Telomere-Associated Changes in Nuclear Architecture of Cancer-Associated Macrophage-like Cells in Liquid Biopsies from Melanoma Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Samples
2.2. May-Grunwald Giemsa Staining, CTCs and CAMLs Isolation
2.3. Cytologic Evaluation of CTCs and CAMLs
2.4. Double Immunocytochemistry (ICC)
2.5. Telomere Three-Dimensional Quantitative Fluorescent in Situ Hybridization (3D-QFISH)
2.6. Imaging & Analysis
2.7. Statistical Analysis
3. Results
3.1. Identification of Circulating Tumor Cells and Circulating Tumor Associated Cells
3.2. 3D Telomere Profiles Identify CAML Cells with Short Telomeres
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Komohara, Y.; Fujiwara, Y.; Ohnishi, K.; Takeya, M. Tumor-associated macrophages: Potential therapeutic targets for anti-cancer therapy. Adv. Drug Deliv. Rev. 2015, 99, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Komohara, Y.; Jinushi, M.; Takeya, M. Clinical significance of macrophage heterogeneity in human malignant tumors. Cancer Sci. 2013, 105, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.R.; Maute, R.L.; Dulken, B.W.; Hutter, G.; George, B.M.; McCracken, M.N.; Gupta, R.; Tsai, J.M.; Sinha, R.; Corey, D.; et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature 2017, 545, 495–499. [Google Scholar] [CrossRef] [PubMed]
- Katsuya, Y.; Horinouchi, H.; Asao, T.; Kitahara, S.; Goto, Y.; Kanda, S.; Fujiwara, Y.; Nokihara, H.; Yamamoto, N.; Watanabe, S.