Abstract
Aspirin resistance (AR) is a pressing problem in current ischemic stroke care. Although the role of genetic variations is widely considered, the data still remain controversial. Our aim was to investigate the contribution of genetic features to laboratory AR measured through platelet aggregation with arachidonic acid (AA) and adenosine diphosphate (ADP) in ischemic stroke patients. A total of 461 patients were enrolled. Platelet aggregation was measured via light transmission aggregometry. Eighteen single-nucleotide polymorphisms (SNPs) in ITGB3, GPIBA, TBXA2R, ITGA2, PLA2G7, HMOX1, PTGS1, PTGS2, ADRA2A, ABCB1 and PEAR1 genes and the intergenic 9p21.3 region were determined using low-density biochips. We found an association of rs1330344 in the PTGS1 gene with AR and AA-induced platelet aggregation. Rs4311994 in ADRA2A gene also affected AA-induced aggregation, and rs4523 in the TBXA2R gene and rs12041331 in the PEAR1 gene influenced ADP-induced aggregation. Furthermore, the effect of rs1062535 in the ITGA2 gene on NIHSS dynamics during 10 days of treatment was found. The best machine learning (ML) model for AR based on clinical and genetic factors was characterized by AUC = 0.665 and F1-score = 0.628. In conclusion, the association study showed that PTGS1, ADRA2A, TBXA2R and PEAR1 polymorphisms may affect laboratory AR. However, the ML model demonstrated the predominant influence of clinical features.
1. Introduction
Aspirin is a key drug widely used for ischemic stroke patients as antiplatelet therapy to prevent recurrent ischemic events []. This drug acts by irreversibly blocking the activity of the cyclooxygenases (COX)-1 and -2 also known as prostaglandin G/H synthases 1 and 2 (PTGS1 and PTGS2), respectively []. While the COX-1 enzyme is produced constitutively, the COX-2 form is highly inducible, mainly by inflammation. The COX-1 enzyme is expressed in mature platelets and catalyzes the conversion of arachidonic acid (AA) to prostaglandins G2 and H2, with a subsequent production of thromboxane A2 (TXA2) [,]. Thromboxane A2 is released into the bloodstream and binds to TXA2 receptors on the surface of neighboring platelets, causing their activation. Additionally, TXA2 acts synergistically with other substances released by activated platelets (adenosine diphosphate (ADP), fibrinogen, factor V) to increase the process. The main antithrombotic effect of low-dose (75–125 mg) aspirin is mediated by selective inhibition of COX-1 []. As a result of aspirin action, the production of TXA2, which is the main compound in platelet activation and aggregation, is suppressed for the lifetime of the platelet (7–10 days) []. The pathway of TXA2 production and the antiplatelet effect of aspirin are shown in Figure S1.
The response to aspirin varies between individuals, and up to 57% of patients show the so-called aspirin resistance (AR) []. AR is classified into clinical and laboratory resistance. Clinical AR is established by the inability of aspirin to prevent the subsequent acute vascular events []. Laboratory AR can be defined as ex vivo high on-treatment platelet reactivity (HTPR) such as the insufficient antiplatelet effect of aspirin measured by different laboratory tests [,]. Tests measure inactive metabolites of TXA2 in serum or urine [,] or analyze platelet aggregation and adhesion. Among the assays that determine platelet function, light transmission aggregometry (LTA) is considered as the gold standard in platelet function testing []. Automated (point-of-care) assays such as VerifyNow®, PFA-100®, Multiplate®, Plateletworks® and others are widely used for monitoring platelet response to antiplatelet agents including aspirin [,,]. HTPR was shown to increase the risk of recurrent vascular events and long-term clinical outcomes for patients with cerebrovascular pathology [,,,]. Nevertheless, platelet function tests differ in their ability to predict the risk of cardiovascular outcomes [].
AR seems to be a complex phenomenon with a number of factors potentially contributing to it, but its causes and mechanisms are still unclear []. One of these factors that might underlie AR is heredity, having a profound impact on the variability in residual platelet function during aspirin therapy []. Genes encoding key platelet aggregation proteins are under the most intense scrutiny.
A number of genetic markers have already been studied to assess their possible contribution to AR [,]. First, single-nucleotide polymorphisms (SNPs) in the genes encoding COX enzymes (PTGS1 and PTGS2) were found to influence AR [,,,,,,,]. Polymorphisms in the TBXA2R gene, encoding the specific TXA2 receptor, were associated with the effect of aspirin in a number of studies [,,]. The genes involved in the COX-independent platelet activation pathways as well as platelet glycoprotein genes might also be involved in AR. The effect of polymorphisms in the genes HMOX1 [], PLA2G7 [], ADRA2A [], ITGB3 [,], GPIBA [], ITGA2 [] and PEAR1 [,,] on inter-individual variations in the aspirin response has been discussed. A locus on chromosome 9p21.3, associated with CVD and ischemic stroke, was also connected with AR [,]. P-glycoprotein (also known as MDR1) plays a crucial role in the intestinal epithelial cell permeability to aspirin [] and might be involved in aspirin absorption. The TT rs1045642 genotype in the gene ABCB1 encoding P-glycoprotein was shown to protect against AR []. Therefore, the molecular changes in the pathways involving various genes appear to influence the AR development. However, the impact of genetic markers on the risk for an individual patient is poorly understood. Implementing the identified genetic risk factors to predict aspirin failure in clinical practice still remains challenging.
One problem lies in the inconsistency of the results from genetic studies. This may be explained by the differences in the diagnoses (ischemic stroke, cardiovascular disease, diabetes mellitus), ethnic groups, platelet function tests, sample sizes, etc. []. There is a noticeable lack of replication studies analyzing AR genetic background in patients with ischemic stroke from the Eastern European populations.
Another problem is the multiplicity of influencing factors that determine the ultimate success or failure of aspirin therapy. The clinical features of the disease, comorbidities, co-medications and non-modifiable risk factors such as age should be taken into account []. Moreover, the interaction of genetic polymorphisms as well as clinical factors may influence sensitivity to aspirin []. Over the past several years, machine learning (ML) models have been proven to be able to solve various problems in the medical and biological fields, including pharmacogenetics [,]. One of the key advantages of the ML approaches lies in their ability to find unobvious relationships and make inferences from the complex data.
The purpose of this study was to investigate genetic features associated with laboratory AR in a cohort of patients with ischemic stroke taking aspirin as antiplatelet therapy to be used in pharmacogenetic testing. We have developed a biochip assay to identify 18 SNPs previously described as markers affecting AR. To establish the connection between the patients’ clinical data, genotype and laboratory response to aspirin treatment, we applied the multiple ML approaches.
2. Materials and Methods
2.1. Patients
The study included 461 Caucasian patients with primary ischemic stroke treated in the Stroke Center of City Clinical Hospital No.1 named N.I. Pirogov. The inclusion criterion was a verified ischemic stroke. Exclusion criteria comprised hemorrhagic transformation, cancer and severe liver disease, as well as other diseases and conditions affecting the parameters of platelet hemostasis. The pathogenetic variant of stroke was established according to the TOAST criteria [] based on the clinical data, computed tomography and magnetic resonance imaging of the brain, Doppler ultrasound of the cerebral arteries and electrocardiography. The study population included 109 patients with cardioembolism, 98 patients with large artery atherosclerosis (LAA, ≥50% stenosis) and 250 patients with undetermined etiology (of which 53 had both LAA and cardioembolism, 197 had neither LAA nor cardioembolism). All patients received the antiplatelet, lipid-lowering, antihypertensive or anticoagulant therapy according to the clinical guidelines. For early prevention of recurrent stroke, all patients took aspirin at a dose of 125 mg daily, starting within 24 h of the stroke onset. Patients with cardioembolic stroke received the anticoagulant treatment starting on day 3, 6 or 12 depending on the stroke severity []. Dynamics of the NIHSS score estimated at admission and after 10 days of aspirin therapy was considered as the short-term clinical outcome.
