Targeting Mitochondrial Function with Chemoptogenetics
Abstract
:1. Mitochondria, OXPHOS and ROS Production
2. MtDNA Packaging and Repair
3. Overview
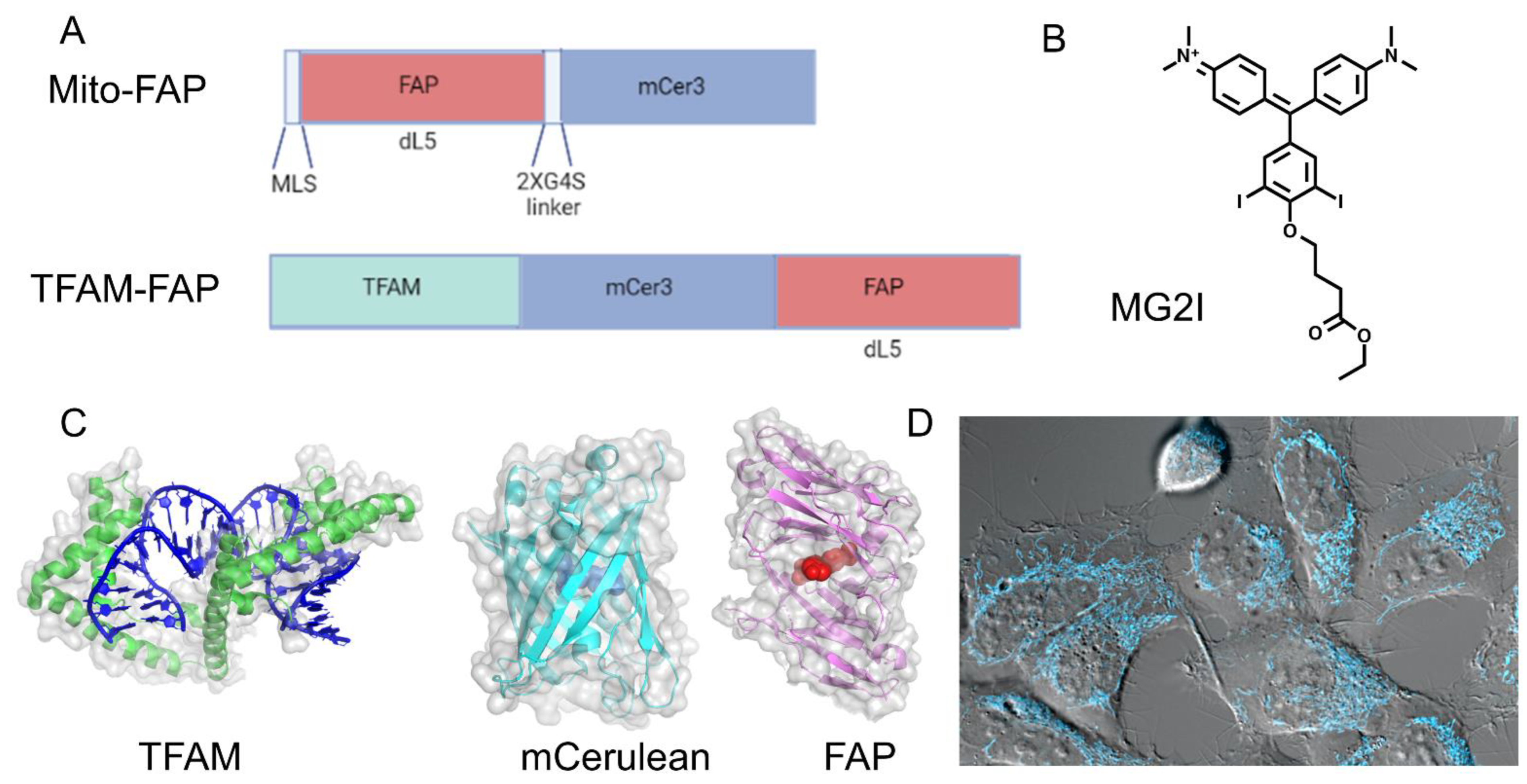
4. Chemoptogenetics and Mitochondrial Injury
5. Chemoptogenetics and MtDNA
6. MtDNA Mutations and Pathologies
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fleming, A.M.; Burrows, C.J. On the irrelevancy of hydroxyl radical to DNA damage from oxidative stress and implications for epigenetics. Chem. Soc. Rev. 2020, 49, 6524–6528. [Google Scholar] [CrossRef]
- Rong, Z.; Tu, P.; Xu, P.; Sun, Y.; Yu, F.; Tu, N.; Guo, L.; Yang, Y. The Mitochondrial Response to DNA Damage. Front. Cell Dev. Biol. 2021, 9, 669379. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Tigano, M.; Sfeir, A. Safeguarding mitochondrial genomes in higher eukaryotes. Nat. Struct. Mol. Biol. 2020, 27, 687–695. [Google Scholar] [CrossRef]
- Tann, A.W.; Boldogh, I.; Meiss, G.; Qian, W.; Van Houten, B.; Mitra, S.; Szczesny, B. Apoptosis induced by persistent single-strand breaks in mitochondrial genome: Critical role of EXOG (5′-EXO/endonuclease) in their repair. J. Biol. Chem. 2011, 286, 31975–31983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simsek, D.; Furda, A.; Gao, Y.; Artus, J.; Brunet, E.; Hadjantonakis, A.K.; Van Houten, B.; Shuman, S.; McKinnon, P.J.; Jasin, M. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature 2011, 471, 245–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaufman, B.A.; Van Houten, B. POLB: A new role of DNA polymerase beta in mitochondrial base excision repair. DNA Repair 2017, 60, A1–A5. [Google Scholar] [CrossRef] [PubMed]
- de Souza-Pinto, N.C.; Eide, L.; Hogue, B.A.; Thybo, T.; Stevnsner, T.; Seeberg, E.; Klungland, A.; Bohr, V.A. Repair of 8-oxodeoxyguanosine lesions in mitochondrial dna depends on the oxoguanine dna glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial dna of OGG1-defective mice. Cancer Res. 2001, 61, 5378–5381. [Google Scholar]
- Yuan, Y.; Ju, Y.S.; Kim, Y.; Li, J.; Wang, Y.; Yoon, C.J.; Yang, Y.; Martincorena, I.; Creighton, C.J.; Weinstein, J.N.; et al. Comprehensive molecular characterization of mitochondrial genomes in human cancers. Nat. Genet. 2020, 52, 342–352. [Google Scholar] [CrossRef] [Green Version]
- Van Remmen, H.; Williams, M.D.; Guo, Z.; Estlack, L.; Yang, H.; Carlson, E.J.; Epstein, C.J.; Huang, T.T.; Richardson, A. Knockout mice heterozygous for Sod2 show alterations in cardiac mitochondrial function and apoptosis. Am. J. Physiol.-Heart Circ. Physiol. 2001, 281, H1422–H1432. [Google Scholar] [CrossRef] [Green Version]
- Yakes, F.M.; Van Houten, B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl. Acad. Sci. USA 1997, 94, 514–519. [Google Scholar] [CrossRef] [Green Version]
- Salazar, J.J.; Van Houten, B. Preferential mitochondrial DNA injury caused by glucose oxidase as a steady generator of hydrogen peroxide in human fibroblasts. Mutat. Res. 1997, 385, 139–149. [Google Scholar] [CrossRef]