-I.; et al. Expression of programmed death 1 (PD-1) and its ligand (PD-L1) in thymic epithelial tumors: Impact on treatment efficacy and alteration in expression after chemotherapy. Lung Cancer 2016, 99, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.W.; Liu, L.; Gong, C.Y.; Shi, H.S.; Zeng, Y.H.; Wang, X.Z.; Zhao, Y.W.; Wei, Y.Q. Prognostic significance of tumor-associated macrophages in solid tumor: A meta-analysis of the literature. PLoS ONE 2012, 7, e50946. [Google Scholar] [CrossRef]
- Zeisberger, S.M.; Odermatt, B.; Marty, C.; Zehnder-Fjällman, A.H.; Ballmer-Hofer, K.; Schwendener, R.A. Clodronate-liposome-mediated depletion of tumour-associated macrophages: A new and highly effective antiangiogenic therapy approach. Br. J. Cancer 2006, 95, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Fritz, J.M.; Tennis, M.A.; Orlicky, D.J.; Lin, H.; Ju, C.; Redente, E.F.; Choo, K.S.; Staab, T.A.; Bouchard, R.J.; Merrick, D.T.; et al. Depletion of tumor-associated macrophages slows the growth of chemically induced mouse lung adenocarcinomas. Front. Immunol. 2014, 5, 587. [Google Scholar] [CrossRef]
- Heusinkveld, M.; van der Burg, S.H. Identification and manipulation of tumor associated macrophages in human cancers. J. Transl. Med. 2011, 16, 216. [Google Scholar] [CrossRef]
- Marsavela, G.; Aya-Bonilla, C.A.; Warkiani, M.E.; Gray, E.S.; Ziman, M. Melanoma circulating tumor cells: Benefits and challenges required for clinical application. Cancer Lett. 2018, 424, 1–8. [Google Scholar] [CrossRef]
- Wark, L.; Quon, H.; Ong, A.; Drachenberg, D.; Rangel-Pozzo, A.; Mai, S. Long-Term Dynamics of Three Dimensional Telomere Profiles in Circulating Tumor Cells in High-Risk Prostate Cancer Patients Undergoing Androgen-Deprivation and Radiation Therapy. Cancers 2019, 11, 1165. [Google Scholar] [CrossRef] [Green Version]
- Balch, C.M. Cutaneous Melanoma, 5th ed.; Quality Medical Pub.: St. Louis, MO, USA, 2009. [Google Scholar]
