The Oncogenic and Tumor Suppressive Long Non-Coding RNA–microRNA–Messenger RNA Regulatory Axes Identified by Analyzing Multiple Platform Omics Data from Cr(VI)-Transformed Cells and Their Implications in Lung Cancer
Abstract
1. Introduction
2. Material and Methods
2.1. Cell Culture and Microarray Analyses
2.2. Gene Set Enrichment (GSE) Analysis
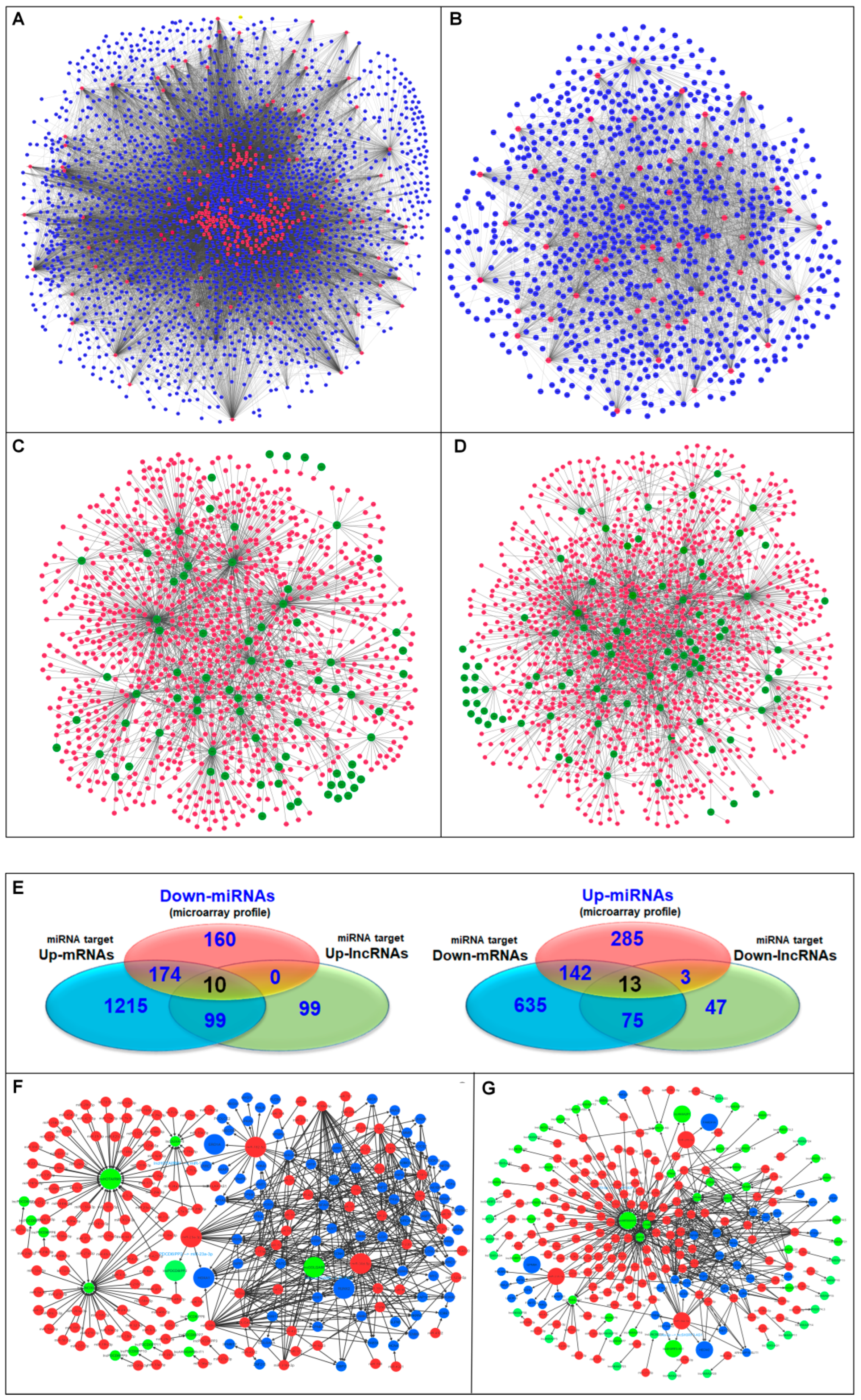
2.3. Construction of the lncRNA/miRNA/mRNA Regulatory ceRNA Module Network
2.4. Cancer Stemness Signature Analysis
2.5. The ceRNA Modules and Survival Analysis Using TCGA Datasets
2.6. The Receiver Operating Characteristic (ROC) Curve Analysis
3. Results
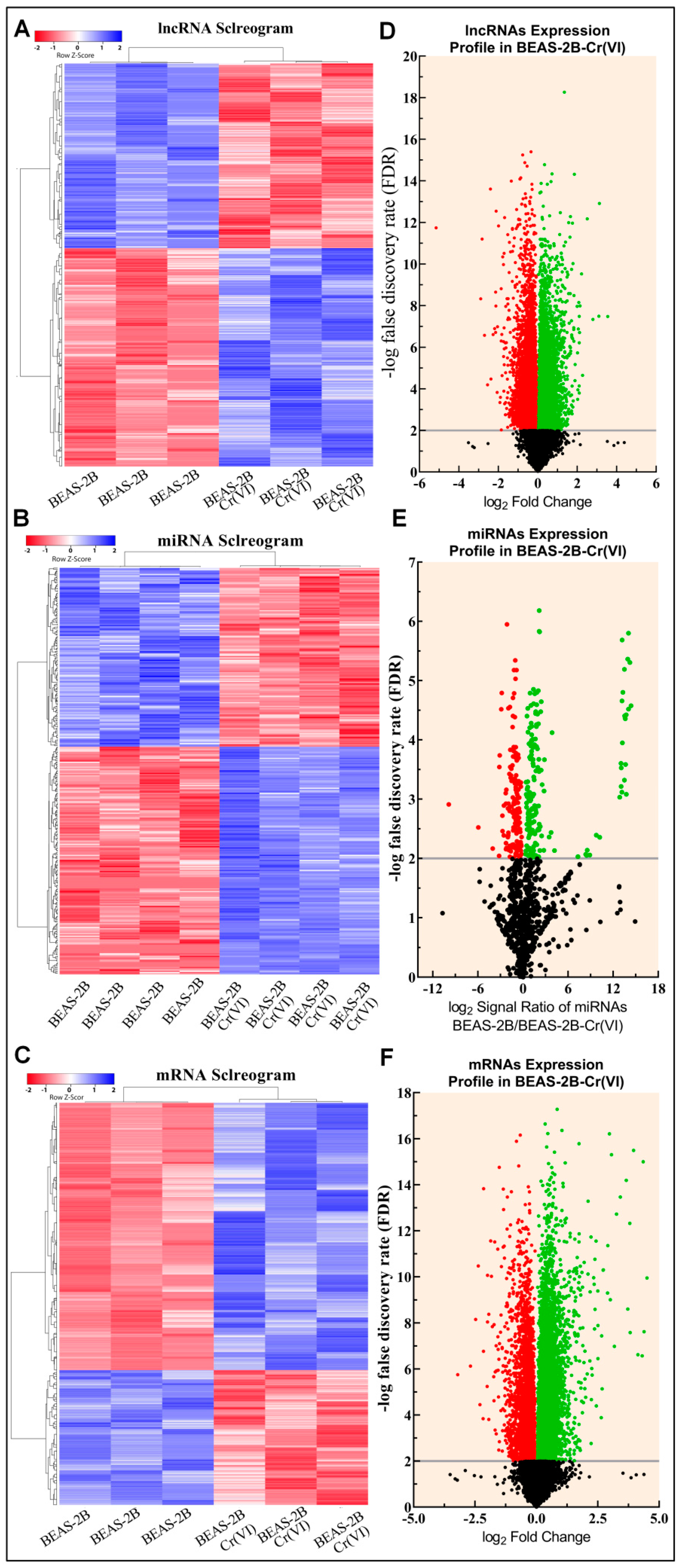
3.1. Microarray Gene Expression Profiling and Identification of DE of lncRNAs, miRNAs, and mRNAs
3.2. Functional Annotation Analysis of DE lncRNA, miRNA, and mRNA
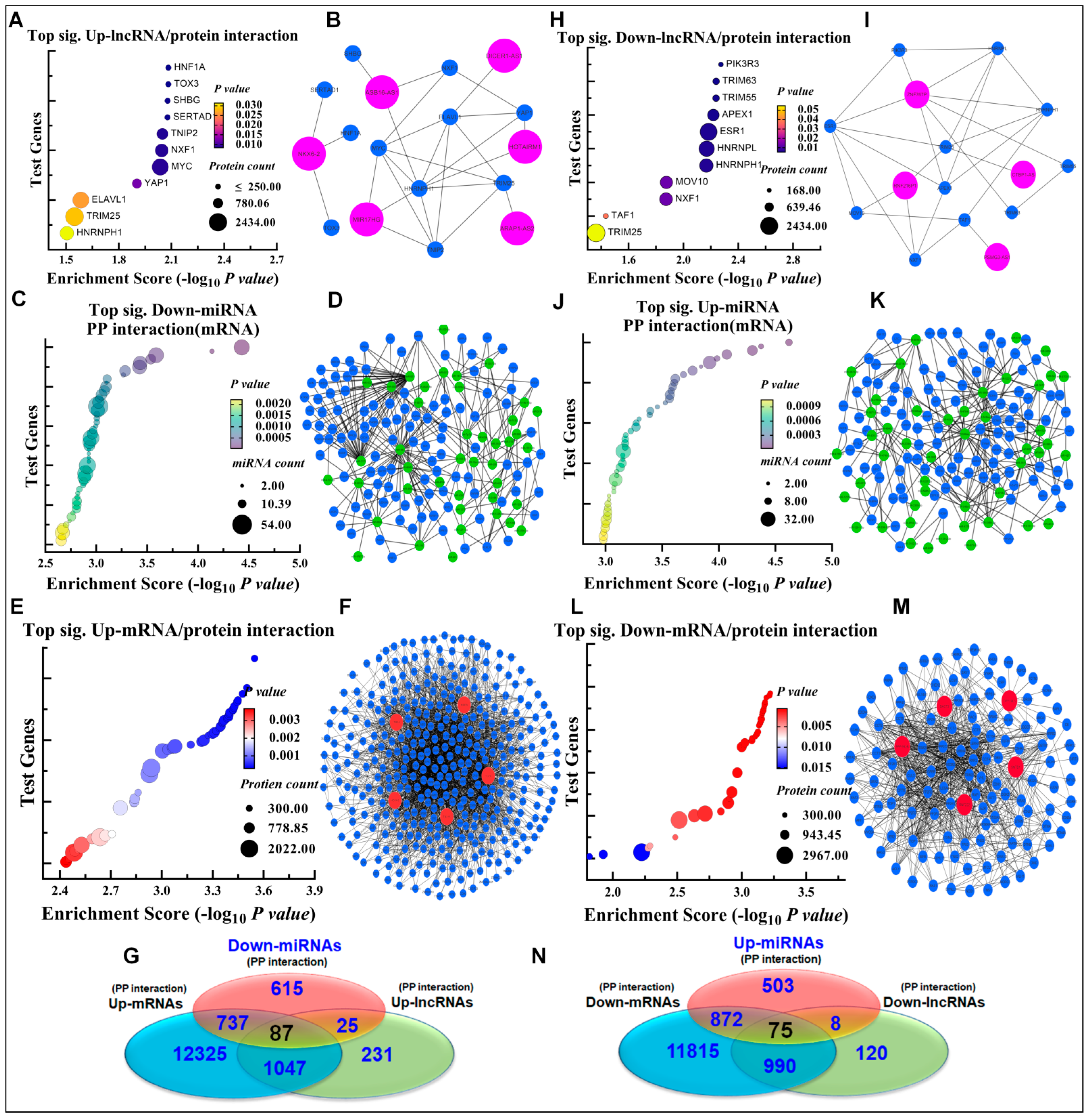
3.3. Oncogenic and Tumor Suppressive ceRNA Module–Protein Interactions
3.4. Identification of Oncogenic and Tumor Suppressor ceRNA Modules
3.5. Validation of the Oncogenic and Tumor Suppressor ceRNA Modules in Human Lung Cancer
3.6. Stemness Signature of ceRNA Modules
3.7. Prognostic Potential Rating of ceRNA Modules in Lung Cancer
3.8. Diagnostic Performance Assessment of Three ceRNA Modules in Lung Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chromium, I. Nickel, and welding. In IARC Monograph on the Evaluation of Carcinogenic Risks to Humans; World Health Organization: Lyon, France, 1990. [Google Scholar]
- ATSDR (Agency for Toxic Substances and Disease Research). Top 20 Hazardous Substances: ATSDR/EPA Priority List for 2017. U.S. Department of Health and Human Services Public Health Service/U.S. Environmental Protection Agency. 2019. Available online: https://www.atsdr.cdc.gov/SPL/index.html#2019spl (accessed on 10 June 2021).