- Santos, J.H.; Hunakova, L.; Chen, Y.; Bortner, C.; Van Houten, B. Cell sorting experiments link persistent mitochondrial DNA damage with loss of mitochondrial membrane potential and apoptotic cell death. J. Biol. Chem. 2003, 278, 1728–1734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furda, A.M.; Marrangoni, A.M.; Lokshin, A.; Van Houten, B. Oxidants and not alkylating agents induce rapid mtDNA loss and mitochondrial dysfunction. DNA Repair 2012, 11, 684–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wisnovsky, S.; Jean, S.R.; Kelley, S.O. Mitochondrial DNA repair and replication proteins revealed by targeted chemical probes. Nat. Chem. Biol. 2016, 12, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Cleaver, J.E.; Brennan-Minnella, A.M.; Swanson, R.A.; Fong, K.W.; Chen, J.; Chou, K.M.; Chen, Y.W.; Revet, I.; Bezrookove, V. Mitochondrial reactive oxygen species are scavenged by Cockayne syndrome B protein in human fibroblasts without nuclear DNA damage. Proc. Natl. Acad. Sci. USA 2014, 111, 13487–13492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Treberg, J.R.; Quinlan, C.L.; Brand, M.D. Evidence for two sites of superoxide production by mitochondrial NADH-ubiquinone oxidoreductase (complex I). J. Biol. Chem. 2011, 286, 27103–27110. [Google Scholar] [CrossRef] [Green Version]
- Przedborski, S.; Jackson-Lewis, V.; Djaldetti, R.; Liberatore, G.; Vila, M.; Vukosavic, S.; Almer, G. The parkinsonian toxin MPTP: Action and mechanism. Restor. Neurol. Neurosci. 2000, 16, 135–142. [Google Scholar]
- Mandavilli, B.S.; Ali, S.F.; Van Houten, B. DNA damage in brain mitochondria caused by aging and MPTP treatment. Brain Res. 2000, 885, 45–52. [Google Scholar] [CrossRef]
- Rodriguez-Nuevo, A.; Torres-Sanchez, A.; Duran, J.M.; De Guirior, C.; Martinez-Zamora, M.A.; Boke, E. Oocytes maintain ROS-free mitochondrial metabolism by suppressing complex I. Nature 2022, 607, 756–761. [Google Scholar] [CrossRef]
- Wisnovsky, S.; Sack, T.; Pagliarini, D.J.; Laposa, R.R.; Kelley, S.O. DNA Polymerase theta Increases Mutational Rates in Mitochondrial DNA. ACS Chem. Biol. 2018, 13, 900–908. [Google Scholar] [CrossRef]
- Horton, K.L.; Stewart, K.M.; Fonseca, S.B.; Guo, Q.; Kelley, S.O. Mitochondria-penetrating peptides. Chem. Biol. 2008, 15, 375–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chamberlain, G.R.; Tulumello, D.V.; Kelley, S.O. Targeted delivery of doxorubicin to mitochondria. ACS Chem. Biol. 2013, 8, 1389–1395. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, S.B.; Pereira, M.P.; Mourtada, R.; Gronda, M.; Horton, K.L.; Hurren, R.; Minden, M.D.; Schimmer, A.D.; Kelley, S.O. Rerouting chlorambucil to mitochondria combats drug deactivation and resistance in cancer cells. Chem. Biol. 2011, 18, 445–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahon, K.P.; Potocky, T.B.; Blair, D.; Roy, M.D.; Stewart, K.M.; Chiles, T.C.; Kelley, S.O. Deconvolution of the cellular oxidative stress response with organelle-specific Peptide conjugates. Chem. Biol. 2007, 14, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Ross, M.F.; Filipovska, A.; Smith, R.A.; Gait, M.J.; Murphy, M.P. Cell-penetrating peptides do not cross mitochondrial membranes even when conjugated to a lipophilic cation: Evidence against direct passage through phospholipid bilayers. Biochem. J. 2004, 383 Pt 3, 457–468. [Google Scholar] [CrossRef] [Green Version]