- Yanagita, M.; Luke, J.J.; Hodi, F.S.; Jänne, P.A.; Paweletz, C.P. Isolation and characterization of circulating melanoma cells by size filtration and fluorescent in-situ hybridization. Melanoma Res. 2018, 28, 89–95. [Google Scholar] [CrossRef]
- Luke, J.J.; Flaherty, K.T.; Ribas, A.; Long, G.V. Targeted agents and immunotherapies: Optimizing outcomes in melanoma. Nat. Rev. Clin. Oncol. 2017, 14, 463–482. [Google Scholar] [CrossRef] [PubMed]
- O’Day, S.J.; Maio, M.; Chiarion-Sileni, V.; Gajewski, T.F.; Pehamberger, H.; Bondarenko, I.N.; Queirolo, P.; Lundgren, L.; Mikhailov, S.; Roman, L.; et al. Efficacy and safety of ipilimumab monotherapy in patients with pretreated advanced melanoma: A multicenter single-arm phase II study. Ann. Oncol. 2010, 21, 1712–1717. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Long, G.V.; Brady, B.; Dutriaux, C.; Maio, M.; Mortier, L.; Hassel, J.C.; Rutkowski, P.; McNeil, C.; Kalinka-Warzocha, E.; et al. Nivolumab in previously untreated melanoma without BRAF mutation. N. Engl. J. Med. 2015, 372, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, activity, and immune correlates of anti–pd-1 antibody in cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef] [PubMed]
- Rizos, H.; Menzies, A.M.; Pupo, G.M.; Carlino, M.S.; Fung, C.; Hyman, J.; Haydu, L.E.; Mijatov, B.; Becker, T.M.; Boyd, S.C.; et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: Spectrum and clinical impact. Clin. Cancer Res. 2014, 20, 1965–1977. [Google Scholar] [CrossRef] [PubMed]
- Roesch, A. Tumor heterogeneity and plasticity as elusive drivers for resistance to MAPK pathway inhibition in melanoma. Oncogene 2014, 34, 2951–2957. [Google Scholar] [CrossRef]
