MicroRNA Profiling Shows a Time-Dependent Regulation within the First 2 Months Post-Birth and after Mild Neonatal Hypoxia in the Hippocampus from Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Mouse Model of Neonatal Hypoxia
2.2. RNA Isolation and Open Array
2.3. Ambulation Score
2.4. RT-PCR of Primary Let-7a Cluster Sequence
2.5. Identification of c-Myc Binding Sites in the Promoter Regions
2.6. Chromatin Immunoprecipitation (ChIP)
2.7. Statistical Analysis
3. Results
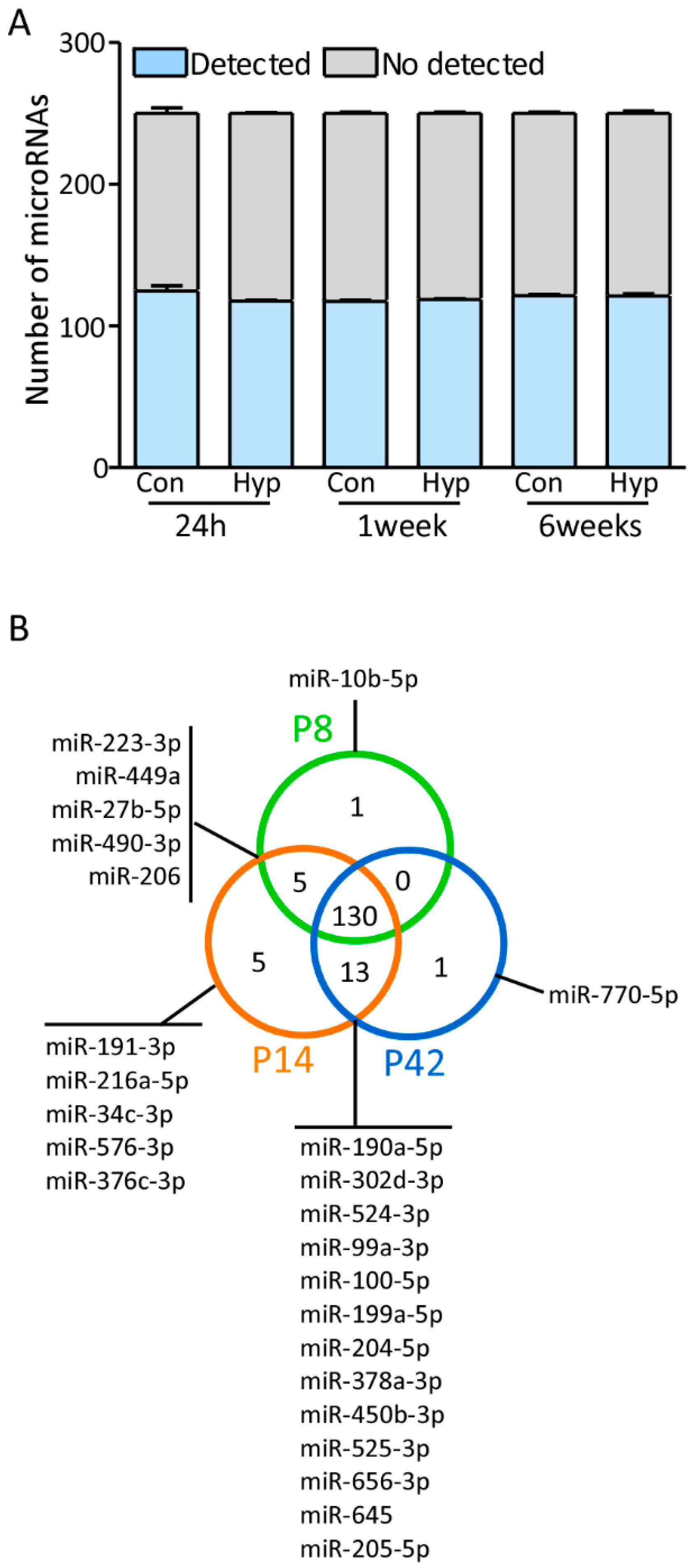
3.1. MicroRNA Expression Profiles in the Postnatal Hippocampus
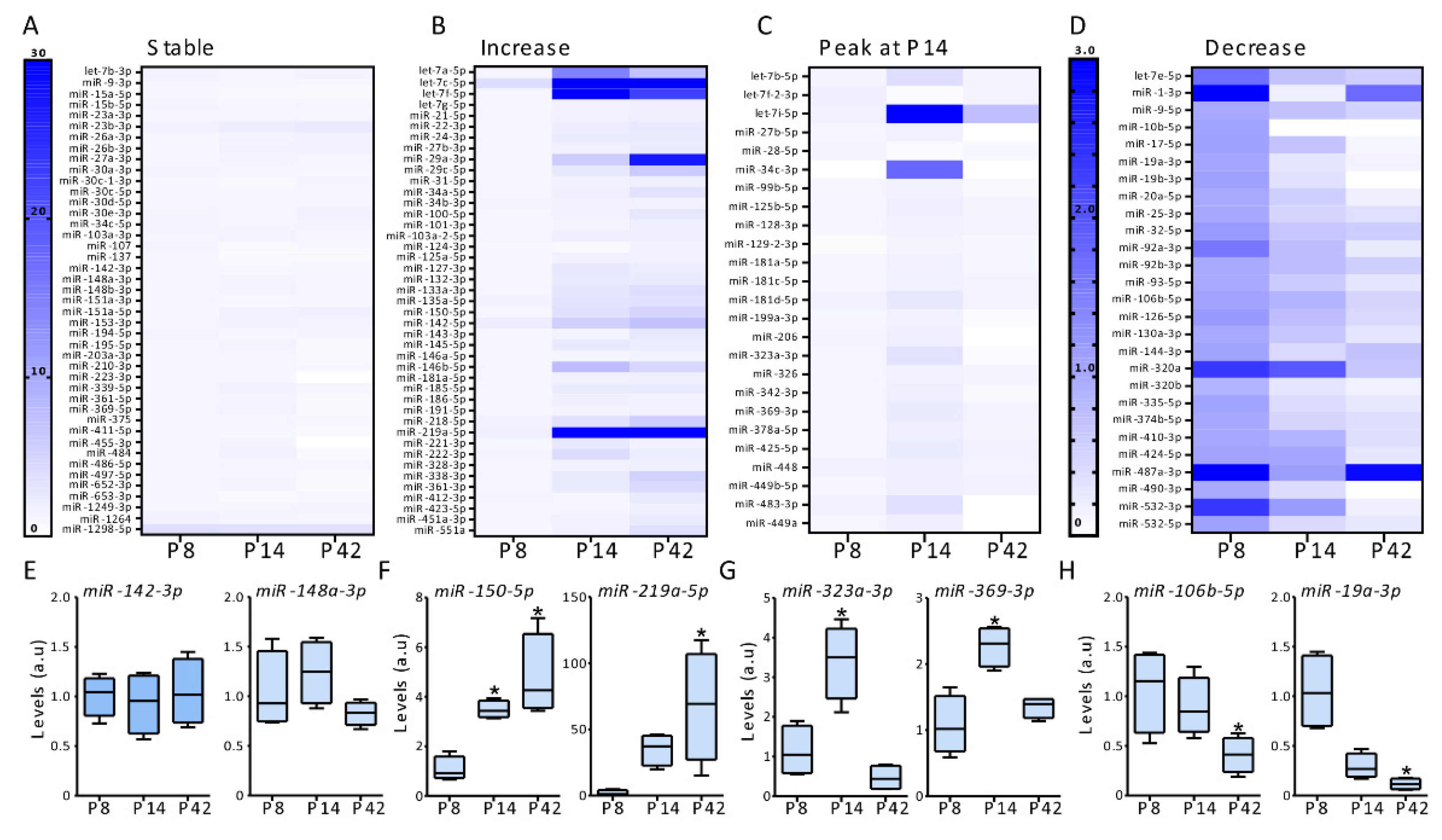
3.2. MicroRNAs Regulation between Ages in the Postnatal Hippocampus in Mice
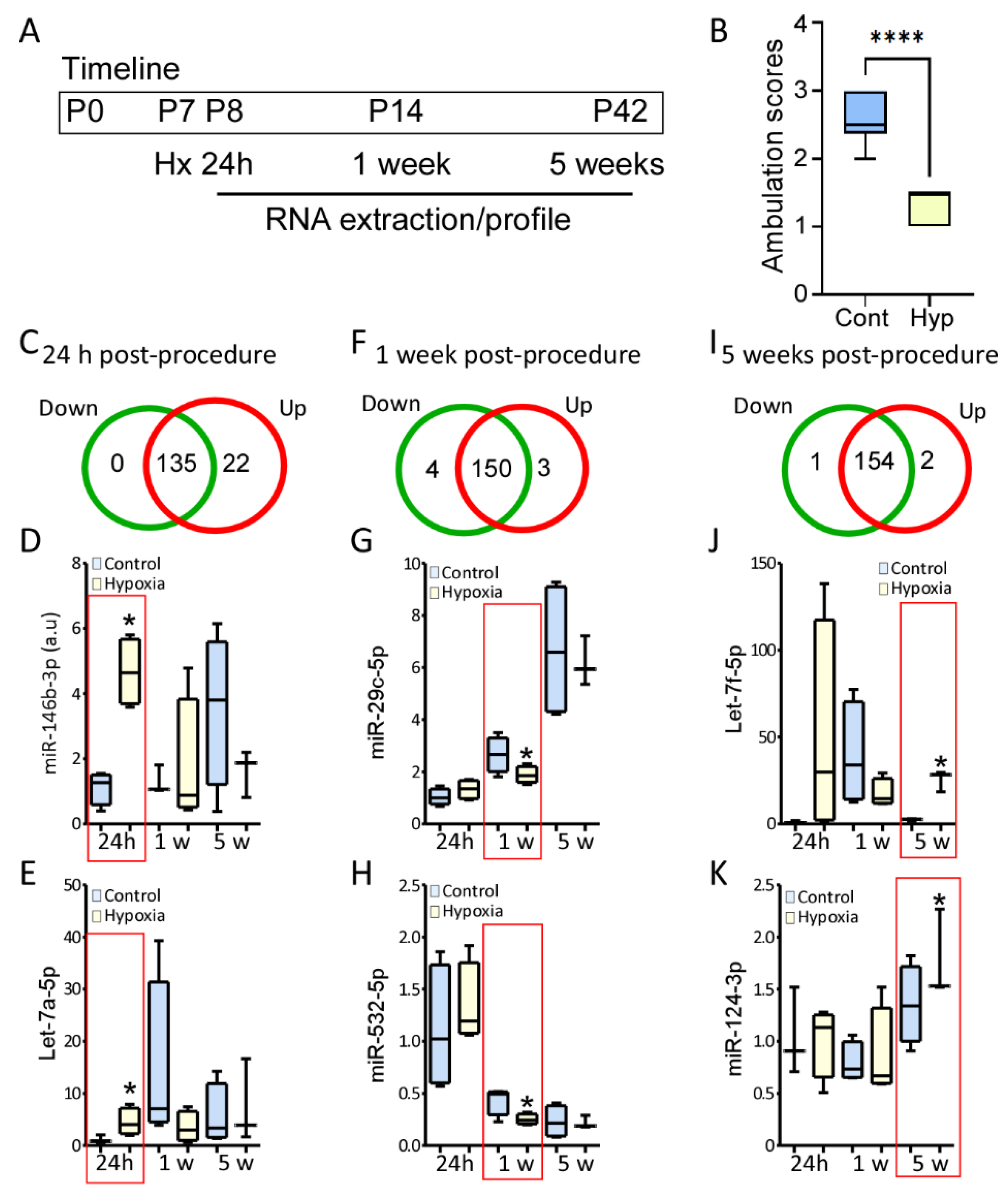
3.3. Differentially Regulated microRNAs after Hypoxia at P7 in Mice
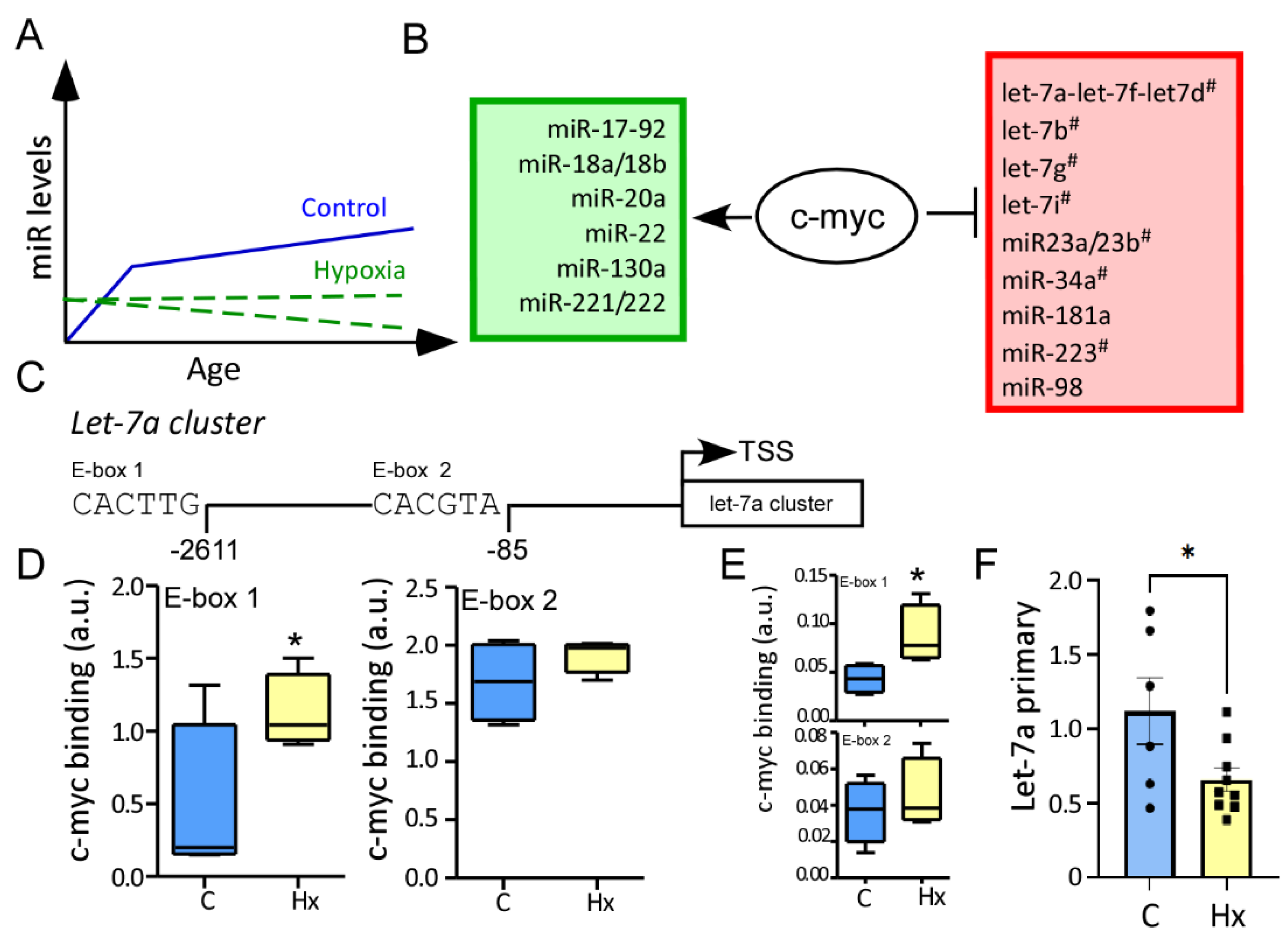
3.4. MYC Has a High Affinity for Let-7a Promoter in the Hippocampus after Hypoxia
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stiles, J. Brain development and the nature versus nurture debate. Prog. Brain. Res. 2011, 189, 3–22. [Google Scholar] [PubMed]
- Paus, T.; Keshavan, M.; Giedd, J.N. Why do many psychiatric disorders emerge during adolescence? Nat. Rev. Neurosci. 2008, 9, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Toga, A.W.; Thompson, P.M.; Sowell, E.R. Mapping brain maturation. Trends Neurosci. 2006, 29, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Stiles, J.; Jernigan, T.L. The basics of brain development. Neuropsychol. Rev. 2010, 20, 327–348. [Google Scholar] [CrossRef]