- DesMarias, T.L.; Costa, M. Mechanisms of chromium-induced toxicity. Curr. Opin. Toxicol. 2019, 14, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Salnikow, K.; Zhitkovich, A. Genetic and epigenetic mechanisms in metal carcinogenesis and cocarcinogenesis: Nickel, arsenic, and chromium. Chem. Res. Toxicol. 2008, 21, 28–44. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, C. Metal carcinogen exposure induces cancer stem cell-like property through epigenetic reprograming: Anovel mechanism of metal carcinogenesis. Semin. Cancer Biol. 2019, 57, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, C. Epigenetic and epitranscriptomic mechanisms of chromium carcinogenesis. Adv. Pharmacol. 2022, 96, 1–25. [Google Scholar] [CrossRef]
- Wang, P.S.; Wang, Z.; Yang, C. Dysregulations of long non-coding RNAs- the emerging “lnc” in environmental carcinogenesis. Semin. Cancer Biol. 2021, 76, 163–172. [Google Scholar] [CrossRef]
- Humphries, B.; Wang, Z.; Yang, C. The role of microRNAs in metal carcinogen-induced cell malignant transformation and tumorigenesis. Food Chem. Toxicol. 2016, 98, 58–65. [Google Scholar] [CrossRef]
- Yan, H.; Bu, P. Non-coding RNA in cancer. Essays Biochem. 2021, 65, 625–639. [Google Scholar]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Ala, U. Competing endogenous RNAs and cancer: How coding and non-coding molecules cross-talk can impinge on disease. Int. J. Biochem. Cell Biol. 2021, 130, 105874. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhang, Y.; Zhang, X.; Dang, Y.; Cheng, Y.; Hua, W.; Teng, M.; Wang, S.; Lu, X. Novel lncRNA-miRNA-mRNA competing endogenous RNA triple networks associated programmed cell death in heart failure. Front. Cardiovasc. Med. 2021, 8, 747449. [Google Scholar] [CrossRef]
- Yin, J.; Zeng, X.; Ai, Z.; Yu, M.; Wu, Y.; Li, S. Construction and analysis of a lncRNA-miRNA-mRNA network based on competitive endogenous RNA reveal functional lncRNAs in oral cancer. BMC Med. Genom. 2020, 13, 84. [Google Scholar] [CrossRef] [PubMed]
- László, B.; Antal, L.; Gyöngyösi, E.; Szalmás, A.; Póliska, S.; Veress, G.; Kónya, J. Coordinated action of human papillomavirus type 16 E6 and E7 oncoproteins on competitive endogenous RNA (ceRNA) network members in primary human keratinocytes. BMC Cancer 2021, 21, 673. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Liu, J.B.; Deng, J.; Zou, D.Z.; Wu, J.J.; Cao, Y.H.; Yin, J.; Ma, Y.S.; Da, F.; Li, W. The role of ceRNA-mediated diagnosis and therapy in hepatocellular carcinoma. Hereditas 2021, 158, 44. [Google Scholar] [CrossRef] [PubMed]