- Shi, H.; Hugo, W.; Kong, X.; Hong, A.; Koya, R.C.; Moriceau, G.; Chodon, T.; Guo, R.; Johnson, D.B.; Dahlman, K.B.; et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014, 4, 80–93. [Google Scholar] [CrossRef]
- Hamilton, G.; Rath, B. Circulating tumor cell interactions with macrophages: Implications for biology and treatment. Transl. Lung Cancer Res. 2017, 6, 418–430. [Google Scholar] [CrossRef]
- Hao, N.B.; Lu, M.H.; Fan, Y.H.; Cao, Y.L.; Zhang, Z.R.; Yang, S.M. Macrophages in tumor microenvironments and the progression of tumors. Clin. Dev. Immunol. 2012, 2012, 948098. [Google Scholar] [CrossRef] [Green Version]
- Medrek, C.; Ponten, F.; Jirstrom, K.; Leandersson, K. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer 2012, 12, 306. [Google Scholar] [CrossRef]
- Kerbel, R.S.; Lagarde, A.E.; Dennis, J.W.; Donaghue, T.P. Spontaneous fusion in vivo between normal host and tumor cells: Possible contribution to tumor progression and metastasis studied with a lectin-resistant mutant tumor. Mol. Cell. Biol. 1983, 3, 523–538. [Google Scholar] [PubMed]
- Chakraborty, A.K.; Sodi, S.; Rachkovsky, M.; Kolesnikova, N.; Platt, J.T.; Bolognia, J.L.; Pawelek, J.M. A spontaneous murine melanoma lung metastasis comprised of host x tumor hybrids. Cancer Res. 2000, 60, 2512–2519. [Google Scholar] [PubMed]
- Rachkovsky, M.; Sodi, S.; Chakraborty, A.; Avissar, Y.; Bolognia, J.; McNiff, J.M.; Platt, J.; Bermudes, D.; Pawelek, J. Melanoma x macrophage hybrids with enhanced metastatic potential. Clin. Exp. Metastasis 1998, 16, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, N.; Yu, X.; Zhang, X.; Li, S.; Lei, Z.; Hu, R.; Li, H.; Mao, Y.; Wang, X.; et al. Tumacrophage: Macrophages transformed into tumor stem-like cells by virulent genetic material from tumor cells. Oncotarget 2017, 8, 82326–82343. [Google Scholar] [CrossRef] [PubMed]