- Lebel, C.; Walker, L.; Leemans, A.; Phillips, L.; Beaulieu, C. Microstructural maturation of the human brain from childhood to adulthood. Neuroimage 2008, 40, 1044–1055. [Google Scholar] [CrossRef] [PubMed]
- Jurkowski, M.P.; Bettio, L.; KWoo, E.; Patten, A.; Yau, S.Y.; Gil-Mohapel, J. Beyond the Hippocampus and the SVZ: Adult Neurogenesis Throughout the Brain. Front. Cell Neurosci. 2020, 14, 576444. [Google Scholar] [CrossRef]
- Cayre, M.; Canoll, P.; Goldman, J.E. Cell migration in the normal and pathological postnatal mammalian brain. Prog. Neurobiol. 2009, 88, 41–63. [Google Scholar] [CrossRef]
- McTigue, D.M.; Tripathi, R.B. The life, death, and replacement of oligodendrocytes in the adult CNS. J. Neurochem. 2008, 107, 1–19. [Google Scholar] [CrossRef]
- Dexheimer, P.J.; Cochella, L. MicroRNAs: From Mechanism to Organism. Front. Cell Dev. Biol. 2020, 8, 409. [Google Scholar] [CrossRef]
- Alvarez-Saavedra, E.; Horvitz, H.R. Many families of C. elegans microRNAs are not essential for development or viability. Curr. Biol. 2010, 20, 367–373. [Google Scholar] [CrossRef]
- Alisch, R.S.; Jin, P.; Epstein, M.; Caspary, T.; Warren, S.T. Argonaute2 is essential for mammalian gastrulation and proper mesoderm formation. PLoS Genet. 2007, 3, e227. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, E.; Kim, S.Y.; Carmell, M.A.; Murchison, E.P.; Alcorn, H.; Li, M.Z.; Mills, A.A.; Elledge, S.J.; Anderson, K.V.; Hannon, G.J. Dicer is essential for mouse development. Nat. Genet. 2003, 35, 215–217. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, Y.; Takeichi, M.; Uemura, T. Developmental roles and molecular characterization of a Drosophila homologue of Arabidopsis Argonaute1, the founder of a novel gene superfamily. Genes Cells 2001, 6, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Vasquez-Rifo, A.; Jannot, G.; Armisen, J.; Labouesse, M.; Bukhari, S.I.; Rondeau, E.L.; Miska, E.A.; Simard, M.J. Developmental characterization of the microRNA-specific C. elegans Argonautes alg-1 and alg-2. PLoS ONE 2012, 7, e33750. [Google Scholar] [CrossRef]
- Hornstein, E.; Shomron, N. Canalization of development by microRNAs. Nat. Genet. 2006, 38, S20–S24. [Google Scholar] [CrossRef]
- Ilbay, O.; Ambros, V. Pheromones and Nutritional Signals Regulate the Developmental Reliance on let-7 Family MicroRNAs in C. elegans. Curr. Biol. 2019, 29, 1735–1745.e4. [Google Scholar] [CrossRef]
- Doubi-Kadmiri, S.; Benoit, C.; Benigni, X.; Beaumont, G.; Vacher, C.M.; Taouis, M.; Baroin-Tourancheau, A.; Amar, L. Substantial and robust changes in microRNA transcriptome support postnatal development of the hypothalamus in rat. Sci. Rep. 2016, 6, 24896. [Google Scholar] [CrossRef]
- Alberti, C.; Cochella, L. A framework for understanding the roles of miRNAs in animal development. Development 2017, 144, 2548–2559. [Google Scholar] [CrossRef]