The study was approved by the local ethics committee of the Pirogov Russian National Research Medical University (protocol no. 181 dated 28 January 2019). All participants provided a written informed consent. The study adhered to the World Medical Association Declaration of Helsinki. With a 95% confidence level, a standard deviation of 0.5 and a confidence interval (margin of error) of ±5%, the sample size was estimated to be 391 patients.
2.2. Platelet Aggregation
Blood samples from the vein of the non-paretic limb were collected in the morning of the third day of aspirin intake. The region of the cubital fossa was usually selected as the venipuncture area. A tourniquet was applied to the middle third of the shoulder, while the pulse was taken on the nearest radial artery. After that, the patient clenched the hand into a fist and unclenched it several times. The skin in the venipuncture area was stretched, fixing the vein. Next, the skin was pierced next to the vein; the needle was moved 1.5 cm deep into the subcutaneous fat, and the vein was punctured. A total of 9 mL of blood was collected in the 14 mL plastic test tubes “Greiner” with 1 mL of 3.8% trisubstituted sodium citrate using 21 G × 1 1/2”/0.8 × 40 mm needles. The blood in the tube was mixed immediately. Stabilized blood was stored at room temperature for no more than 30 min prior to centrifugation. The samples were centrifuged at 200× g for 7 min. Then, 2.5 mL of the supernatant containing platelet-rich plasma was carefully taken for analysis in the aggregometer. Platelet aggregation was measured by LTA using the laser analyzer of platelet aggregation ALAT-2 (Biola Scientific, Moscow, Russia) based on the method of Born and O’Brien.
To identify a group of patients with AR, we relied on the criteria proposed by Gum et al. []. AR was defined as aggregation of ≥70% with 10 μm ADP and aggregation of ≥20% with 0.5 mM AA. Aspirin semi-resistance (ASR) was defined as aggregation of ≥70% with 10 μM ADP or aggregation of ≥20% with 0.5 mM AA []. The patients with AR and ASR were pooled into the AR group. Patients with ADP-induced aggregation <70% and AA-induced aggregation <20% were considered aspirin-sensitive (AS) and were assigned to the AS group [].
2.3. DNA Extraction
Genomic DNA was extracted from the blood collected into the EDTA-containing tubes using the QIamp DNA Mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. DNA was isolated from 200 μL of the whole blood. The procedure included cell lysis, sorption on the silica gel membrane of the column, washing and elution (in 100 µL of elution buffer). The DNA concentration was measured using the NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). DNA samples were subjected to further analysis if DNA concentration was at least 10 ng/µL and its 260/280 ratio was in the range of 1.75 to 1.95.
2.4. Selection of SNPs and Genotyping
Genetic markers in ten genes (ITGB3, GPIBA, TBXA2R, ITGA2, PLA2G7, HMOX1, PTGS1, PTGS2, ADRA2A, ABCB1, PEAR1) and one intergenic region (9p21.3) were selected (Table 1).

Table 1.
A list of studied genetic markers.
Genotyping involved the multiplex one-step PCR followed by allele-specific hybridization on a biochip as described before []. 2′-deoxyuridine 5′-triphosphate (dUTP) derivatives containing the Cy7 cyanine dye were used as fluorophores []. The sequences of primers and allele-specific oligonucleotide probes are listed in the Supplementary Tables S1 and S2. The biochip scheme and an example of the hybridization picture are shown in Figure S2. Genotyping results were verified by direct sequencing and high-resolution melting analysis.
2.5. Statistical Analysis
The online service SNPStats (https://www.snpstats.net/ (accessed on 26 April 2022)) [] was used to evaluate the association of genotypes with aspirin resistance and aggregation with AA and ADP as well as the NIHSS score dynamics (adjusted by clinical variables). We used individual SNPs’ data for co-dominant, dominant, recessive and log-additive models. Comparison of baseline characteristics in groups with different genotypes was performed using the Kruskal–Wallis test and the chi-square test. Allele frequencies between AS and AR groups were compared using the two-sided Fisher exact test. Statistical analysis was performed in R (version 4.1.1; R Foundation for Statistical Computing, Vienna, Austria). The differences were considered statistically significant if the p-value was below 0.05. The boxplots display the median, two hinges which correspond to the first and third quartiles and two whiskers. The upper and lower whiskers extend from the hinges to the largest value no further than 1.5×IQR from the corresponding hinge (where IQR is the inter-quartile range). Points beyond the whiskers indicate the outliers.
2.6. Machine Learning
To build a predictive machine learning model, several approaches have been tested using the following Python 3.8 libraries: sklearn.linear_model.LogisticRegression, sklearn.svm.SVC, sklearn.ensemble.RandomForestClassifier [], XGBoost [] and CatBoost []. All models were trained in a five-fold cross validation (CV) setting with folds stratified to keep the proportion of studies similar to the whole data set. Each model parameter was optimized in order to increase the classification metrics: accuracy, AUC and F1-score, paying the most attention to the latter metric. The array of features consisted of all 16 genetic markers along with the age, gender, NHISS score at admission, body mass index (BMI), atrial fibrillation (AF), stenosis, high-density lipoproteins (HDLs), low-density lipoproteins (LDLs), cholesterol and triglycerides. Feature importance ranking was obtained using Shapley additive explanations (SHAP) values, a game theoretic approach to explain the output of any machine learning model []. The sequence of the ML procedure pipeline is shown in Figure 1.
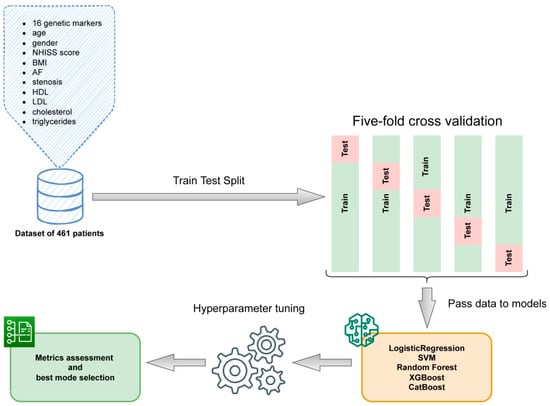
Figure 1.
The machine learning pipeline.
3. Results
3.1. Baseline Characteristics of AR and AS Patients
The baseline clinical characteristics of patients are shown in Table 2. A total of 461 patients were included in the analysis. Full AR and ASR were established in 28 patients (6.1%) and 192 patients (41.6%), respectively, and these two groups were pooled into one AR group. Another 241 patients (52.3%) were AS.

Table 2.
The clinical characteristics and laboratory parameters in the AS and AR groups.
The AS and AR groups differed in some clinical parameters (Table 2). AR patients were significantly older: the mean age of 73.21 years in the AR group vs. 68.72 years in the AS group (p < 0.001). AR patients had a more severe stroke: the mean NHISS score at admission was 12.35 and 10.45 in the AR and AS group, respectively (p < 0.001). Moreover, atrial fibrillation was more frequent in the AR group (41.74%) compared to the AS group (29.58%) (p = 0.0088).