- Poliseno, L.; Pandolfi, P.P. PTEN ceRNA networks in human cancer. Methods 2015, 77, 41–50. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, J.; Humphries, B.; Kondo, K.; Jiang, Y.; Shi, X.; Yang, C. Upregulation of histone-lysine methyltransferases plays a causal role in hexavalent chromium-induced cancer stem cell-like property and cell transformation. Toxicol. Appl. Pharmacol. 2018, 342, 22–30. [Google Scholar] [CrossRef]
- Yamanashi, K.; Okumura, N.; Takahashi, A.; Nakashima, T.; Matsuoka, T. Surgical and survival outcomes of lung cancer patients with intratumoral lung abscesses. J. Cardiothorac. Surg. 2017, 12, 1–6. [Google Scholar] [CrossRef]
- Goodall, G.J.; Wickramasinghe, V.O. RNA in cancer. Nat. Rev. Cancer 2021, 21, 22–36. [Google Scholar] [CrossRef]
- Zheng, Z.Q.; Li, Z.X.; Zhou, G.Q.; Lin, L.; Zhang, L.L.; Lv, J.W.; Huang, X.D.; Liu, R.Q.; Chen, F.; He, X.J.; et al. Long noncoding RNA FAM225A promotes nasopharyngeal carcinoma tumorigenesis and metastasis by acting as ceRNA to sponge miR-590–3p/miR-1275 and upregulate ITGB3. Cancer Res. 2019, 79, 4612–4626. [Google Scholar] [CrossRef]
- Sun, L.; Liu, L.; Yang, J.; Li, H.; Zhang, C. SATB1 3-UTR and lncRNA-UCA1 competitively bind to miR-495–3p and together regulate the proliferation and invasion of gastric cancer. J. Cell. Biochem. 2019, 120, 6671–6682. [Google Scholar] [CrossRef]
- Zhang, Y.; Han, P.; Guo, Q.; Hao, Y.; Qi, Y.; Xin, M.; Zhang, Y.; Cui, B.; Wang, P. Oncogenic landscape of somatic mutations perturbing pan-cancer lncRNA-ceRNA regulation. Front. Cell Dev. Biol. 2021, 9, 1193. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.H.; Wang, S.; Zhang, Y.; Huang, T.; Cai, Y.D. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS ONE 2017, 12, e0184129. [Google Scholar] [CrossRef] [PubMed]
- Silver, D.P.; Livingston, D.M. Mechanisms of BRCA1 tumor suppression. Cancer Discov. 2012, 2, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2017, 36, 1461–1473. [Google Scholar] [CrossRef] [PubMed]
- Sommer, G.; Heise, T. Role of the RNA-binding protein La in cancer pathobiology. RNA Biol. 2021, 18, 218–236. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, L.; Li, X.; Zhang, Y.; Xu, S.; Huang, X. Prediction of miRNA targets by learning from interaction sequences. PLoS ONE 2020, 15, e0232578. [Google Scholar] [CrossRef]
- Zhou, R.S.; Zhang, E.X.; Sun, Q.F.; Ye, Z.J.; Liu, J.W.; Zhou, D.H.; Tang, Y. Integrated analysis of lncRNA-miRNA-mRNA ceRNA network in squamous cell carcinoma of tongue. BMC Cancer 2019, 19, 779. [Google Scholar] [CrossRef]
- Xiong, F.; Yin, H.; Zhang, H.; Zhu, C.; Zhang, B.; Chen, S.; Ling, C.; Chen, X. Clinicopathologic features and the prognostic implications of long noncoding RNA HOTAIRM1 in non-small cell lung cancer. Genet. Test. Mol. Biomark. 2020, 24, 47–53. [Google Scholar] [CrossRef]