- Zaytseva, O.; Kim, N.H.; Quinn, L.M. MYC in Brain Development and Cancer. Int. J. Mol. Sci. 2020, 21, 7742. [Google Scholar] [CrossRef]
- Wang, Z.; Lin, S.; Li, J.J.; Xu, Z.; Yao, H.; Zhu, X.; Xie, D.; Shen, Z.; Sze, J.; Li, K.; et al. MYC protein inhibits transcription of the microRNA cluster MC-let-7a-1~let-7d via noncanonical E-box. J. Biol. Chem. 2011, 286, 39703–39714. [Google Scholar] [CrossRef]
- Lin, S.T.; Huang, Y.; Zhang, L.; Heng, M.Y.; Ptacek, L.J.; Fu, Y.H. MicroRNA-23a promotes myelination in the central nervous system. Proc. Natl. Acad. Sci. USA 2013, 110, 17468–17473. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, S.; Merino-Serrais, P.; Di Grande, A.; Dussmann, H.; Prehn, J.H.M.; Ni Chonghaile, T.; Henshall, D.C.; Jimenez-Mateos, E.M. The Anti-inflammatory Compound Candesartan Cilexetil Improves Neurological Outcomes in a Mouse Model of Neonatal Hypoxia. Front. Immunol. 2019, 10, 1752. [Google Scholar] [CrossRef] [PubMed]
- Engel, T.; Brennan, G.P.; Sanz-Rodriguez, A.; Alves, M.; Beamer, E.; Watters, O.; Henshall, D.C.; Jimenez-Mateos, E.M. A calcium-sensitive feed-forward loop regulating the expression of the ATP-gated purinergic P2X7 receptor via specificity protein 1 and microRNA-22. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 255–266. [Google Scholar] [CrossRef]