3.2. The Association of SNPs with Aspirin Resistance in Whole Cohort of Patients
A total of 461 samples were genotyped for selected SNPs (Table 3). Genotype frequencies in the total sample, AR and AS groups conformed to the Hardy–Weinberg equilibrium (data not shown). The rs1126643 and rs1062535 markers in the ITGA2 gene as well as rs1051931 and rs7756935 in the PLA2G7 gene were in strong linkage disequilibrium (D’ = 1.0, R2 = 1.0), and only one of them in each pair was included in the analysis.

Table 3.
Genotype and allele frequencies in the AS and AR groups.
Allele and genotype frequencies for sixteen SNPs in the AS and AR groups are listed in Table 3. The frequency of the minor allele C for rs1330344 PTGS1 was significantly higher in the AR group than in the AS group (27% vs. 21%, p = 0.044).
The association of genotypes with the response to aspirin was investigated using the SNPStats online service. We included age, AF and the NHISS score at admission as covariates in the analysis, since they showed a different distribution between AR and AS groups (Table 2). The results for all studied markers are in Supplementary Table S1 (for AR and AS groups) and Supplementary Table S2 (for AA- and ADP-induced aggregation). We revealed the following associations.
The CC genotype of rs1330344 in the PTGS1 gene was more frequent in the AR group than in the AS group (OR = 2.75, 95% CI = 1.14–6.63, p = 0.019). Data are shown in Supplementary Table S3.
We compared the association of different genotypes with AA- and ADP-induced aggregation. For PTGS1 rs1330344, mean AA-induced aggregation was 40.5% higher in the CC genotype compared to the TT + CT genotypes (p = 0.038). For rs4311994 in the ADRA2A gene, mean AA-induced aggregation was 72.7% higher in patients with the TT genotype compared to the CC + CT genotypes (p = 0.043). Data are shown in Figure 2 and in Supplementary Table S4.
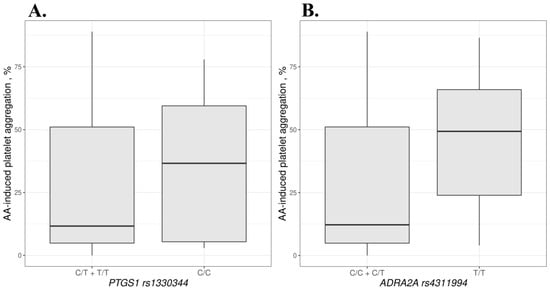
Figure 2.
AA-induced aggregation based on the PTGS1 rs1330344 (A) and ADRA2A rs4311994 (B) genotypes.
Mean ADP-induced aggregation was 9.2% higher in the TT + CT genotypes of TBXA2R rs4523 compared to the CC genotype (p = 0.043). For rs12041331 in the PEAR1 gene, mean ADP-induced aggregation was 59.5% lower in AA homozygotes compared to the GG + GA genotypes (p = 0.017). Data are shown in Figure 3 and in Supplementary Table S2.
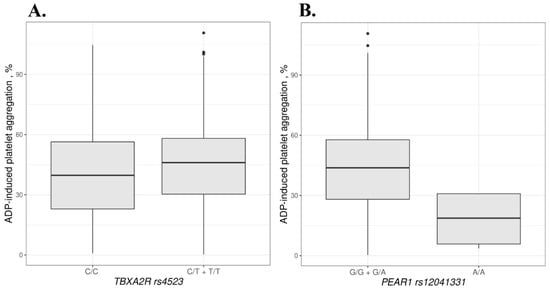
Figure 3.
ADP-induced aggregation based on the TBXA2R rs4523 (A) and PEAR1 rs12041331 (B) genotypes. Dots beyond the whiskers indicate the outliers.
3.3. The Association of SNPs with AR and Platelet Reactivity in Patients with Noncardioembolic Ischemic Stroke
In total, 296 patients had noncardioembolic ischemic stroke, with 127 (43%) and 169 (57%) patients being assigned to the AR and AS groups, respectively. We analyzed the frequency of different genotypes for sixteen SNPs in the AR and AS groups. Although the CC genotype of rs1330344 in the PTGS1 gene was more frequent in the AR group than in the AS group (OR = 2.48, 95% CI = 0.93–6.60, p = 0.062), the difference is not statistically significant.
The CC homozygotes of PTGS1 rs1330344 had 55.4% higher mean AA-induced aggregation compared to the TT + CT genotypes (p = 0.026). Mean ADP-induced aggregation was 14.8% higher in the TT + CT genotypes of rs4523 TBXA2R than in the CC genotypes (p = 0.031) and 11.6% lower in the AA + GA genotypes of ITGA2 rs1062535 comparing to GG homozygotes (p = 0.051).
Note that p-values are given before the correction for multiple comparisons; after the Bonferroni correction, all p-values were >0.05.
3.4. Clinical Outcome Evaluation
We evaluated the clinical outcomes of patients with noncardioembolic ischemic stroke within the first 10 days after admission and their association with aspirin resistance. Five patients died and were excluded from the analysis. The NIHSS score on day 10 was compared with the admission NIHSS score. We found no statistically significant association of the NIHSS score dynamics analyzed with the groups of AR and AS patients.
Furthermore, the NIHSS score dynamics was evaluated in patients with different genotypes adjusted by age and the NIHSS score at admission. The AA + GA genotypes of rs1062535 in the ITGA2 gene had worse dynamics in the NIHSS score compared to the GG genotype (5.3 vs. 6.57, p = 0.0008).
3.5. Machine Learning Model
To investigate the contribution of clinical and genetic features to AR, we created ML models. The overall best performance was achieved after utilizing CatBoost algorithm, high-performance open-source library for gradient boosting on decision trees. The parameters of the model that showed best performance in CV are listed in the Appendix A. For ML model generation, the total cohort of patients with ischemic stroke was included.
The ML models did not have enough predictive power if they were based only on genetic features. To overcome this limitation, we included anthropometric and clinical data in the model.
After training several models in a five-fold cross-validation setting, we compared the output metrics in order to choose the classification method with the best performance. As expected, the gradient boosting on the decision tree algorithm, CatBoost, outperformed logistic regression, the support vector machine and random forest classifiers since it was designed to leverage the information gained from categorical features. The average values of classification metrics were as follows: AUC = 0.665, F1-score = 0.628, specificity = 0.773, sensitivity = 0.60, precision = 0.63. The ML model is in the Supplementary Files (Model S1).
To assess the impact of each feature on the model performance and identify the most important factors, we conducted the Shapley additive explanations analysis (Figure 4), which allowed us to study the relationships between variables for the predicted case and their contribution to the final score. Shapley values indicate the importance of a feature by comparing model predictions with and without this feature.
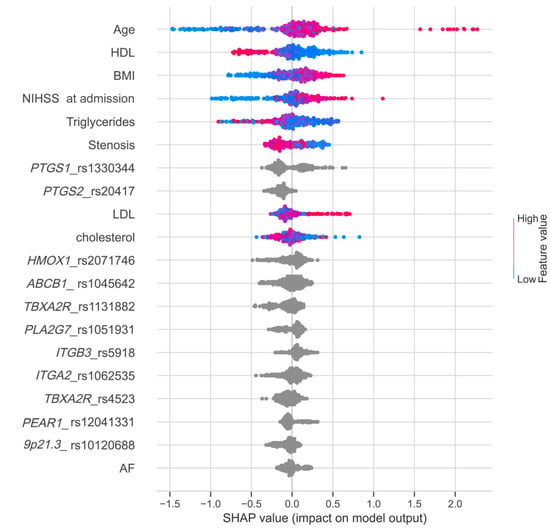
Figure 4.
Feature importance ranking obtained using SHAP values. Variables are listed in order of significance from top to bottom on the y-axis. Each point represents a patient, and its color indicates the value of corresponding variable. The position of the points on the x-axis represents SHAP values, indicating the changes in log odds, and the probability of success can be extracted from this value.