- Rangel-Pozzo, A.; Corrêa de Souza, D.; Schmid-Braz, A.T.; de Azambuja, A.P.; Ferraz-Aguiar, T.; Borgonovo, T.; Mai, S. 3D Telomere Structure Analysis to Detect Genomic Instability and Cytogenetic Evolution in Myelodysplastic Syndromes. Cells 2019, 8, 304. [Google Scholar] [CrossRef]
- Drachenberg, D.; Awe, J.A.; Rangel-Pozzo, A.; Saranchuk, J.; Mai, S. Advancing Risk Assessment of Intermediate Risk Prostate Cancer Patients. Cancers 2019, 11, 855. [Google Scholar] [CrossRef]
- Rachakonda, S.; Srinivas, N.; Mahmoudpour, S.H.; Garcia-Casado, Z.; Requena, C.; Traves, V.; Soriano, V.; Cardelli, M.; Pjanova, D.; Molven, A.; et al. Telomere length and survival in primary cutaneous melanoma patients. Sci. Rep. 2018, 8, 10947. [Google Scholar] [CrossRef]
- Adams, D.L.; Martin, S.S.; Alpaugh, R.K.; Charpentier, M.; Tsai, S.; Bergan, R.C.; Ogden, I.M.; Catalona, W.; Chumsri, S.; Tang, C.M.; et al. Circulating giant macrophages as a potential biomarker of solid tumors. Proc. Natl. Acad. Sci. USA 2014, 111, 3514–3519. [Google Scholar] [CrossRef]
- Mu, Z.; Wang, C.; Ye, Z.; Rossi, G.; Sun, C.; Li, L.; Zhu, Z.; Yang, H.; Cristofanilli, M. Prognostic values of cancer associated macrophage-like cells (CAML) enumeration in metastatic breast cancer. Breast Cancer Res. Treat. 2017, 165, 733–741. [Google Scholar] [CrossRef]