- Quinlan, S.M.M.; Rodriguez-Alvarez, N.; Molloy, E.J.; Madden, S.F.; Boylan, G.B.; Henshall, D.C.; Jimenez-Mateos, E.M. Complex spectrum of phenobarbital effects in a mouse model of neonatal hypoxia-induced seizures. Sci. Rep. 2018, 8, 9986. [Google Scholar] [CrossRef] [PubMed]
- Feather-Schussler, D.N.; Ferguson, T.S. A Battery of Motor Tests in a Neonatal Mouse Model of Cerebral Palsy. J. Vis. Exp. 2016, e53569. [Google Scholar] [CrossRef]
- Olsen, L.C.; O’Reilly, K.C.; Liabakk, N.B.; Witter, M.P.; Saetrom, P. MicroRNAs contribute to postnatal development of laminar differences and neuronal subtypes in the rat medial entorhinal cortex. Brain. Struct. Funct. 2017, 222, 3107–3126. [Google Scholar] [CrossRef]
- Kole, A.J.; Swahari, V.; Hammond, S.M.; Deshmukh, M. miR-29b is activated during neuronal maturation and targets BH3-only genes to restrict apoptosis. Genes Dev. 2011, 25, 125–130. [Google Scholar] [CrossRef]
- Lippi, G.; Steinert, J.R.; Marczylo, E.L.; D’Oro, S.; Fiore, R.; Forsythe, I.; Schratt, G.; Zoli, M.; Nicotera, P.; Young, K.W. Targeting of the Arpc3 actin nucleation factor by miR-29a/b regulates dendritic spine morphology. J. Cell Biol. 2011, 194, 889–904. [Google Scholar] [CrossRef]
- Bruinsma, I.B.; Van Dijk, M.; Bridel, C.; Van De Lisdonk, T.; Haverkort, S.Q.; Runia, T.F.; Steinman, L.; Hintzen, R.Q.; Killestein, J.; Verbeek, M.M.; et al. Regulator of oligodendrocyte maturation, miR-219, a potential biomarker for MS. J. Neuroinflamm. 2017, 14, 235. [Google Scholar] [CrossRef]