4. Discussion
In the present study, we used a biochip-based assay to analyze 18 SNPs in patients with acute ischemic stroke and variable response to aspirin treatment. The SNPs were selected based on the literature data. All of them are involved in platelet activation and aggregation, and their contribution to aspirin resistance is discussed in numerous studies [,,,,,,,,,,,,,,,,,]. We evaluated the distribution of 16 genetic markers in the AS and AR groups in a cohort of 461 patients with acute ischemic stroke.
The aspirin resistance was associated with the following clinical parameters: age, the NIHSS score at admission and atrial fibrillation (Table 2). Aging is known to be associated with an elevated platelet activity [] as well as aspirin resistance [,], which is consistent with our results. The initial NIHSS score was also higher in the AR patients []. In several studies concerning AR, ischemic stroke patients with atrial fibrillation (cardioembolism) were excluded from the analysis since they had anticoagulant therapy prescribed earlier [,]. In our study, all patients with all stroke variants received aspirin at least for the first 3 days, while laboratory AR was estimated during this period. This allowed us to enroll all the patients in the study, which aimed at identifying the associations between genetic markers and aspirin non-sensitivity. In addition, we performed the association studies in a cohort of non-embolic patients and evaluated clinical recovery for 10 days based on the NIHSS score dynamics. Determining the prognostic genetic markers of AR in this group can be very helpful given that the long-term aspirin treatment is recommended for these patients.
Among 16 SNPs studied, four genetic variants showed a significant association with aspirin non-sensitivity in the whole cohort: PTGS1 (rs1330344), ADRA2A (rs4311994), TBXA2R (rs4523) and PEAR1 (rs12041331).
The C allele and CC genotype (rs1330344) of the PTGS1 gene encoding COX-1 were associated with AR and a higher level of AA-induced aggregation. A similar observation was made in the study by Li et.al. []. The CC genotype was associated with poor functional outcomes in Chinese patients with a stroke during aspirin therapy [,]. However, the obtained results were not always consistent [,,,]. This polymorphism is located in the regulatory region (T-1676C), and this substitution may lead to an increase in COX-1 activity and contribute to a decreased or absent response to aspirin []. The C allele was also found to be associated with an increased risk of ischemic stroke in the Chinese population []. However, in our study, rs10306114 in this gene, most frequently associated with AR [], showed no association with AR.
Another polymorphism that demonstrated an association with AA-induced aggregation in our study, rs4311994, is also located in the regulatory region downstream (63 kb) of the 3’ end of the ADRA2A gene; its effect may arise from the regulation of gene expression or linkage disequilibrium with the other variants. In our study, the minor allele T of the ADRA2A gene (rs4311994) was associated with a higher level of AA-induced aggregation. The ADRA2A gene encodes the alpha-2A-adrenergic receptor involved in epinephrine-induced platelet aggregation and shear-dependent platelet function. This allele was associated with increased platelet reactivity to aspirin in the population with type 2 diabetes mellitus. []. However, these results were not always reproducible [].
Notably, the alleles of the PTGS1 and ADRA2A genes, associated with AR and/or high AA-induced aggregation in our study, correlated with a reduced risk of complications from the gastrointestinal tract when taking aspirin in other studies [,,]. This may indicate a role in stimulating platelet activity in carriers of these alleles.
The TBXA2R rs4523 (T924C) affected ADP-induced aggregation: the aggregation was higher in the TT and CT genotypes than in the CC genotype. In other studies, the TT homozygotes also showed increased platelet reactivity [,]. It is a synonymous nucleotide change that can affect splicing or mRNA stabilization and translation efficiency. Otherwise, this SNP may be in linkage disequilibrium with other clinically relevant polymorphisms [,]. The other SNP (rs1131882) in the TBXA2R gene showed no association with AR in our study.
The ADP-induced aggregation was affected by intronic rs12041331 in the PEAR1 gene being lower in the AA homozygote as compared to the GG and GA genotypes. These data are consistent with some other studies [,,,]. The PEAR1 gene encodes the type 1 membrane protein expressed in platelets and endothelial cells. Its phosphorylation appears to promote platelet aggregation [,]. The rs12041331 polymorphism results in a G to A substitution in intron 1 and was previously shown to be implicated in reducing PEAR1 expression []. According to Faraday et al. [], the major G allele of rs12041331 was associated with a higher platelet aggregation both in the presence and absence of aspirin treatment. Thus, the influence of the PEAR1 gene may not be specific to the aspirin action. The AA genotype of PEAR1 rs12041331 was shown to be associated with an increased response to ticagrelor in healthy people []. However, some studies revealed no such association for this SNP [].
In patients with noncardioembolic stroke, the polymorphism PTGS1 rs1330344 showed a significant association with AA-induced aggregation. Thus, PTGS1 rs1330344 might be considered as the strongest predictor of laboratory AR among the analyzed SNPs, both in the whole cohort of ischemic stroke and noncardioembolic patients. The second genetic marker associated with laboratory AR in both cohorts was rs4523 in TBXA2R gene. The T allele acted as a risk factor for increased ADP-induced aggregation during aspirin treatment.
An ambiguous association for ITGA2 rs1062535 was revealed in noncardioembolic patients. The ITGA2 gene encodes the alpha chain of the platelet collagen receptor integrin α2β1 (glycoprotein IA/IIa, GPIa/IIa), which promotes an initial interaction between platelets and collagen with further platelet activation and aggregation. The A allele of rs1062535 was suggested to stimulate the protein expression and increase affinity to collagen, which in turn facilitated platelet reactivity. The A allele of ITGA2 rs1062535 was significantly associated with reduced post-operative bleeding after cardiac surgery []. In our study, on the contrary, the AA + GA genotypes correlated with lower ADP-induced aggregation. In contrast, in previously published data, the A allele was considered as a possible risk factor for thromboischaemic events []. This suggestion is in agreement with our findings implying a strong relationship between the A allele and negative NIHSS dynamics in noncardioembolic patients.
Thus, the role of genetic factors underlying the inter-individual differences in aspirin action is of immense interest, but further research is required to understand how genetic data can be efficiently applied to personalized therapy. Different approaches, such as general multifactor dimensionality reduction (GMDR), were employed to study the potential contribution of multiple genetic factors along with the single-locus analysis [].
We applied the ML method to predict the risk of AR development using clinical and genetic factors. This is the first attempt to bring in the ML approach to the analysis of genetics of AR. We obtained an AUC = 0.665 for our best model (Model S1, Figure 4). On the one hand, this value seems to be modest, but on the other hand, it is in agreement with the parameters of other models based on ML for multifactorial processes. For example, similar sensitivity and specificity values were obtained for antidepressants [,]. However, in those studies, these parameters were obtained only from the genetic factors, whereas in our study they mainly depended on clinical factors. The developed ML model may be considered as a first approximation aimed at dealing with the problem of AR prediction. The relevance of the developed model for clinical practice is still to be confirmed. We assume that further studies involving larger and more clinically uniform cohorts of patients are required to shed light on the genetic background contributing to the resistance to aspirin treatment. Another approach relies on searching for more relevant genetic markers utilizing throughput methods of genetic analysis such as the next-generation sequencing. The assessment of polygenic risk score might prove promising as well.
As there is an alternative to aspirin for secondary stroke prevention, such as dual antiplatelet therapy or ticagrelor [], identifying patients with a predisposition to AR can be used for personalized therapy to reduce the risk of adverse events. However, it is possible that the risk alleles for AR might also be associated with platelet aggregation when taking other antiplatelet drugs requiring special attention for such patients.
