition in Hamsters. Appl. Environ. Microbiol. 2013, 79, 516–524. [Google Scholar] [CrossRef]
- Cox, L.M.; Yamanishi, S.; Sohn, J.; Alekseyenko, A.V.; Leung, J.M.; Cho, I.; Kim, S.G.; Li, H.; Gao, Z.; Mahana, D.; et al. Altering the Intestinal Microbiota during a Critical Developmental Window Has Lasting Metabolic Consequences. Cell 2014, 158, 705–721. [Google Scholar] [CrossRef]
- Lin, Y.-W.; Montassier, E.; Knights, D.; Wei, L.-N. Gut microbiota from metabolic disease-resistant, macrophage-specific RIP140 knockdown mice improves metabolic phenotype and gastrointestinal integrity. Sci. Rep. 2016, 6, 38599. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, B.-H.; Rim, C.-Y.; Lee, S.; Kim, T.-Y.; Joo, S.-S.; Lee, S.-J.; Park, H.-K.; Kim, M. Alteration of Gut Immunity and Microbiome in Mixed Granulocytic Asthma. Biomedicines 2022, 10, 2946. https://doi.org/10.3390/biomedicines10112946
Gu B-H, Rim C-Y, Lee S, Kim T-Y, Joo S-S, Lee S-J, Park H-K, Kim M. Alteration of Gut Immunity and Microbiome in Mixed Granulocytic Asthma. Biomedicines. 2022; 10(11):2946. https://doi.org/10.3390/biomedicines10112946
Chicago/Turabian StyleGu, Bon-Hee, Chae-Yun Rim, Sangjin Lee, Tae-Yong Kim, Sang-Seok Joo, Sang-Jin Lee, Han-Ki Park, and Myunghoo Kim. 2022. "Alteration of Gut Immunity and Microbiome in Mixed Granulocytic Asthma" Biomedicines 10, no. 11: 2946. https://doi.org/10.3390/biomedicines10112946
APA StyleGu, B.-H., Rim, C.-Y., Lee, S., Kim, T.-Y., Joo, S.-S., Lee, S.-J., Park, H.-K., & Kim, M. (2022). Alteration of Gut Immunity and Microbiome in Mixed Granulocytic Asthma. Biomedicines, 10(11), 2946. https://doi.org/10.3390/biomedicines10112946