- Adebayo Awe, J.; Xu, M.C.; Wechsler, J.; Benali-Furet, N.; Cayre, Y.E.; Saranchuk, J.; Drachenberg, D.; Mai, S. Three-Dimensional Telomeric Analysis of Isolated Circulating Tumor Cells (CTCs) Defines CTC Subpopulations. Transl. Oncol. 2013, 6, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Desitter, I.; Guerrouahen, B.S.; Benali-Furet, N.; Wechsler, J.; Jänne, P.A.; Kuang, Y.; Yanagita, M.; Wang, L.; Berkowitz, J.A.; Distel, R.J.; et al. A new device for rapid isolation by size and characterization of rare circulating tumor cells. Anticancer Res. 2011, 31, 427–441. [Google Scholar] [PubMed]
- Freidin, M.B.; Tay, A.; Freydina, D.V.; Chudasama, D.; Nicholson, A.G.; Rice, A.; Anikin, V.; Lim, E. An assessment of diagnostic performance of a filter-based antibody-independent peripheral blood circulating tumour cell capture paired with cytomorphologic criteria for the diagnosis of cancer. Lung Cancer 2014, 85, 182–185. [Google Scholar] [CrossRef]
- Sawabata, N.; Susaki, Y.; Nakamura, T.; Kawaguchi, T.; Yasukawa, M.; Taniguchi, S. Cluster circulating tumor cells in surgical cases of lung cancer. Gen. Thorac. Cardiovasc. Surg. 2020, 68, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, L.H.; Schuster, D.; Herz, H. Generalized approach for accelerated maximum likelihood based image restoration applied to three-dimensional fluorescence microscopy. J. Microsc. 2002, 204 Pt 2, 99–107. [Google Scholar] [CrossRef]
- Vermolen, B.J.; Garini, Y.; Mai, S.; Mougey, V.; Fest, T.; Chuang, T.C.; Chuang, A.Y.; Wark, L.; Young, I.T. Characterizing the three–dimensional organization of telomeres. Cytom. Part A 2005, 67, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.L.; Adams, D.K.; Alpaugh, R.K.; Cristofanilli, M.; Martin, S.S.; Chumsri, S.; Tang, C.M.; Marks, J.R. Circulating cancer-associated macrophage-like cells differentiate malignant breast cancer and benign breast conditions. Cancer Epidemiol. Biomark. Prev. 2016, 25, 1037–1042. [Google Scholar] [CrossRef]