- Arzhanov, I.; Sintakova, K.; Romanyuk, N. The Role of miR-20 in Health and Disease of the Central Nervous System. Cells 2022, 11, 1525. [Google Scholar] [CrossRef]
- Kim, J.W.; Gao, P.; Liu, Y.C.; Semenza, G.L.; Dang, C.V. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol. Cell. Biol. 2007, 27, 7381–7393. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leavy, A.; Brennan, G.P.; Jimenez-Mateos, E.M. MicroRNA Profiling Shows a Time-Dependent Regulation within the First 2 Months Post-Birth and after Mild Neonatal Hypoxia in the Hippocampus from Mice. Biomedicines 2022, 10, 2740. https://doi.org/10.3390/biomedicines10112740
Leavy A, Brennan GP, Jimenez-Mateos EM. MicroRNA Profiling Shows a Time-Dependent Regulation within the First 2 Months Post-Birth and after Mild Neonatal Hypoxia in the Hippocampus from Mice. Biomedicines. 2022; 10(11):2740. https://doi.org/10.3390/biomedicines10112740
Chicago/Turabian StyleLeavy, Aisling, Gary P. Brennan, and Eva M. Jimenez-Mateos. 2022. "MicroRNA Profiling Shows a Time-Dependent Regulation within the First 2 Months Post-Birth and after Mild Neonatal Hypoxia in the Hippocampus from Mice" Biomedicines 10, no. 11: 2740. https://doi.org/10.3390/biomedicines10112740
APA StyleLeavy, A., Brennan, G. P., & Jimenez-Mateos, E. M. (2022). MicroRNA Profiling Shows a Time-Dependent Regulation within the First 2 Months Post-Birth and after Mild Neonatal Hypoxia in the Hippocampus from Mice. Biomedicines, 10(11), 2740. https://doi.org/10.3390/biomedicines10112740