- Ross, M.F.; Murphy, M.P. Cell-penetrating peptides are excluded from the mitochondrial matrix. Biochem. Soc. Trans. 2004, 32 Pt 6, 1072–1074. [Google Scholar] [CrossRef]
- Lovell, J.F.; Liu, T.W.; Chen, J.; Zheng, G. Activatable photosensitizers for imaging and therapy. Chem. Rev. 2010, 110, 2839–2857. [Google Scholar] [CrossRef]
- He, J.; Wang, Y.; Missinato, M.A.; Onuoha, E.; Perkins, L.A.; Watkins, S.C.; St Croix, C.M.; Tsang, M.; Bruchez, M.P. A genetically targetable near-infrared photosensitizer. Nat. Methods 2016, 13, 263–268. [Google Scholar] [CrossRef]
- Shibuya, T.; Tsujimoto, Y. Deleterious effects of mitochondrial ROS generated by KillerRed photodynamic action in human cell lines and C. elegans. J. Photochem. Photobiol. B: Biol. 2012, 117, 1–12. [Google Scholar] [CrossRef]
- Lee, A.; Mathuru, A.S.; Teh, C.; Kibat, C.; Korzh, V.; Penney, T.B.; Jesuthasan, S. The habenula prevents helpless behavior in larval zebrafish. Curr. Biol. 2010, 20, 2211–2216. [Google Scholar] [CrossRef] [Green Version]
- Teh, C.; Chudakov, D.M.; Poon, K.L.; Mamedov, I.Z.; Sek, J.Y.; Shidlovsky, K.; Lukyanov, S.; Korzh, V. Optogenetic in vivo cell manipulation in KillerRed-expressing zebrafish transgenics. BMC Dev. Biol. 2010, 10, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.Y.; Yang, W.Y. Spatiotemporally controlled initiation of Parkin-mediated mitophagy within single cells. Autophagy 2011, 7, 1230–1238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, S.; Chisholm, A.D. Highly efficient optogenetic cell ablation in C. elegans using membrane-targeted miniSOG. Sci. Rep. 2016, 6, 21271. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Kumar, N.; Roginskaya, V.; Fouquerel, E.; Opresko, P.L.; Shiva, S.; Watkins, S.C.; Kolodieznyi, D.; Bruchez, M.P.; Van Houten, B. Chemoptogenetic damage to mitochondria causes rapid telomere dysfunction. Proc. Natl. Acad. Sci. USA 2019, 116, 18435–18444. [Google Scholar] [CrossRef] [Green Version]
- Henle, E.S.; Han, Z.; Tang, N.; Rai, P.; Luo, Y.; Linn, S. Sequence-specific DNA cleavage by Fe2+-mediated fenton reactions has possible biological implications. J. Biol. Chem. 1999, 274, 962–971. [Google Scholar] [CrossRef] [Green Version]
- Patel, J.; Baptiste, B.A.; Kim, E.; Hussain, M.; Croteau, D.L.; Bohr, V.A. DNA damage and mitochondria in cancer and aging. Carcinogenesis 2020, 41, 1625–1634. [Google Scholar] [CrossRef]
- Xie, W.; Jiao, B.; Bai, Q.; Ilin, V.A.; Sun, M.; Burton, C.E.; Kolodieznyi, D.; Calderon, M.J.; Stolz, D.B.; Opresko, P.L.; et al. Chemoptogenetic ablation of neuronal mitochondria in vivo with spatiotemporal precision and controllable severity. eLife 2020, 9, e51845. [Google Scholar] [CrossRef]
- Garrido, N.; Griparic, L.; Jokitalo, E.; Wartiovaara, J.; van der Bliek, A.M.; Spelbrink, J.N. Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell 2003, 14, 1583–1596. [Google Scholar] [CrossRef] [Green Version]