- Mai, S. Genomic instability and circulating tumor cells in prostate cancer. Transl. Cancer Res. 2018, 7 (Suppl. 2), S192–S196. [Google Scholar] [CrossRef]
- Passerini, V.; Ozeri-Galai, E.; de Pagter, M.S.; Donnelly, N.; Schmalbrock, S.; Kloosterman, W.P.; Kerem, B.; Storchová, Z. The presence of extra chromosomes leads to genomic instability. Nat. Commun. 2016, 7, 10754. [Google Scholar] [CrossRef]
- Lin, Y.; Xu, J.; Lan, H. Tumor-associated macrophages in tumor metastasis: Biological roles and clinical therapeutic applications. J. Hematol. Oncol. 2019, 12, 76. [Google Scholar] [CrossRef]
- Reduzzi, C.; Vismara, M.; Gerratana, L.; Silvestri, M.; De Braud, F.; Raspagliesi, F.; Verzoni, E.; Di Cosimo, S.; Locati, L.D.; Cristofanilli, M.; et al. The curious phenomenon of dual-positive circulating cells: Longtime overlooked tumor cells. Semin. Cancer Biol. 2019, 60, 344–350. [Google Scholar] [CrossRef] [PubMed]

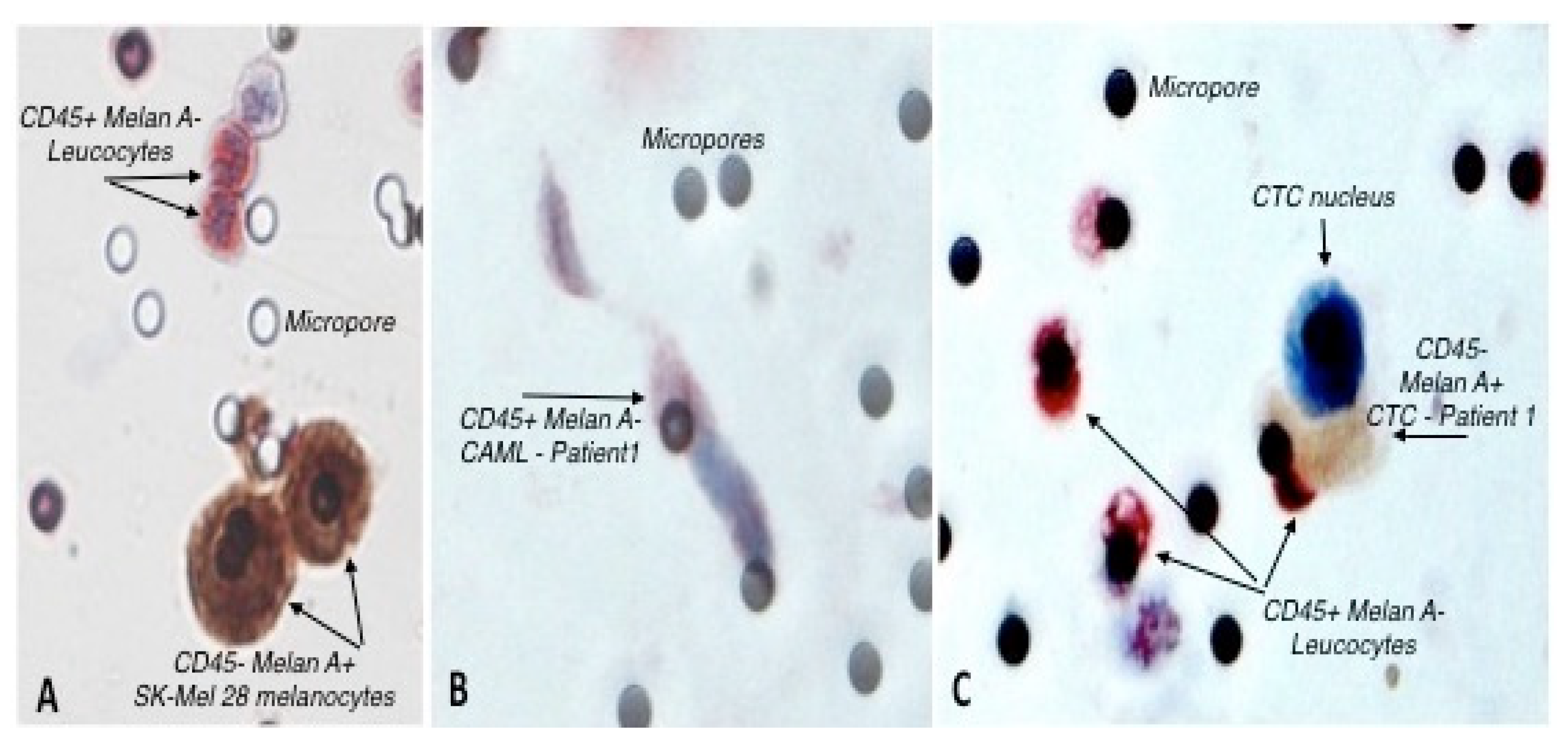
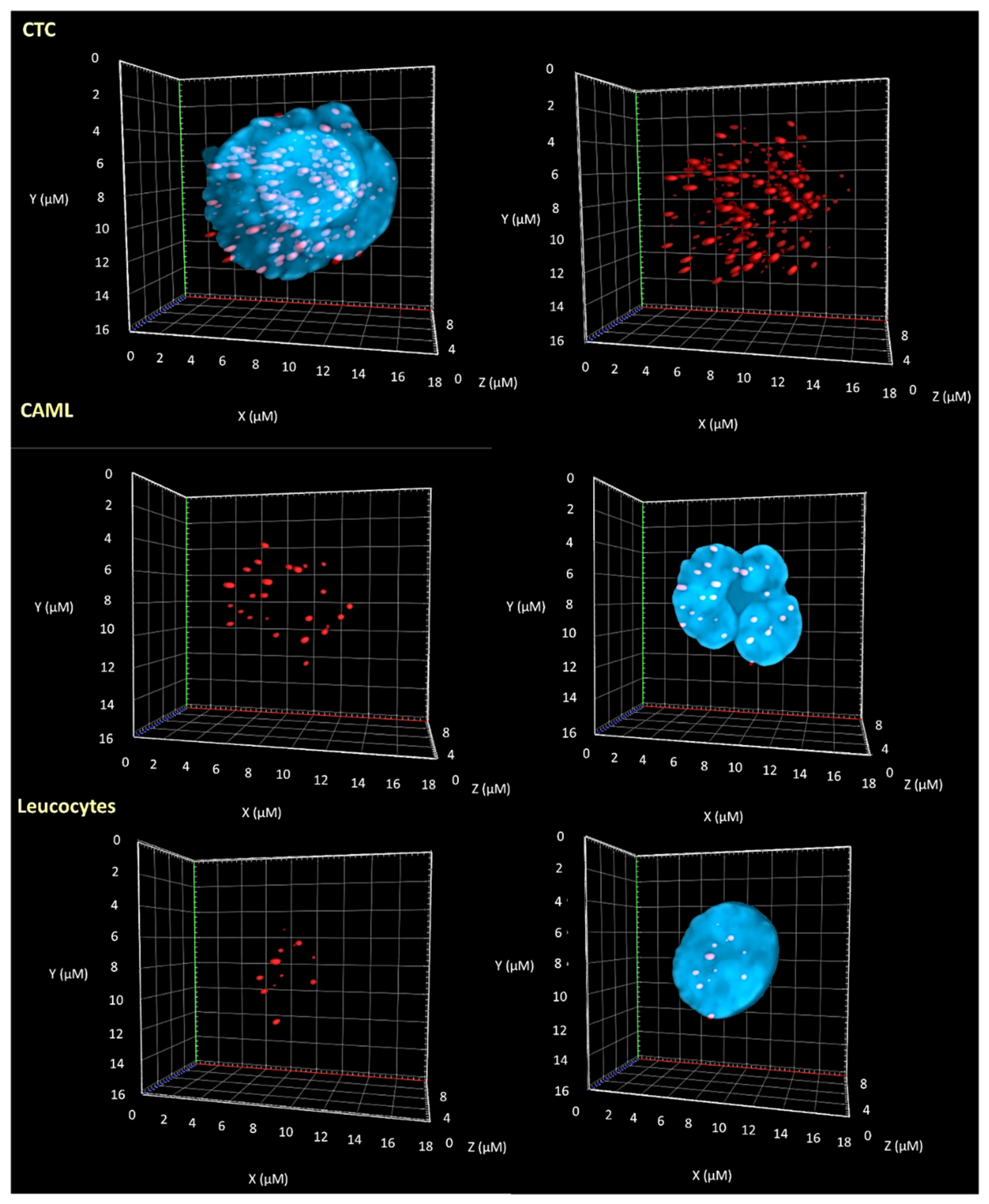

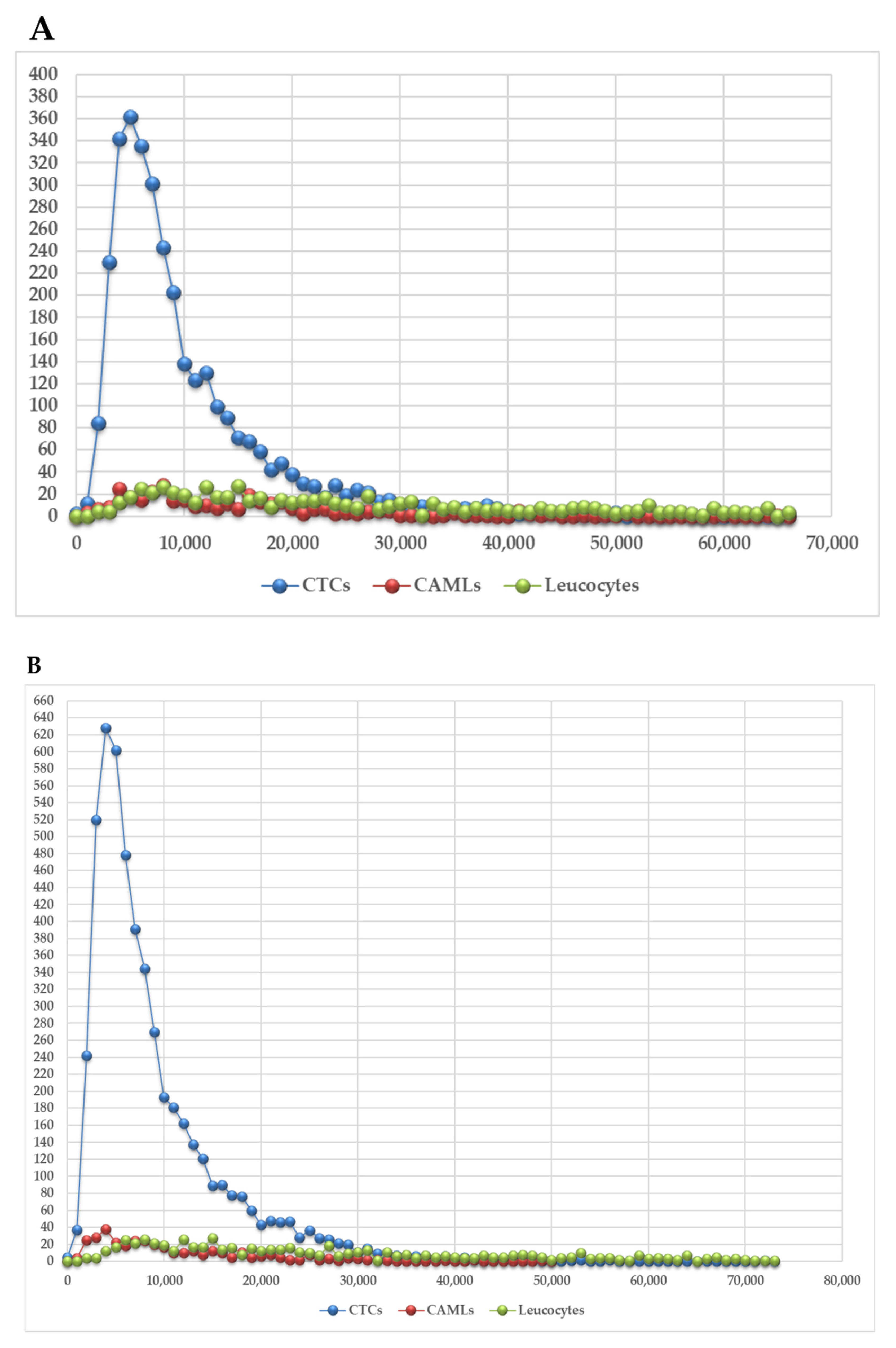
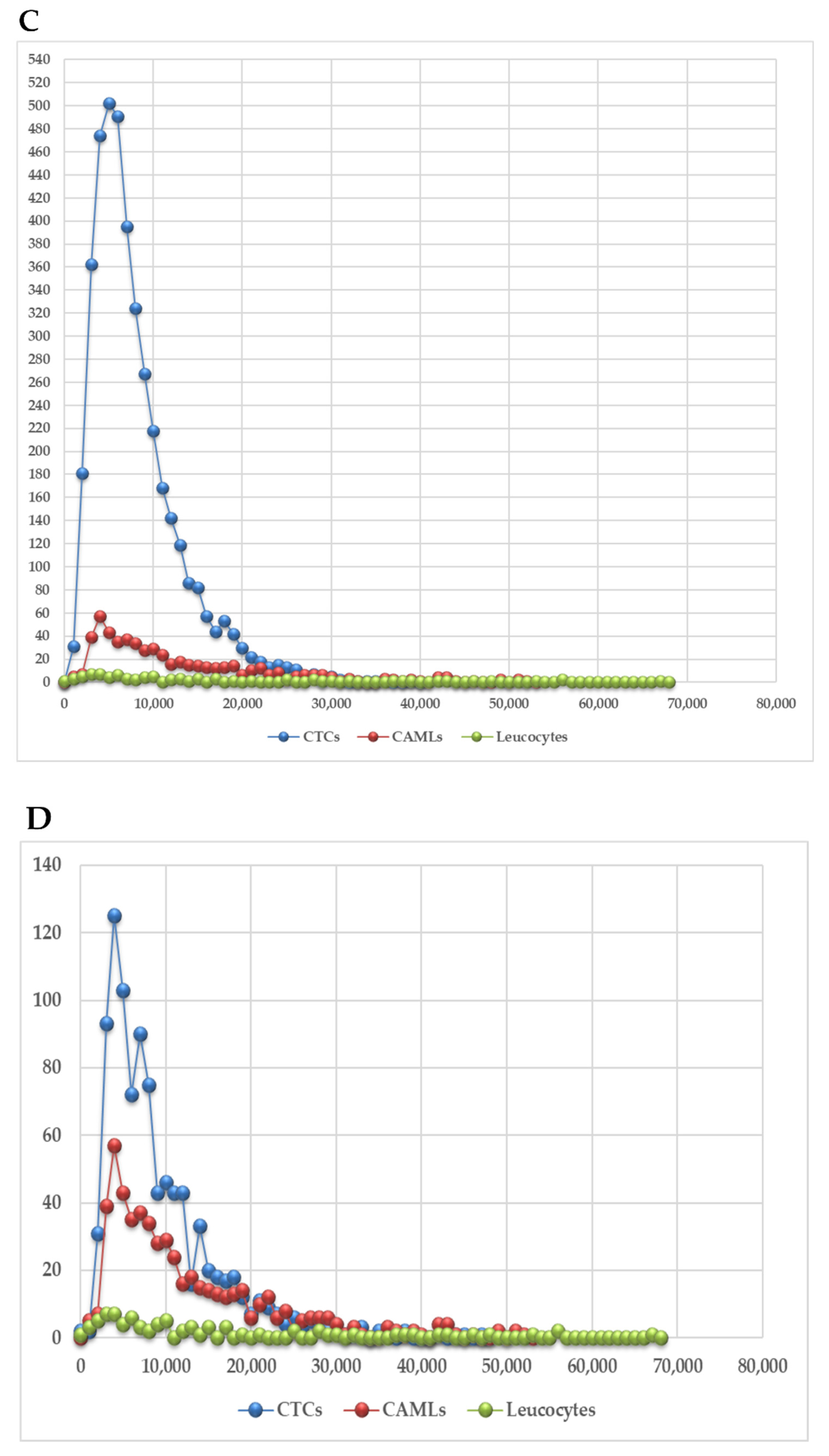
| Patient Number | Reference of Sample | Blood Sample Volume | CAML Nb | CTC Nb | CTC-Cluster Nb |
|---|---|---|---|---|---|
| 1 | VS-1-018 (ET3) 16AB0304 | 3 ml | 2 | 56 | 0 |
| 1.1 | VS-1-018_M1-3-17AA1313 | 3 ml | 7 | 62 | 4 |
| 2 | TM-1-019_M1- 16AB0258 | 3 ml | 2 | 11 | 0 |
| 3 | LM-1-039_CL2-17AA0828 | 3 ml | 1 | 33 | 0 |
| 4 | BJ-1-071_ET4-16AA8528 | 3 ml | 1 | 22 | 0 |
| 5 | DA-1-075_ET2-16AA7280 | 3 ml | 3 | 26 | 0 |