- Wu, X.; Wang, W.; Wu, G.; Peng, C.; Liu, J. miR-182–5p Serves as an Oncogene in Lung Adenocarcinoma through Binding to STARD13. Comput. Math. Methods Med. 2021, 2021, 7074343. [Google Scholar] [CrossRef]
- Yan, W.; Wang, X.; Liu, T.; Chen, L.; Han, L.; Xu, J.; Jin, G.; Harada, K.; Lin, Z.; Ren, X. Expression of endoplasmic reticulum oxidoreductase 1-α in cholangiocarcinoma tissues and its effects on the proliferation and migration of cholangiocarcinoma cells. Cancer Manag. Res. 2019, 11, 6727. [Google Scholar] [CrossRef]
- Chen, D.; Li, Y.; Wang, Y.; Xu, J. LncRNA HOTAIRM1 knockdown inhibits cell glycolysis metabolism and tumor progression by miR-498/ABCE1 axis in non-small cell lung cancer. Genes Genom. 2021, 43, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Wang, H.; Chen, X.; Liang, C.; Shang, W.; Wang, L.; Li, J.; Xu, D. MiR-182–5p inhibits colon cancer tumorigenesis, angiogenesis, and lymphangiogenesis by directly downregulating VEGF-C. Cancer Lett. 2020, 488, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.M.; An, A.R.; Park, H.S.; Jang, K.Y.; Moon, W.S.; Kang, M.J.; Lee, Y.C.; Ku, J.H.; Chung, M.J. Combined expression of protein disulfide isomerase and endoplasmic reticulum oxidoreductin 1-α is a poor prognostic marker for non-small cell lung cancer. Oncol. Lett. 2018, 16, 5753–5760. [Google Scholar] [CrossRef]
- Li, Z.; Li, Y.; Li, N.; Shen, L.; Liu, A. Silencing GOLGA8B inhibits cell invasion and metastasis by suppressing STAT3 signaling pathway in lung squamous cell carcinoma. Clin. Sci. 2022, 136, 895–909. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Hou, Y.; Qi, L. miR-30d-5p represses the proliferation, migration, and invasion of lung squamous cell carcinoma via targeting DBF4. J. Environ. Sci. Health Part C 2021, 39, 251–268. [Google Scholar] [CrossRef]
- Herreño, A.M.; Ramírez, A.C.; Chaparro, V.P.; Fernandez, M.J.; Cañas, A.; Morantes, C.F.; Moreno, O.M.; Brugés, R.E.; Mejía, J.A.; Bustos, F.J.; et al. Role of RUNX2 transcription factor in epithelial mesenchymal transition in non-small cell lung cancer lung cancer: Epigenetic control of the RUNX2 P1 promoter. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2019, 41, 1010428319851014. [Google Scholar] [CrossRef]
- Ou, H.; Liu, Q.; Lin, J.; He, W.; Zhang, W.; Ma, J.; Wang, W. Pseudogene annexin a2 pseudogene 1 contributes to hepatocellular carcinoma progression by modulating its parental gene ANXA2 via miRNA-376a-3p. Dig. Dis. Sci. 2021, 66, 3903–3915. [Google Scholar] [CrossRef]
- Ding, H.; Luo, Y.; Hu, K.; Liu, P.; Xiong, M. Linc00467 promotes lung adenocarcinoma proliferation via sponging miR-20b-5p to activate CCND1 expression. OncoTargets Ther. 2019, 12, 6733. [Google Scholar] [CrossRef]