- Legros, F.; Malka, F.; Frachon, P.; Lombes, A.; Rojo, M. Organization and dynamics of human mitochondrial DNA. J. Cell Sci. 2004, 117 Pt 13, 2653–2662. [Google Scholar] [CrossRef] [Green Version]
- Bender, A.; Krishnan, K.J.; Morris, C.M.; Taylor, G.A.; Reeve, A.K.; Perry, R.H.; Jaros, E.; Hersheson, J.S.; Betts, J.; Klopstock, T.; et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat. Genet. 2006, 38, 515–517. [Google Scholar] [CrossRef]
- Egensperger, R.; Kosel, S.; Schnopp, N.M.; Mehraein, P.; Graeber, M.B. Association of the mitochondrial tRNA(A4336G) mutation with Alzheimer’s and Parkinson’s diseases. Neuropathol. Appl. Neurobiol. 1997, 23, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Castellani, R.; Hirai, K.; Aliev, G.; Drew, K.L.; Nunomura, A.; Takeda, A.; Cash, A.D.; Obrenovich, M.E.; Perry, G.; Smith, M.A. Role of mitochondrial dysfunction in Alzheimer’s disease. J. Neurosci. Res. 2002, 70, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Cottrell, D.A.; Borthwick, G.M.; Johnson, M.A.; Ince, P.G.; Turnbull, D.M. The role of cytochrome c oxidase deficient hippocampal neurones in Alzheimer’s disease. Neuropathol. Appl. Neurobiol. 2002, 28, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Coskun, P.E.; Beal, M.F.; Wallace, D.C. Alzheimer’s brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc. Natl. Acad. Sci. USA 2004, 101, 10726–10731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, L.; Soares, P.; Maximo, V.; Samuels, D.C. Somatic mitochondrial DNA mutations in cancer escape purifying selection and high pathogenicity mutations lead to the oncocytic phenotype: Pathogenicity analysis of reported somatic mtDNA mutations in tumors. BMC Cancer 2012, 12, 53. [Google Scholar] [CrossRef]
- Wallace, D.C. Mitochondria and cancer. Nat. Rev. Cancer 2012, 12, 685–698. [Google Scholar] [CrossRef] [Green Version]
- Ishikawa, K.; Hayashi, J. Trading mtDNA uncovers its role in metastasis. Cell Adhes. Migr. 2009, 3, 11–13. [Google Scholar] [CrossRef] [Green Version]
- Ishikawa, K.; Takenaga, K.; Akimoto, M.; Koshikawa, N.; Yamaguchi, A.; Imanishi, H.; Nakada, K.; Honma, Y.; Hayashi, J. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science 2008, 320, 661–664. [Google Scholar] [CrossRef] [Green Version]
- Imanishi, H.; Hattori, K.; Wada, R.; Ishikawa, K.; Fukuda, S.; Takenaga, K.; Nakada, K.; Hayashi, J. Mitochondrial DNA mutations regulate metastasis of human breast cancer cells. PLoS ONE 2011, 6, e23401. [Google Scholar] [CrossRef] [Green Version]
- Ishikawa, K.; Imanishi, H.; Takenaga, K.; Hayashi, J. Regulation of metastasis; mitochondrial DNA mutations have appeared on stage. J. Bioenerg. Biomembr. 2012, 44, 639–644. [Google Scholar] [CrossRef]