The current study has several limitations. First, when choosing genetic polymorphisms, we relied on the published studies focusing on certain candidate genetic markers. Searching for more relevant genetic markers using such high-throughput methods of genetic analysis as next-generation sequencing may prove promising. Moreover, given the complex nature of aspirin resistance, the polygenic risk score may be introduced for identifying patients with a high risk of aspirin treatment failure. The second limitation is related to the size of the studied population. It seems to be large enough compared with other studies in the field [,,,,,,,,,,,]. However, clarifying the genetic background of aspirin treatment failure, which is affected by numerous clinical parameters and studied SNPs, requires further studies including larger and more clinically uniform cohorts of patients. The third limitation may be related to clinical outcome assessment. A number of studies confirmed an increased risk of adverse outcomes in patients with laboratory AR [,,,]. However, the underlying mechanism of a poor response to aspirin is still unclear. The ex vivo platelet reactivity tests do not always clearly correlate with the therapeutic effect of the drug []. Our study focused on analyzing laboratory AR, but the most important results can be obtained from the long-term follow-up of patients and assessing the influence of genetic and clinical factors and laboratory measurements of AR on clinical outcomes. Finally, in our study, we were not always able to take into account the potential impact of other drugs used by our patients, such as anticoagulants or statins, which are usually prescribed for the secondary prevention of a stroke. The drug–drug interactions as well as malabsorption or renal dysfunction could also affect aspirin pharmacokinetics or pharmacodynamics and thus lead to a number of subsequent pharmacological effects [,].
5. Conclusions
Early detection of aspirin resistance in ischemic stroke patients is important for timely prescription of other antiaggregant drugs when possible. Therefore, searching for predictive markers of aspirin treatment failure is of great importance. In our study, we revealed the association between clinical parameters (age, NIHSS score, atrial fibrillation), as well as SNPs in the PTGS1, ADRA2A, TBXA2R and PEAR1 genes, and laboratory indicators of platelet activity in ischemic stroke patients taking aspirin for secondary stroke prevention. The ML model of AR in the studied cohort of patients showed the prevailing contribution of clinical parameters. However, we assume that the genetic factors are a promising predictor of aspirin resistance. The ML approach revealed the prospective future directions of predicting the risk of AR development. Further replication studies including more homogeneous groups of patients, the implementation of high-throughput genotyping technologies and development of risk-predictive models based both on clinical and genetic features may be considered as key steps towards better understanding aspirin resistance in patients with an ischemic stroke.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biomedicines10102564/s1. Figure S1: The pathway of TXA2 production and the antiplatelet effect of aspirin.; Figure S2: The biochip scheme and an example of analysis.; Table S1: Primer sequences.; Table S2: Allele-specific oligonucleotide probe sequences.; Table S3: Distribution of genetic markers in AS- and AR-groups.; Table S4: AA- and ADP-induced aggregation based on genotypes.; Model S1: ML model for aspirin resistance.
Author Contributions
Conceptualization, A.A. and T.N.; methodology, A.I., S.G., A.G., Z.A., M.F., S.S., V.S. and L.S.; validation, A.I., L.S., V.S. and Z.A.; formal analysis, A.I., S.G., Z.A. and A.G.; investigation, A.I., S.G., A.A. and T.N; resources, A.A. and S.G.; writing—original draft preparation, A.I., M.A. and S.G.; writing—review and editing, A.A., T.N. and A.Z.; visualization, A.I.; supervision, A.Z.; project administration, A.A., T.N. and A.Z.; funding acquisition, T.N. and A.Z. All authors have read and agreed to the published version of the manuscript.
Funding
The work was carried out with the support of the state program 0103-2018-0003.
Institutional Review Board Statement
The study was approved by the local ethics committee of the Pirogov Russian National Research Medical University (protocol number 181 dated 28 January 2019).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A. Parameters of the Best-Performing Model
| loss_function = ‘Logloss’ |
| learning_rate = 0.01 |
| Iterations = 500 |
| depth = 10 |
| grow_policy = ‘Lossguide’ |
| max_leaves = 30 |
| od_type = ‘IncToDec’ |
| od_pval = 0.05 |
| od_wait = 10 |
References
- Rothwell, P.M.; Algra, A.; Chen, Z.; Diener, H.-C.; Norrving, B.; Mehta, Z. Effects of aspirin on risk and severity of early recurrent stroke after transient ischaemic attack and ischaemic stroke: Time-course analysis of randomised trials. Lancet 2016, 388, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Capodanno, D.; Angiolillo, D.J. Aspirin for Primary Cardiovascular Risk Prevention and Beyond in Diabetes Mellitus. Circulation 2016, 134, 1579–1594. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zheng, L.; Fei, Q.; Fu, Y.; Weng, Y.; Wu, H.; Li, H.; Jun, Q.; Shao, J.; Xu, Y. Association of thromboxane A2 receptor gene polymorphisms with cerebral infarction in a Chinese population. Neurol. Sci. 2013, 34, 1791–1796. [Google Scholar] [CrossRef] [PubMed]
- Goodman, T.; Ferro, A.; Sharma, P. Pharmacogenetics of aspirin resistance: A comprehensive systematic review. Br. J. Clin. Pharmacol. 2008, 66, 222–232. [Google Scholar] [CrossRef] [PubMed]
- SSantos-Gallego, C.G.; Badimon, J. Overview of Aspirin and Platelet Biology. Am. J. Cardiol. 2021, 144, S2–S9. [Google Scholar] [CrossRef]
- Ferreira, M.; Freitas-Silva, M.; Assis, J.; Pinto, R.; Nunes, J.P.; Medeiros, R. The emergent phenomenon of aspirin resistance: Insights from genetic association studies. Pharmacogenomics 2020, 21, 125–140. [Google Scholar] [CrossRef]