- Han, C.; Li, H.; Ma, Z.; Dong, G.; Wang, Q.; Wang, S.; Fang, P.; Li, X.; Chen, H.; Liu, T.; et al. MIR99AHG is a noncoding tumor suppressor gene in lung adenocarcinoma. Cell Death Dis. 2021, 12, 424. [Google Scholar] [CrossRef]
- Chen, G.; Wang, Q.; Wang, K. MicroRNA-218–5p affects lung adenocarcinoma progression through targeting endoplasmic reticulum oxidoreductase 1 alpha. Bioengineered 2022, 13, 10061–10070. [Google Scholar] [CrossRef]
- Ye, J.; Li, H.; Wei, J.; Luo, Y.; Liu, H.; Zhang, J.; Luo, X. Risk scoring system based on lncRNA expression for predicting survival in hepatocellular carcinoma with cirrhosis. Asian Pac. J. Cancer Prev. APJCP 2020, 21, 1787. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Pan, T.; Jiang, W.; Zhao, H. HCG18/miR-34a-5p/HMMR axis accelerates the progression of lung adenocarcinoma. Biomed. Pharmacother. 2020, 129, 110217. [Google Scholar] [CrossRef]
- Yin, X.; Wang, P.; Yang, T.; Li, G.; Teng, X.; Huang, W.; Yu, H. Identification of key modules and genes associated with breast cancer prognosis using WGCNA and ceRNA network analysis. Aging 2021, 13, 2519. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Sun, Y.; Zhang, N.; Qiu, X.; Wang, L.; Luo, Q. By regulating miR-182–5p/BCL10/CYCS, sufentanil reduces the apoptosis of umbilical cord mesenchymal stem cells caused by ropivacaine. Biosci. Trends 2019, 13, 49–57. [Google Scholar] [CrossRef]
- Wang, F.; Wu, D.; Xu, Z.; Chen, J.; Zhang, J.; Li, X.; Chen, S.; He, F.; Xu, J.; Su, L.; et al. miR-182–5p affects human bladder cancer cell proliferation, migration and invasion through regulating Cofilin 1. Cancer Cell Int. 2019, 19, 42. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, M.; Kidoaki, S.; Fujie, H.; Miyoshi, H. Designing Elastic Modulus of Cell Culture Substrate to Regulate YAP and RUNX2 Localization for Controlling Differentiation of Human Mesenchymal Stem Cells. Anal. Sci. 2021, 37, 447–453. [Google Scholar] [CrossRef] [PubMed]
- De Kumar, B.; Parker, H.J.; Parrish, M.E.; Lange, J.J.; Slaughter, B.D.; Unruh, J.R.; Paulson, A.; Krumlauf, R. Dynamic regulation of Nanog and stem cell-signaling pathways by Hoxa1 during early neuro-ectodermal differentiation of ES cells. Proc. Natl. Acad. Sci. 2017, 114, 5838–5845. [Google Scholar] [CrossRef]
- Tang, D.; Yang, Z.; Long, F.; Luo, L.; Yang, B.; Zhu, R.; Sang, X.; Cao, G.; Wang, K. Long noncoding RNA MALAT1 mediates stem cell-like properties in human colorectal cancer cells by regulating miR-20b-5p/Oct4 axis. J. Cell. Physiol. 2019, 234, 20816–20828. [Google Scholar] [CrossRef]
- Michibata, H.; Okuno, T.; Konishi, N.; Kyono, K.; Wakimoto, K.; Aoki, K.; Kondo, Y.; Takata, K.; Kitamura, Y.; Taniguchi, T. Human GPM6A is associated with differentiation and neuronal migration of neurons derived from human embryonic stem cells. Stem Cells Dev. 2009, 18, 629–640. [Google Scholar] [CrossRef]