- Santidrian, A.F.; Matsuno-Yagi, A.; Ritland, M.; Seo, B.B.; LeBoeuf, S.E.; Gay, L.J.; Yagi, T.; Felding-Habermann, B. Mitochondrial complex I activity and NAD+/NADH balance regulate breast cancer progression. J. Clin. Investig. 2013, 123, 1068–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Y.; Wang, W.; Li, H.; Yu, Y.; Tao, J.; Huang, S.; Zeng, Z. Nonsense and missense mutation of mitochondrial ND6 gene promotes cell migration and invasion in human lung adenocarcinoma. BMC Cancer 2015, 15, 346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papa, L.; Germain, D. SirT3 regulates the mitochondrial unfolded protein response. Mol. Cell Biol. 2014, 34, 699–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gitschlag, B.L.; Kirby, C.S.; Samuels, D.C.; Gangula, R.D.; Mallal, S.A.; Patel, M.R. Homeostatic Responses Regulate Selfish Mitochondrial Genome Dynamics in C. elegans. Cell Metab. 2016, 24, 91–103. [Google Scholar] [CrossRef] [Green Version]
- Kenny, T.C.; Germain, D. mtDNA, Metastasis, and the Mitochondrial Unfolded Protein Response (UPR(mt)). Front. Cell Dev. Biol. 2017, 5, 37. [Google Scholar] [CrossRef] [Green Version]
- Filograna, R.; Mennuni, M.; Alsina, D.; Larsson, N.G. Mitochondrial DNA copy number in human disease: The more the better? FEBS Lett. 2021, 595, 976–1002. [Google Scholar] [CrossRef]
- Dolle, C.; Flones, I.; Nido, G.S.; Miletic, H.; Osuagwu, N.; Kristoffersen, S.; Lilleng, P.K.; Larsen, J.P.; Tysnes, O.B.; Haugarvoll, K.; et al. Defective mitochondrial DNA homeostasis in the substantia nigra in Parkinson disease. Nat. Commun. 2016, 7, 13548. [Google Scholar] [CrossRef] [Green Version]
- Grunewald, A.; Rygiel, K.A.; Hepplewhite, P.D.; Morris, C.M.; Picard, M.; Turnbull, D.M. Mitochondrial DNA Depletion in Respiratory Chain-Deficient Parkinson Disease Neurons. Ann. Neurol. 2016, 79, 366–378. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Santiago, B.; Casademont, J.; Nunes, V. Is mitochondrial DNA depletion involved in Alzheimer’s disease? Eur. J. Hum. Genet. 2001, 9, 279–285. [Google Scholar] [CrossRef]
- Leon, J.; Sakumi, K.; Castillo, E.; Sheng, Z.; Oka, S.; Nakabeppu, Y. 8-Oxoguanine accumulation in mitochondrial DNA causes mitochondrial dysfunction and impairs neuritogenesis in cultured adult mouse cortical neurons under oxidative conditions. Sci. Rep. 2016, 6, 22086. [Google Scholar] [CrossRef] [Green Version]
- Torres-Gonzalez, M.; Gawlowski, T.; Kocalis, H.; Scott, B.T.; Dillmann, W.H. Mitochondrial 8-oxoguanine glycosylase decreases mitochondrial fragmentation and improves mitochondrial function in H9C2 cells under oxidative stress conditions. Am. J. Physiol. Cell Physiol. 2014, 306, C221–C229. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romesberg, A.; Van Houten, B. Targeting Mitochondrial Function with Chemoptogenetics. Biomedicines 2022, 10, 2459. https://doi.org/10.3390/biomedicines10102459
Romesberg A, Van Houten B. Targeting Mitochondrial Function with Chemoptogenetics. Biomedicines. 2022; 10(10):2459. https://doi.org/10.3390/biomedicines10102459
Chicago/Turabian StyleRomesberg, Amy, and Bennett Van Houten. 2022. "Targeting Mitochondrial Function with Chemoptogenetics" Biomedicines 10, no. 10: 2459. https://doi.org/10.3390/biomedicines10102459
APA StyleRomesberg, A., & Van Houten, B. (2022). Targeting Mitochondrial Function with Chemoptogenetics. Biomedicines, 10(10), 2459. https://doi.org/10.3390/biomedicines10102459