- Kuliczkowski, W.; Witkowski, A.; Polonski, L.; Watala, C.; Filipiak, K.; Budaj, A.; Golanski, J.; Sitkiewicz, D.; Pregowski, J.; Gorski, J.; et al. Interindividual variability in the response to oral antiplatelet drugs: A position paper of the Working Group on antiplatelet drugs resistance appointed by the Section of Cardiovascular Interventions of the Polish Cardiac Society, endorsed by the Working Group on Thrombosis of the European Society of Cardiology. Eur. Heart J. 2008, 30, 426–435. [Google Scholar] [CrossRef]
- Grinstein, J.; Cannon, C.P. Aspirin resistance: Current status and role of tailored therapy. Clin. Cardiol. 2012, 35, 673–680. [Google Scholar] [CrossRef]
- Lordkipanidzé, M.; Pharand, C.; Schampaert, E.; Turgeon, J.; Palisaitis, D.A.; Diodati, J.G. A comparison of six major platelet function tests to determine the prevalence of aspirin resistance in patients with stable coronary artery disease. Eur. Heart J. 2007, 28, 1702–1708. [Google Scholar] [CrossRef]
- Van Oosterom, N.; Barras, M.; Cottrell, N.; Bird, R. Platelet Function Assays for the Diagnosis of Aspirin Resistance. Platelets 2021, 33, 329–338. [Google Scholar] [CrossRef]
- Le Blanc, J.; Mullier, F.; Vayne, C.; Lordkipanidzé, M. Advances in Platelet Function Testing—Light Transmission Aggregometry and Beyond. J. Clin. Med. 2020, 9, 2636. [Google Scholar] [CrossRef]
- Venketasubramanian, N.; Agustin, S.J.; Padilla, J.L.; Yumul, M.P.; Sum, C.; Lee, S.H.; Ponnudurai, K.; Gan, R.N. Comparison of Different Laboratory Tests to Identify “Aspirin Resistance” and Risk of Vascular Events among Ischaemic Stroke Patients: A Double-Blind Study. J. Cardiovasc. Dev. Dis. 2022, 9, 156. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.; Jain, S.; Gallant, R.C.; Syed, M.H.; Zamzam, A.; Al-Omran, M.; Rand, M.L.; Ni, H.; Abdin, R.; Qadura, M. Plateletworks® as a Point-of-Care Test for ASA Non-Sensitivity. J. Pers. Med. 2021, 11, 813. [Google Scholar] [CrossRef] [PubMed]
- Fiolaki, A.; Katsanos, A.H.; Kyritsis, A.P.; Papadaki, S.; Kosmidou, M.; Moschonas, I.C.; Tselepis, A.D.; Giannopoulos, S. High on treatment platelet reactivity to aspirin and clopidogrel in ischemic stroke: A systematic review and meta-analysis. J. Neurol. Sci. 2017, 376, 112–116. [Google Scholar] [CrossRef]
- Lv, H.; Yang, Z.; Wu, H.; Liu, M.; Mao, X.; Liu, X.; Ding, H.; Shi, Z.; Zhou, Y.; Liu, Q.; et al. High On-Treatment Platelet Reactivity as Predictor of Long-term Clinical Outcomes in Stroke Patients with Antiplatelet Agents. Transl. Stroke Res. 2021, 13, 391–398. [Google Scholar] [CrossRef]
- Sikora, J.; Karczmarska-Wódzka, A.; Bugieda, J.; Sobczak, P. The Importance of Platelets Response during Antiplatelet Treatment after Ischemic Stroke—Between Benefit and Risk: A Systematic Review. Int. J. Mol. Sci. 2022, 23, 1043. [Google Scholar] [CrossRef]
- Wiśniewski, A.; Filipska, K.; Sikora, J.; Kozera, G. Aspirin Resistance Affects Medium-Term Recurrent Vascular Events after Cerebrovascular Incidents: A Three-Year Follow-up Study. Brain Sci. 2020, 10, 179. [Google Scholar] [CrossRef]
- Wiśniewski, A. Multifactorial Background for a Low Biological Response to Antiplatelet Agents Used in Stroke Prevention. Medicina 2021, 57, 59. [Google Scholar] [CrossRef]
- Faraday, N.; Yanek, L.R.; Mathias, R.; Herrera-Galeano, J.E.; Vaidya, D.; Moy, T.F.; Fallin, M.D.; Wilson, A.F.; Bray, P.F.; Becker, L.C.; et al. Heritability of platelet responsiveness to aspirin in activation pathways directly and indirectly related to cyclooxygenase-1. Circulation 2007, 115, 2490–2496. [Google Scholar] [CrossRef]
- Strisciuglio, T.; Franco, D.; Di Gioia, G.; De Biase, C.; Morisco, C.; Trimarco, B.; Barbato, E. Impact of genetic polymorphisms on platelet function and response to anti platelet drugs. Cardiovasc. Diagn. Ther. 2018, 8, 610–620. [Google Scholar] [CrossRef]
- Yi, X.; Cheng, W.; Lin, J.; Zhou, Q.; Wang, C. Interaction between COX-1 and COX-2 Variants Associated with Aspirin Resistance in Chinese Stroke Patients. J. Stroke Cerebrovasc. Dis. 2016, 25, 2136–2144. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Wang, C.; Zhou, Q.; Lin, J. Interaction among COX-2, P2Y1 and GPIIIa gene variants is associated with aspirin resistance and early neurological deterioration in Chinese stroke patients. BMC Neurol. 2017, 17, 4. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-L.; Cao, J.; Fan, L.; Wang, Q.; Ye, L.; Cui, C.-P.; Wang, Y.-Z.; Liu, L.; Li, B.; Wu, R.-J.; et al. Genetic polymorphisms of ho-1 and cox-1 are associated with aspirin resistance defined by light transmittance aggregation in chinese han patients. Clin. Appl. Thromb. 2012, 19, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.-H.; Jiao, J.-R.; Yang, R.; Luo, B.-Y.; Wang, X.-F.; Wu, F. Aspirin resistance: Clinical significance and genetic polymorphism. J. Int. Med. Res. 2012, 40, 282–292. [Google Scholar] [CrossRef]
- Halushka, M.; Walker, L.P. Genetic variation in cyclooxygenase 1: Effects on response to aspirin. Clin. Pharmacol. Ther. 2003, 73, 122–130. [Google Scholar] [CrossRef]
- Kunicki, T.J.; Williams, S.A.; Nugent, D.J.; Harrison, P.; Segal, H.C.; Syed, A.; Rothwell, P.M. Lack of association between aspirin responsiveness and seven candidate gene haplotypes in patients with symptomatic vascular disease. Thromb. Haemost. 2009, 101, 123–133. [Google Scholar]
- Ulehlova, J.; Slavik, L.; Kucerova, J.; Krcova, V.; Vaclavik, J.; Indrak, K. Genetic polymorphisms of platelet receptors in patients with acute myocardial infarction and resistance to antiplatelet therapy. Genet. Test. Mol. Biomark. 2014, 18, 599–604. [Google Scholar] [CrossRef]
- Maree, A.O.; Curtin, R.J.; Chubb, A.; Dolan, C.; Cox, D.; O’Brien, J.; Crean, P.; Shields, D.C.; Fitzgerald, D.J. Cyclooxygenase-1 haplotype modulates platelet response to aspirin. J. Thromb. Haemost. 2005, 3, 2340–2345. [Google Scholar] [CrossRef]
- Peng, L.-L.; Zhao, Y.-Q.; Zhou, Z.-Y.; Jin, J.; Zhao, M.; Chen, X.-M.; Chen, L.; Cai, Y.-F.; Li, J.-L.; Huang, M. Associations of MDR1, TBXA2R, PLA2G7, and PEAR1 genetic polymorphisms with the platelet activity in Chinese ischemic stroke patients receiving aspirin therapy. Acta Pharmacol. Sin. 2016, 37, 1442–1448. [Google Scholar] [CrossRef]
- Postula, M.; Kaplon-Cieslicka, A.; Rosiak, M.; Kondracka, A.; Serafin, A.; Filipiak, K.J.; Członkowski, A.; Opolski, G.; Janicki, P.K. Genetic determinants of platelet reactivity during acetylsalicylic acid therapy in diabetic patients: Evaluation of 27 polymorphisms within candidate genes. J. Thromb. Haemost. 2011, 9, 2291–2301. [Google Scholar] [CrossRef]