- Dong, Y.; Fan, X.; Wang, Z.; Zhang, L.; Guo, S. Circ_HECW2 functions as a miR-30e-5p sponge to regulate LPS-induced endothelial-mesenchymal transition by mediating NEGR1 expression. Brain Res. 2020, 1748, 147114. [Google Scholar] [CrossRef]
- Kamarudin, A.N.; Cox, T.; Kolamunnage-Dona, R. Time-dependent ROC curve analysis in medical research: Current methods and applications. BMC Med. Res. Methodol. 2017, 17, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Spruance, S.L.; Reid, J.E.; Grace, M.; Samore, M. Hazard ratio in clinical trials. Antimicrob. Agents Chemother. 2004, 48, 2787–2792. [Google Scholar] [CrossRef]
- Engelhardt, A.; Shen, Y.M.; Mansmann, U. Constructing an ROC curve to assess a treatment-predictive continuous biomarker. Stud Health Technol Inform. 2016, 228, 745–749. [Google Scholar] [PubMed]
- Wan, L.; Kong, J.; Tang, J.; Wu, Y.; Xu, E.; Lai, M.; Zhang, H. HOTAIRM 1 as a potential biomarker for diagnosis of colorectal cancer functions the role in the tumour suppressor. J. Cell. Mol. Med. 2016, 20, 2036–2044. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, C.; Li, S.; Qu, Y.; Xue, P.; Ma, Z.; Zhang, X.; Bai, H.; Wang, J. ERO1L is a novel and potential biomarker in lung adenocarcinoma and shapes the immune-suppressive tumor microenvironment. Front. Immunol. 2021, 12, 677169. [Google Scholar] [CrossRef]
- Cheng, Y.; Li, L.; Qin, Z.; Li, X.; Qi, F. Identification of castration-resistant prostate cancer-related hub genes using weighted gene co-expression network analysis. J. Cell. Mol. Med. 2020, 24, 8006–8017. [Google Scholar] [CrossRef]
- Zheng, M.; Hou, L.; Ma, Y.; Zhou, L.; Wang, F.; Cheng, B.; Wang, W.; Lu, B.; Liu, P.; Lu, W.; et al. Exosomal let-7d-3p and miR-30d-5p as diagnostic biomarkers for non-invasive screening of cervical cancer and its precursors. Mol. Cancer 2019, 18, 76. [Google Scholar] [CrossRef]
- Liang, S.; Li, Y.; Wang, B. The cancer-related transcription factor Runx2 combined with osteopontin: A novel prognostic biomarker in resected osteosarcoma. Int. J. Clin. Oncol. 2021, 26, 2347–2354. [Google Scholar] [CrossRef]
- Dai, Y.; Zheng, C.; Li, H. Inhibition of miR-23a-3p promotes osteoblast proliferation and differentiation. J. Cell. Biochem. 2019, 10, 1002. [Google Scholar]
- Li, H.; Wang, X.; Zhang, M.; Wang, M.; Zhang, J.; Ma, S. Identification of HOXA1 as a novel biomarker in prognosis of head and neck squamous cell carcinoma. Front. Mol. Biosci. 2021, 7, 602068. [Google Scholar] [CrossRef]