| 6 | GF-1-080_ET1-16AA7266 | 3 ml | 2 | 32 | 30 |
| 7 | LM-1-081_ET1-16AA7320 | 3 ml | 2 | 20 | 2 |
| 8 | AR-1-092_ET1-16AA2042 | 3 ml | 2 | 19 | 12 |
| 9 | KJ-1-084_ET1-17AA0845 | 3 ml | 2 | 12 | 0 |
| Patient Number | Reference of Sample | Telomere Profile |
|---|---|---|
| 1 | VS-1-018 (ET3) 16AB0304 | Profile 2 |
| 1.1 | VS-1-018_M1-3-17AA1313 | Profile 2 |
| 2 | TM-1-019_M1- 16AB0258 | Profile 2 |
| 3 | LM-1-039_CL2-17AA0828 | Profile 1 |
| 4 | BJ-1-071_ET4-16AA8528 | Profile 2 |
| 5 | DA-1-075_ET2-16AA7280 | Profile 2 |
| 6 | GF-1-080_ET1-16AA7266 | Profile 1 |
| 7 | LM-1-081_ET1-16AA7320 | Profile 1 |
| 8 | AR-1-092_ET1-16AA2042 | Profile 1 |
| 9 | KJ-1-084_ET1-17AA0845 | Profile 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rangel-Pozzo, A.; Wechsler, J.; Groult, J.; Da Meda, L.; Lebbe, C.; Mai, S. Telomere-Associated Changes in Nuclear Architecture of Cancer-Associated Macrophage-like Cells in Liquid Biopsies from Melanoma Patients. Biomedicines 2022, 10, 2391. https://doi.org/10.3390/biomedicines10102391
Rangel-Pozzo A, Wechsler J, Groult J, Da Meda L, Lebbe C, Mai S. Telomere-Associated Changes in Nuclear Architecture of Cancer-Associated Macrophage-like Cells in Liquid Biopsies from Melanoma Patients. Biomedicines. 2022; 10(10):2391. https://doi.org/10.3390/biomedicines10102391
Chicago/Turabian StyleRangel-Pozzo, Aline, Janine Wechsler, Jessica Groult, Laetitia Da Meda, Celeste Lebbe, and Sabine Mai. 2022. "Telomere-Associated Changes in Nuclear Architecture of Cancer-Associated Macrophage-like Cells in Liquid Biopsies from Melanoma Patients" Biomedicines 10, no. 10: 2391. https://doi.org/10.3390/biomedicines10102391