- Milanowski, L.; Pordzik, J.; Janicki, P.K.; Kaplon-Cieslicka, A.; Rosiak, M.; Peller, M.; Tyminska, A.; Ozieranski, K.; Filipiak, K.J.; Opolski, G.; et al. New single-nucleotide polymorphisms associated with differences in platelet reactivity and their influence on survival in patients with type 2 diabetes treated with acetylsalicylic acid: An observational study. Geol. Rundsch. 2016, 54, 343–351. [Google Scholar] [CrossRef][Green Version]
- Szczeklik, A.; Undas, A.; Sanak, M.; Frołow, M.; Węgrzyn, W. Relationship between bleeding time, aspirin and the PlA1/A2 polymorphism of platelet glycoprotein IIIa. Br. J. Haematol. 2000, 110, 965–967. [Google Scholar] [CrossRef]
- Al-Azzam, S.I.; Alzoubi, K.H.; Khabour, O.F.; Tawalbeh, D.; Al-Azzeh, O. The contribution of platelet glycoproteins (GPIa C807T and GPIba C-5T) and cyclooxygenase 2 (COX-2G-765C) polymorphisms to platelet response in patients treated with aspirin. Gene 2013, 526, 118–121. [Google Scholar] [CrossRef]
- Wang, H.; Sun, X.; Dong, W.; Cai, X.; Zhou, Y.; Zhang, Y.; Jiang, W.; Fang, Q. Association of GPIa and COX-2 gene polymorphism with aspirin resistance. J. Clin. Lab. Anal. 2017, 32, e22331. [Google Scholar] [CrossRef]
- Xiang, Q.; Zhou, S.; Lewis, J.P.; Shuldiner, A.R.; Ren, G.; Cui, Y. Genetic Variants of PEAR1 are Associated with Platelet Function and Antiplatelet Drug Efficacy: A Systematic Review and Meta-Analysis. Curr. Pharm. Des. 2018, 23, 6815–6827. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jiang, H.; Ding, Y.; Zhang, D.; Zhang, X.; Xue, J.; Ma, R.; Hu, L.; Yue, Y. Platelet Endothelial Aggregation Receptor 1 Polymorphism Is Associated With Functional Outcome in Small-Artery Occlusion Stroke Patients Treated With Aspirin. Front. Cardiovasc. Med. 2021, 8, 664012. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.P.; Ryan, K.; O’Connell, J.R.; Horenstein, R.B.; Damcott, C.M.; Gibson, Q.; Pollin, T.I.; Mitchell, B.D.; Beitelshees, A.L.; Pakzy, R.; et al. Genetic variation in PEAR1 is associated with platelet aggregation and cardiovascular outcomes. Circ. Cardiovasc. Genet. 2013, 6, 184–192. [Google Scholar] [CrossRef]
- Musunuru, K.; Post, W.S.; Herzog, W.; Shen, H.; O’Connell, J.R.; McArdle, P.F.; Ryan, K.A.; Gibson, Q.; Cheng, Y.-C.; Clearfield, E.; et al. Association of single nucleotide polymorphisms on chromosome 9p21.3 with platelet reactivity: A potential mechanism for increased vascular disease. Circ Cardiovasc Genet. Circ. Cardiovasc. Genet. 2010, 3, 445–453. [Google Scholar] [CrossRef]
- Kugai, M.; Uchiyama, K.; Tsuji, T.; Yoriki, H.; Fukui, A.; Qin, Y.; Higashimura, Y.; Mizushima, K.; Yoshida, N.; Katada, K.; et al. MDR1 is Related to intestinal epithelial injury induced by acetylsalicylic acid. Cell. Physiol. Biochem. 2013, 32, 942–950. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Cai, B.; Sun, L.; Zhang, H.; Zhou, S.; Cao, L.; Guo, H.; Sun, W.; Yan, B.; Davis, S.M.; et al. Association between PTGS1 polymorphisms and functional outcomes in Chinese patients with stroke during aspirin therapy: Interaction with smoking. J. Neurol. Sci. 2017, 376, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Cilluffo, G.; Fasola, S.; Ferrante, G.; Malizia, V.; Montalbano, L.; La Grutta, S. Machine Learning: An Overview and Applications in Pharmacogenetics. Genes 2021, 12, 1511. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Zhao, J.; Roden, D.M.; Peterson, J.F. Machine Learning Challenges in Pharmacogenomic Research. Clin. Pharmacol. Ther. 2021, 110, 552–554. [Google Scholar] [CrossRef]
- Madden, K.P.; Karanjia, P.N.; Adams, H.P.; Clarke, W.R. Accuracy of initial stroke subtype diagnosis in the TOAST study. Trial of ORG 10172 in Acute Stroke Treatment. Neurology. Neurology 1995, 45, 1975–1979. [Google Scholar] [CrossRef]
- Kirchhof, P.; Benussi, S.; Kotecha, D.; Ahlsson, A.; Atar, D.; Casadei, B.; Castella, M.; Diener, H.-C.; Heidbuchel, H.; Hendriks, J.; et al. 2016 ESC Guidelines for the management of atrial fibrillation developed in collaboration with EACTS. Eur. Heart J. 2016, 37, 2893–2962. [Google Scholar] [CrossRef]
- Gum, P.A.; Kottke-Marchant, K.; Welsh, P.A.; White, J.; Topol, E.J. A prospective, blinded determination of the natural history of aspirin resistance among stable patients with cardiovascular disease. J. Am. Coll. Cardiol. 2003, 41, 961–965. [Google Scholar] [CrossRef]
- Ikonnikova, A.Y.; Filippova, M.A.; Surzhikov, S.A.; Pozhitnova, V.O.; Kazakov, R.E.; Lisitsa, T.S.; Belkov, S.A.; Nasedkina, T.V. Biochip-based approach for comprehensive pharmacogenetic testing. Drug Metab. Drug Interact. 2020, 36, 33–40. [Google Scholar] [CrossRef]
- Shershov, V.E.; Ikonnikova, A.Y.; Vasiliskov, V.A.; Lapa, S.A.; Miftakhov, R.A.; Kuznetsova, V.E.; Chudinov, A.V.; Nasedkina, T.V. The Efficiency of DNA Labeling with Near-Infrared Fluorescent Dyes. Biophysics 2020, 65, 736–741. [Google Scholar] [CrossRef]
- Solé, X.; Guinó, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Duchesnay, E. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016. [Google Scholar]
- Dorogush; Veronika, A.; Ershov, V.; Gulin, A. CatBoost: Gradient boosting with categorical features support. arXiv 2018, arXiv:1810.11363. [Google Scholar]
- Lundberg; Scott, M.; Lee, S. A unified approach to interpreting model predictions. Adv. Neural Inf. Process. Syst. 2017, 30, 4768–4777. [Google Scholar]
- Le Blanc, J.; Lordkipanidzé, M. Platelet Function in Aging. Front. Cardiovasc. Med. 2019, 6, 109. [Google Scholar] [CrossRef]
- Oh, M.S.; Yu, K.-H.; Lee, J.-H.; Jung, S.; Kim, C.; Jang, M.U.; Lee, J.; Lee, B.C. Aspirin resistance is associated with increased stroke severity and infarct volume. Neurology 2016, 86, 1808–1817. [Google Scholar] [CrossRef]
- Ghorbani-Shirkouhi, S.; Ashouri, F.; Neshin, S.A.S.; Saberi, A.; Hasanzadeh, B.; Shahshahani, P. The prevalence and associated factors of aspirin resistance among prophylactic aspirin users. Rom. J. Neurol. 2021, 20, 50–56. [Google Scholar] [CrossRef]
- Cao, L.; Zhang, Z.; Sun, W.; Bai, W.; Sun, W.; Zhang, Y.; Wang, X.; Cai, B.; Xie, X.; Duan, Z.; et al. Impacts of COX-1 gene polymorphisms on vascular outcomes in patients with ischemic stroke and treated with aspirin. Gene 2014, 546, 172–176. [Google Scholar] [CrossRef]