- Li, S.; Zou, H.; Shao, Y.Y.; Mei, Y.; Cheng, Y.; Hu, D.L.; Tan, Z.R.; Zhou, H.H. Pseudogenes of annexin A2, novel prognosis biomarkers for diffuse gliomas. Oncotarget 2017, 8, 106962. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hu, S.; Li, Z.; Lutz, H.; Huang, K.; Su, T.; Cores, J.; Dinh, P.U.C.; Cheng, K. Dermal exosomes containing miR-218–5p promote hair regeneration by regulating ß-catenin signaling. Sci. Adv. 2020, 6, eaba1685. [Google Scholar] [CrossRef] [PubMed]
- Cai, G.; Sun, M.; Li, X.; Zhu, J. Construction and characterization of rectal cancer-related lncRNA-mRNA ceRNA network reveals prognostic biomarkers in rectal cancer. IET Syst. Biol. 2021, 15, 192–204. [Google Scholar] [CrossRef]
- Misir, S.; Hepokur, C.; Oksasoglu, B.; Yildiz, C.; Yanik, A.; Aliyazicioglu, Y. Circulating serum miR-200c and miR-34a-5p as diagnostic biomarkers for endometriosis. J. Gynecol. Obs. Hum. Reprod. 2021, 50, 102092. [Google Scholar] [CrossRef] [PubMed]

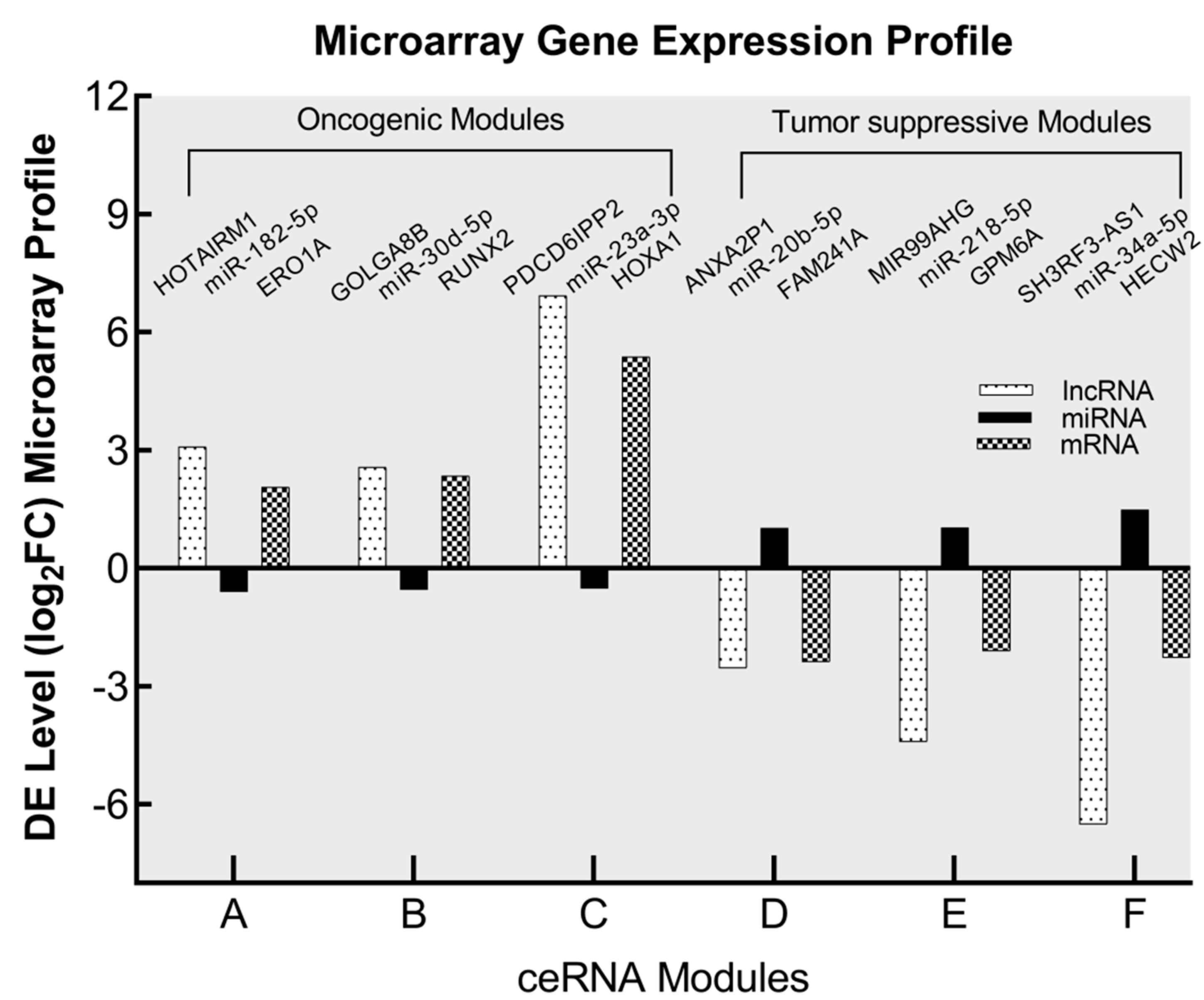
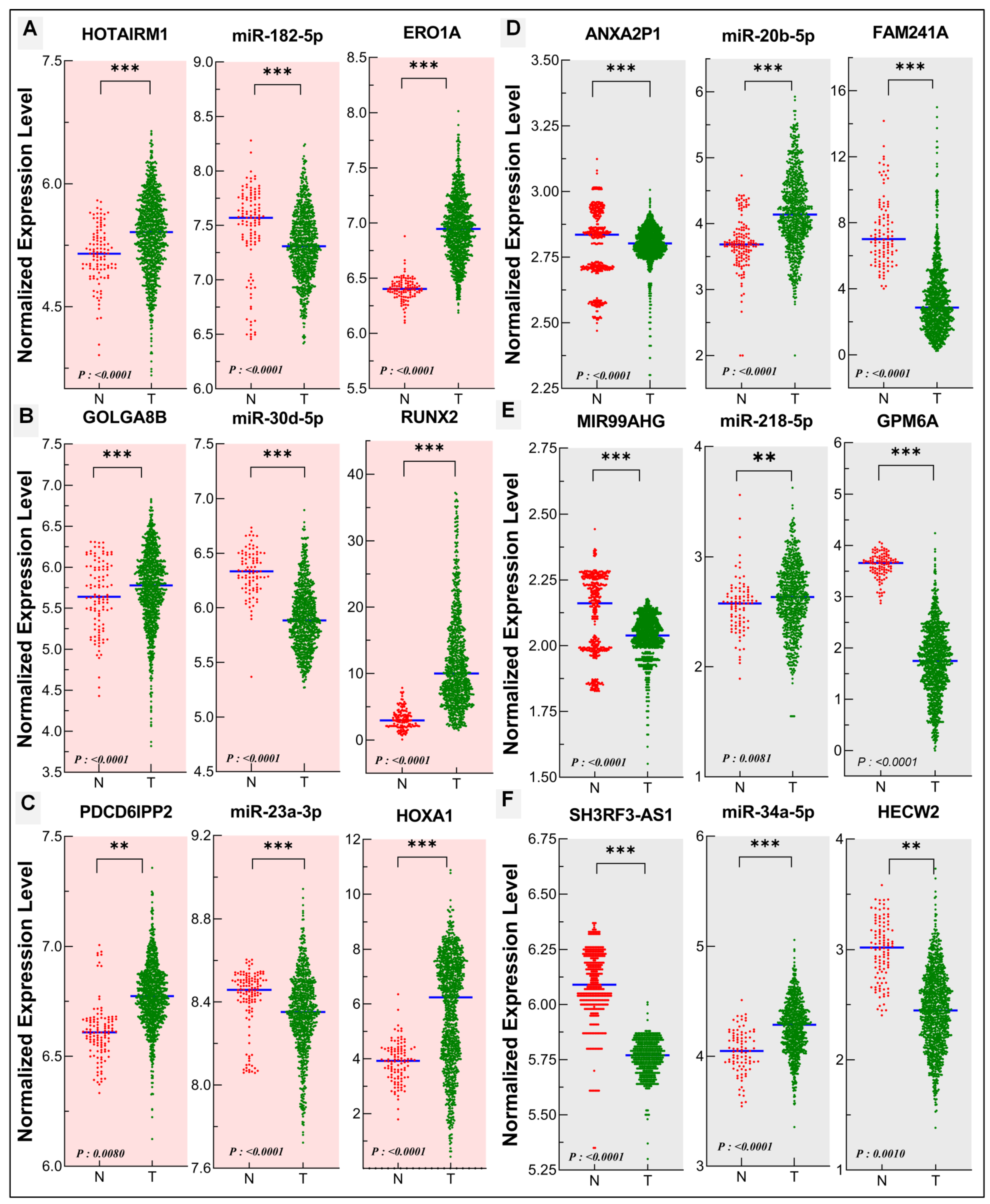
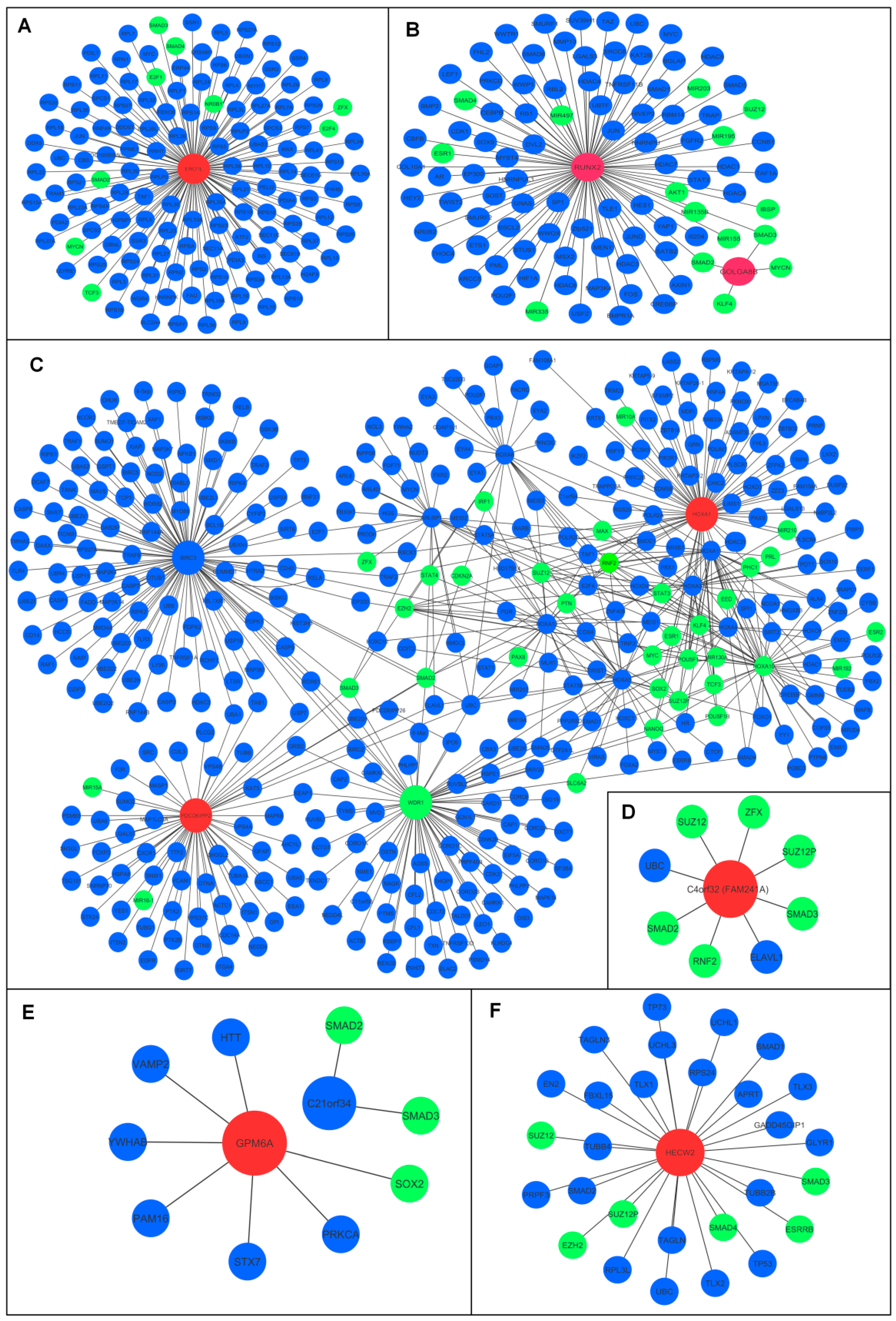
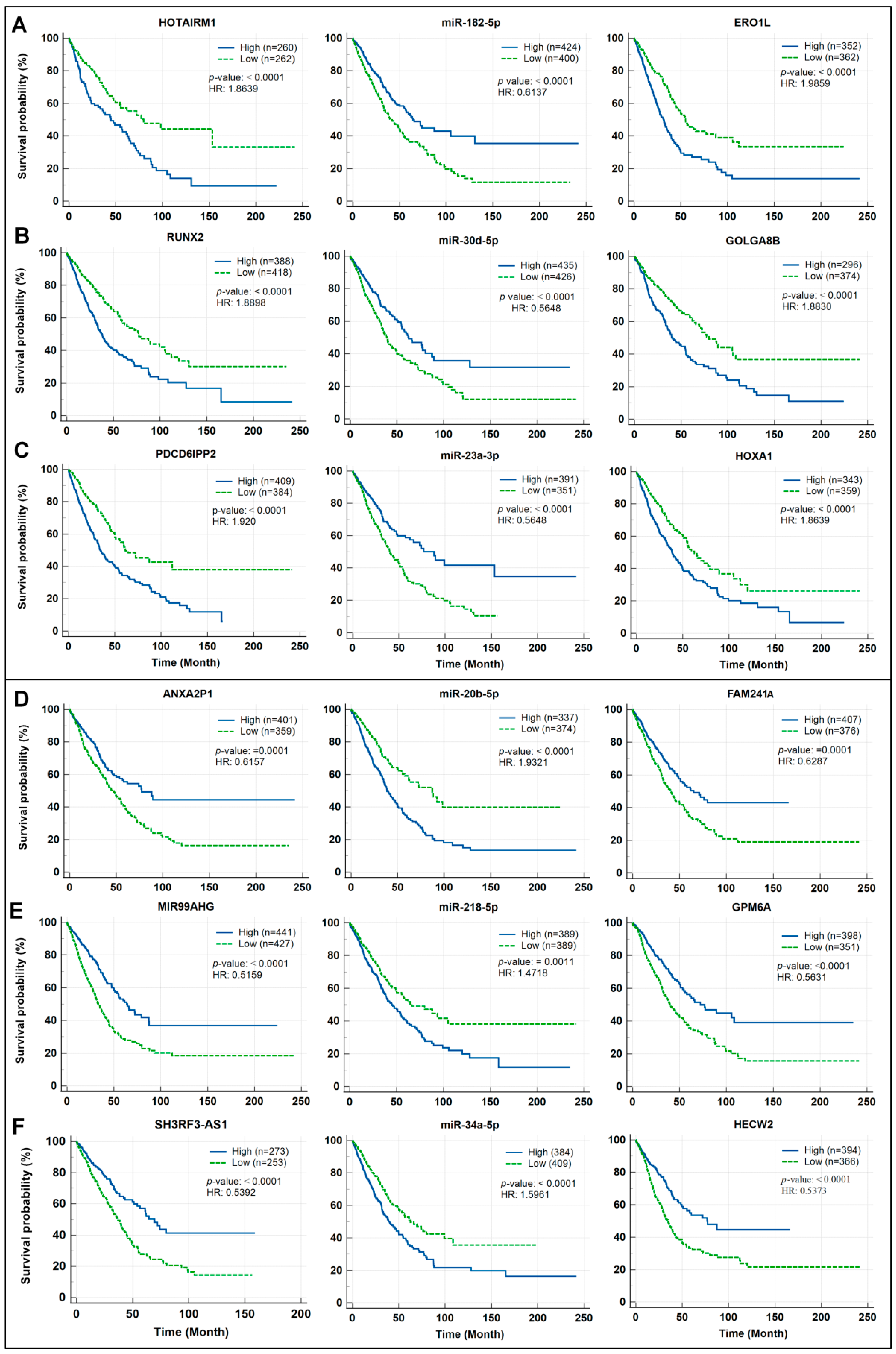
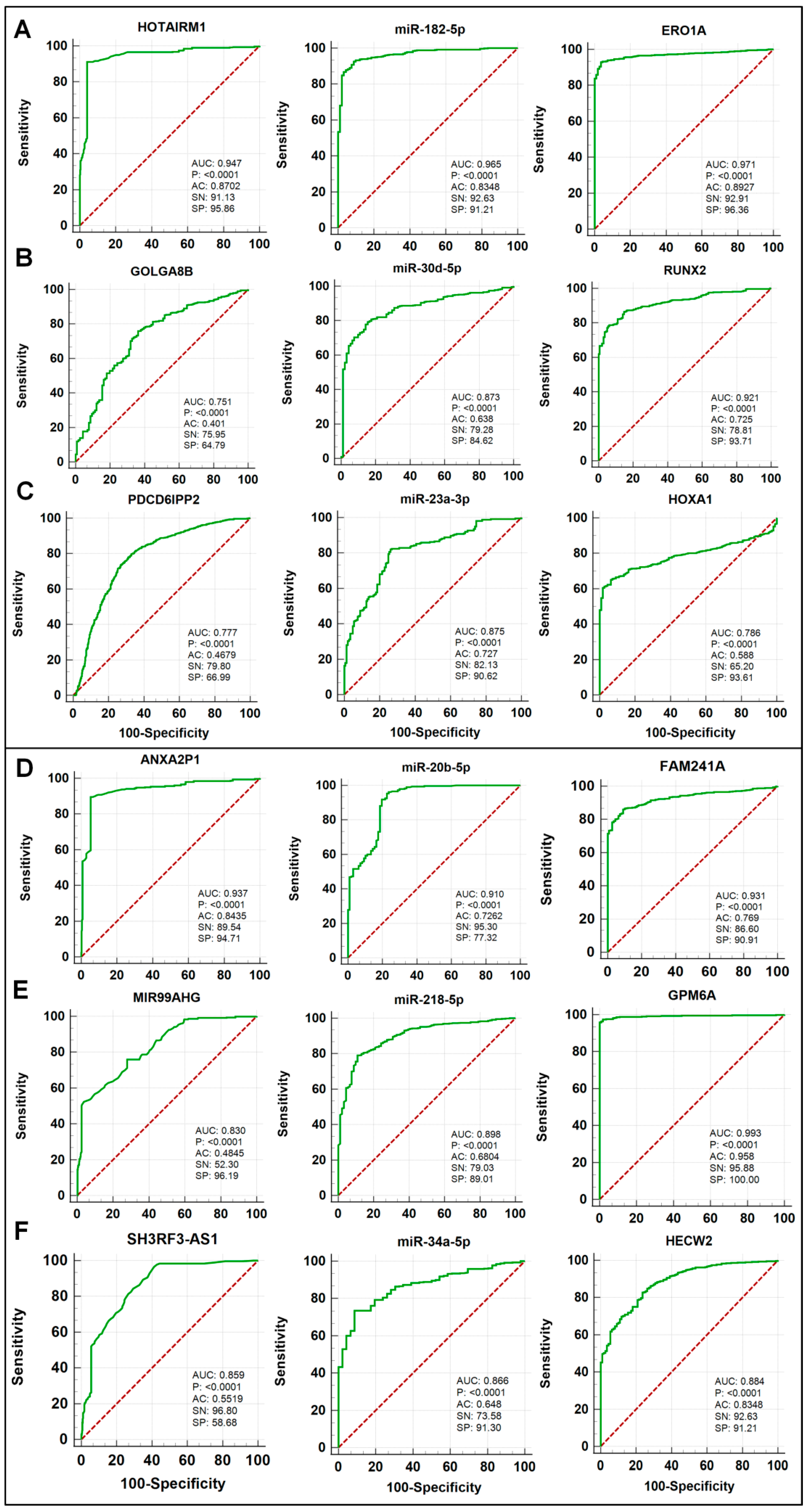
| lncRNA | miRNA | mRNA | |
|---|---|---|---|
| Oncogenic ceRNA modules | |||
| A | HOTAIRM1 | miR-182-5p | ERO1A |
| B | GOLGA8B | miR-30d-5p | RUNX2 |
| C | PDCD6IPP2 | miR-23a-3p | HOXA1 |
| Tumor suppressor ceRNA modules | |||
| D | ANXA2P1 | miR-20b-5p | FAM241A (C4orf32) |
| E | MIR99AHG | miR-218-5p | GPM6A |
| F | SH3RF3-AS1 | miR-34a-5p | HECW2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sweef, O.; Yang, C.; Wang, Z. The Oncogenic and Tumor Suppressive Long Non-Coding RNA–microRNA–Messenger RNA Regulatory Axes Identified by Analyzing Multiple Platform Omics Data from Cr(VI)-Transformed Cells and Their Implications in Lung Cancer. Biomedicines 2022, 10, 2334. https://doi.org/10.3390/biomedicines10102334
Sweef O, Yang C, Wang Z. The Oncogenic and Tumor Suppressive Long Non-Coding RNA–microRNA–Messenger RNA Regulatory Axes Identified by Analyzing Multiple Platform Omics Data from Cr(VI)-Transformed Cells and Their Implications in Lung Cancer. Biomedicines. 2022; 10(10):2334. https://doi.org/10.3390/biomedicines10102334
Chicago/Turabian StyleSweef, Osama, Chengfeng Yang, and Zhishan Wang. 2022. "The Oncogenic and Tumor Suppressive Long Non-Coding RNA–microRNA–Messenger RNA Regulatory Axes Identified by Analyzing Multiple Platform Omics Data from Cr(VI)-Transformed Cells and Their Implications in Lung Cancer" Biomedicines 10, no. 10: 2334. https://doi.org/10.3390/biomedicines10102334
APA StyleSweef, O., Yang, C., & Wang, Z. (2022). The Oncogenic and Tumor Suppressive Long Non-Coding RNA–microRNA–Messenger RNA Regulatory Axes Identified by Analyzing Multiple Platform Omics Data from Cr(VI)-Transformed Cells and Their Implications in Lung Cancer. Biomedicines, 10(10), 2334. https://doi.org/10.3390/biomedicines10102334