- Pettinella, C.; Romano, M.; Stuppia, L.; Santilli, F.; Liani, R.; Davì, G. Cyclooxygenase-1 haplotype C50T/A-842G does not affect platelet response to aspirin. Thromb. Haemost. 2009, 101, 687–690. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chen, B.-L.; Ozdemir, V.; Ji, W.; Mao, Y.-M.; Wang, L.-C.; Lei, H.-P.; Fan, L.; Zhang, W.; Liu, J.; et al. Frequency of genetic polymorphisms of COX1, GPIIIa and P2Y1 in a Chinese population and association with attenuated response to aspirin. Pharmacogenomics 2007, 8, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Fang, J.; Zhou, M.; Zhou, J.; Yu, L.; Chen, N.; He, L. Interaction between COX-1 and COX-2 increases susceptibility to ischemic stroke in a Chinese population. BMC Neurol. 2019, 19, 291. [Google Scholar] [CrossRef]
- Johnson, A.D.; Yanek, L.R.; Chen, M.-H.; Faraday, N.; Larson, M.; Tofler, G.; Lin, S.J.; Kraja, A.T.; Province, M.A.; Yang, Q.; et al. Genome-wide meta-analyses identifies seven loci associated with platelet aggregation in response to agonists. Nat. Genet. 2010, 42, 608–613. [Google Scholar] [CrossRef] [PubMed]
- Forgerini, M.; Urbano, G.; de Nadai, T.R.; Batah, S.S.; Fabro, A.T.; Mastroianni, P.D.C. Genetic Variants in PTGS1 and NOS3 Genes Increase the Risk of Upper Gastrointestinal Bleeding: A Case-Control Study. Front. Pharmacol. 2021, 12, 671835. [Google Scholar] [CrossRef] [PubMed]
- Mallah, N.; Zapata-Cachafeiro, M.; Aguirre, C.; Ibarra-García, E.; Palacios–Zabalza, I.; Macías-García, F.; Domínguez-Muñoz, J.E.; Piñeiro-Lamas, M.; Ibáñez, L.; Vidal, X.; et al. Influence of Polymorphisms Involved in Platelet Activation and Inflammatory Response on Aspirin-Related Upper Gastrointestinal Bleeding: A Case-Control Study. Front. Pharmacol. 2020, 11, 860. [Google Scholar] [CrossRef]
- Wu, Y.; Hu, Y.; You, P.; Chi, Y.-J.; Zhou, J.-H.; Zhang, Y.-Y.; Liu, Y.-L. Study of Clinical and Genetic Risk Factors for Aspirin-induced Gastric Mucosal Injury. Chin. Med. J. 2016, 129, 174–180. [Google Scholar] [CrossRef]
- Fujiwara, T.; Ikeda, M.; Esumi, K.; Fujita, T.D.; Kono, M.; Tokushige, H.; Hatoyama, T.; Maeda, T.; Asai, T.; Ogawa, T.; et al. Exploratory aspirin resistance trial in healthy Japanese volunteers (J-ART) using platelet aggregation as a measure of thrombogenicity. Pharm. J. 2007, 7, 395–403. [Google Scholar] [CrossRef]
- Wang, Z.; Gao, F.; Men, J.; Yang, J.; Modi, P.; Wei, M. Polymorphisms and high on-aspirin platelet reactivity after off-pump coronary artery bypass grafting. Scand. Cardiovasc. J. 2013, 47, 194–199. [Google Scholar] [CrossRef]
- Würtz, M.; Nissen, P.H.; Grove, E.L.; Kristensen, S.D.; Hvas, A.-M. Genetic determinants of on-aspirin platelet reactivity: Focus on the influence of pear1. PLoS ONE 2014, 9, e111816. [Google Scholar] [CrossRef]
- Hu, X.; Liu, C.; Zhang, M.; Zhang, W. Impact of the PEAR 1 polymorphism on clinical outcomes in Chinese patients receiving dual antiplatelet therapy after percutaneous coronary intervention. Pharmacogenomics 2022, 23, 639–648. [Google Scholar] [CrossRef]
- Nanda, N.; Bao, M.; Lin, H.; Clauser, K.; Komuves, L.; Quertermous, T.; Conley, P.B.; Phillips, D.R.; Hart, M.J. Platelet endothelial aggregation receptor 1 (pear1), a novel epidermal growth factor repeat-containing transmembrane receptor, participates in platelet contact-induced activation. J. Biol. Chem. 2005, 280, 24680–24689. [Google Scholar] [CrossRef]
- Kauskot, A.; Di Michele, M.; Loyen, S.; Freson, K.; Verhamme, P.; Hoylaerts, M.F. A novel mechanism of sustained platelet αIIbβ3 activation via PEAR1. Blood 2012, 119, 4056–4065. [Google Scholar] [CrossRef]
- Li, M.; Hu, Y.; Wen, Z.; Li, H.; Hu, X.; Zhang, Y.; Zhang, Z.; Xiao, J.; Tang, J.; Chen, X. Association of PEAR1 rs12041331 polymorphism and pharmacodynamics of ticagrelor in healthy Chinese volunteers. Xenobiotica 2017, 47, 1130–1138. [Google Scholar] [CrossRef]
- Lewis, J.P.; Riaz, M.; Xie, S.; Polekhina, G.; Wolfe, R.; Nelson, M.; Tonkin, A.M.; Reid, C.M.; Murray, A.M.; McNeil, J.J.; et al. Genetic Variation in PEAR1, Cardiovascular Outcomes and Effects of Aspirin in a Healthy Elderly Population. Clin. Pharmacol. Ther. 2020, 108, 1289–1298. [Google Scholar] [CrossRef]
- Greiff, G.; Pleym, H.; Stenseth, R.; Wahba, A.; Videm, V. Genetic variation influences the risk of bleeding after cardiac surgery: Novel associations and validation of previous findings. Acta Anaesthesiol. Scand. 2015, 59, 796–806. [Google Scholar] [CrossRef] [PubMed]
- Rath, D.; Schaeffeler, E.; Winter, S.; Levertov, S.; Müller, K.; Droppa, M.; Stimpfle, F.; Langer, H.F.; Gawaz, M.; Schwab, M.; et al. GPla Polymorphisms Are Associated with Outcomes in Patients at High Cardiovascular Risk. Front. Cardiovasc. Med. 2017, 4, 52. [Google Scholar] [CrossRef] [PubMed]
- Maciukiewicz, M.; Marshe, V.S.; Hauschild, A.-C.; Foster, J.A.; Rotzinger, S.; Kennedy, J.L.; Kennedy, S.H.; Müller, D.J.; Geraci, J. GWAS-based machine learning approach to predict duloxetine response in major depressive disorder. J. Psychiatr. Res. 2018, 99, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, C.; Corponi, F.; Albani, D.; Raimondi, I.; Forloni, G.; Schruers, K.; Kasper, S.; Kautzky, A.; Zohar, J.; Souery, D.; et al. Pleiotropic genes in psychiatry: Calcium channels and the stress-related FKBP5 gene in antidepressant resistance. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2018, 81, 203–210. [Google Scholar] [CrossRef]
- Shah, J.; Liu, S.; Yu, W. Contemporary antiplatelet therapy for secondary stroke prevention: A narrative review of current literature and guidelines. Stroke Vasc. Neurol. 2022; ahead of print. [Google Scholar] [CrossRef]
- Agayeva, N.; Gungor, L.; Topcuoglu, M.A.; Arsava, E.M. Pathophysiologic, rather than laboratory-defined resistance drives aspirin failure in ischemic stroke. J. Stroke Cerebrovasc. Dis. 2015, 24, 745–750. [Google Scholar] [CrossRef]
- Rocca, B.; Petrucci, G. Variability in the Responsiveness to low-dose aspirin: Pharmacological and disease-related mechanisms. Thrombosis 2012, 2012, 376721. [Google Scholar] [CrossRef]
- Gonzalez-Conejero, R.; Rivera, J.; Corral, J.; Acuña, C.; Guerrero, J.A.; Vicente, V. Biological assessment of aspirin efficacy on healthy individuals: Heterogeneous response or aspirin failure? Stroke 2005, 36, 276–280. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).