DNA Damage Repair: Predictor of Platinum Efficacy in Ovarian Cancer?
Abstract
1. Ovarian Cancer
2. Treatment of Advanced Ovarian Cancer
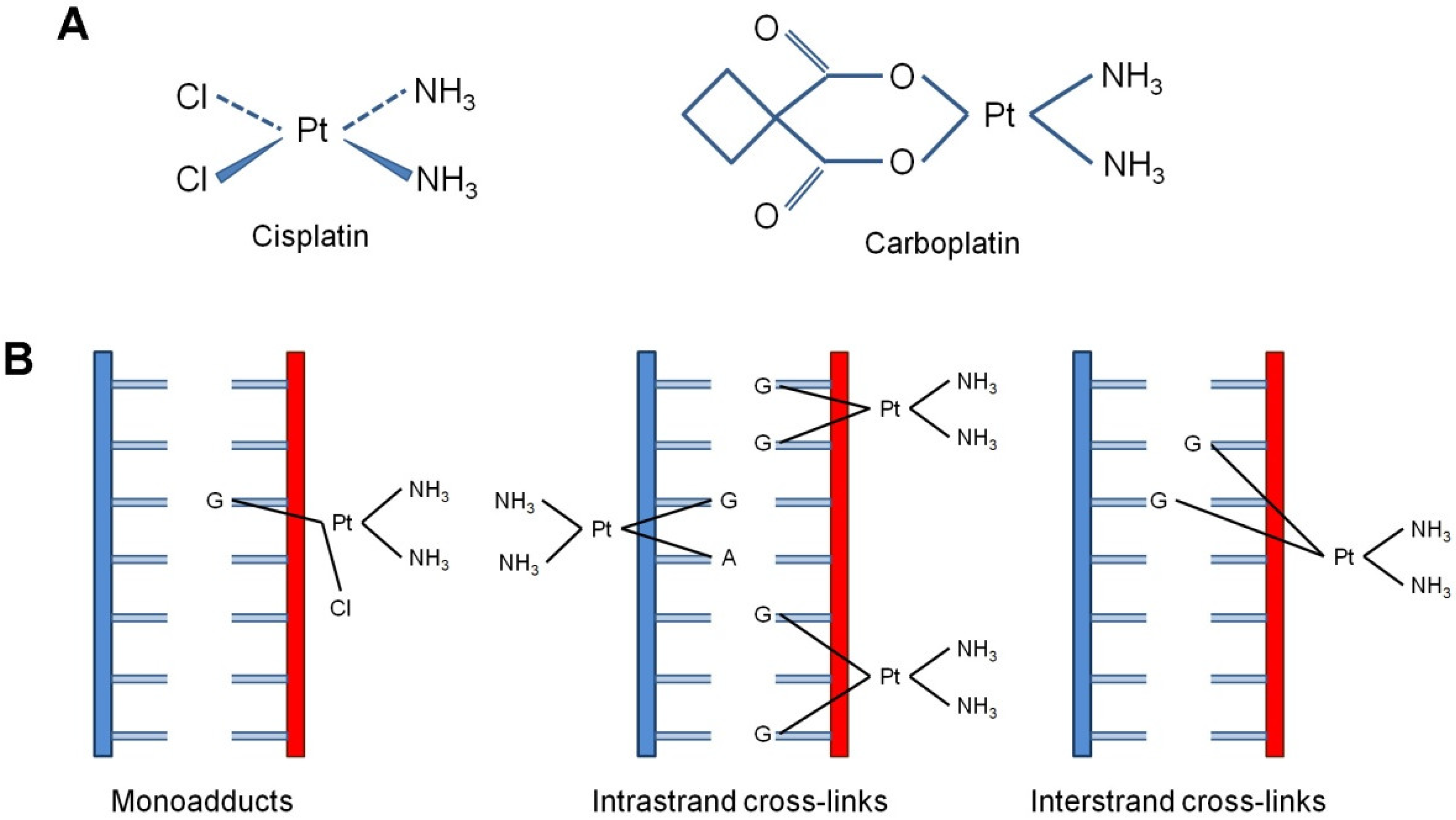
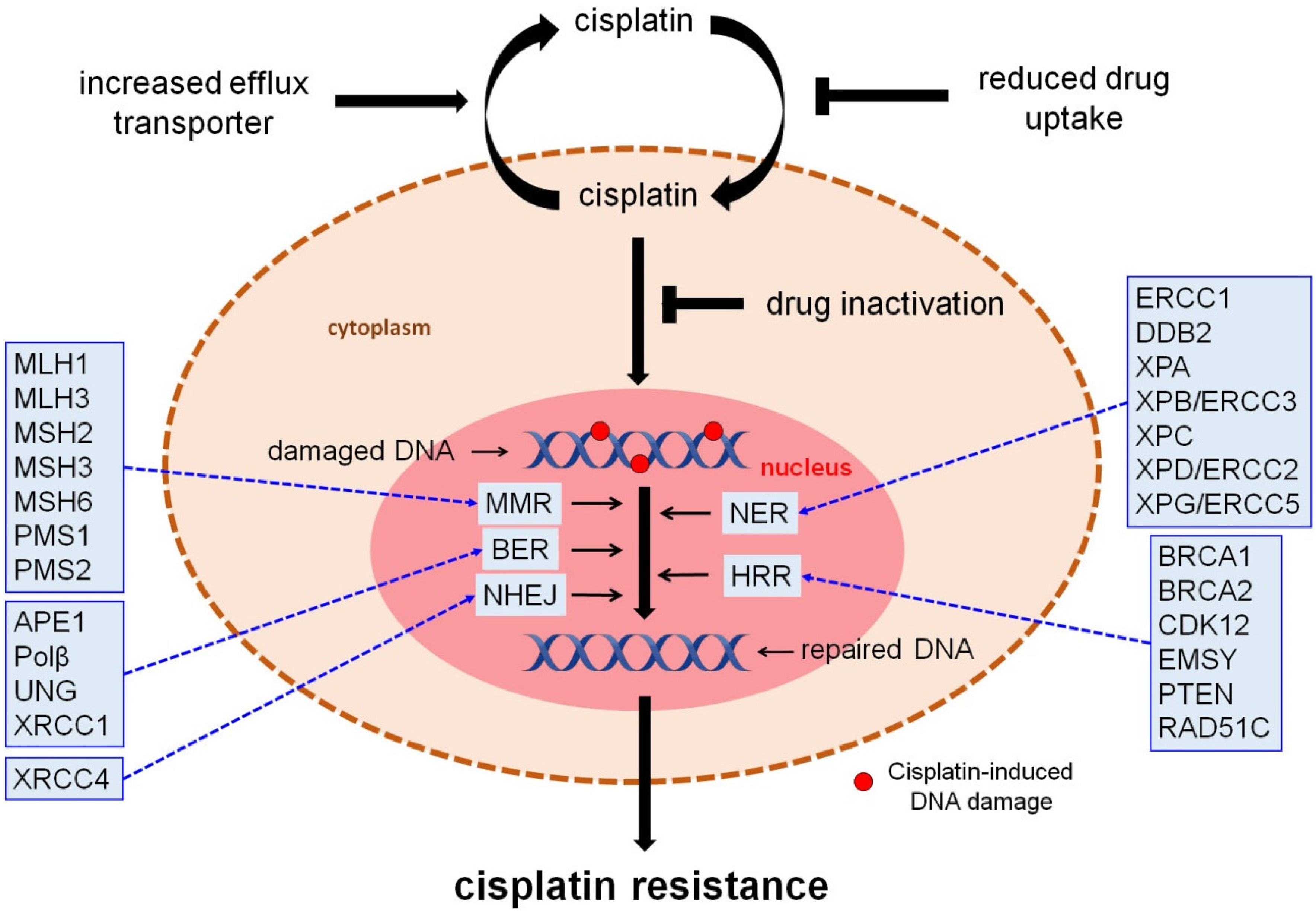
3. DNA Repair Responses to Cisplatin-Induced DNA Damage
3.1. Homologous Recombination Repair (HRR)
3.2. Nucleotide Excision Repair (NER)
3.3. Mismatch Repair (MMR)
3.4. Non-Homologous End-Joining (NHEJ)
3.5. Base Excision Repair (BER)
| DNA Repair Pathway | Symbol | Description | Reference |
|---|---|---|---|
| Homologous recombination repair (HRR) | BRCA1 | Breast cancer type 1 susceptibility protein | Pietragalla et al. [6] |
| BRCA2 | Breast cancer type 2 susceptibility protein | Pietragalla et al. [6] | |
| CDK12 | Cyclin-dependent kinase 12 | Joshi et al. [25] | |
| EMSY | BRCA2-interacting transcriptional repressor EMSY | Hughes-Davies et al. [24] | |
| PTEN | Phosphatase and tensin homolog | The Cancer Genome Atlas Research Network [23] | |
| RAD51C | RAD51 homolog C | Hurley et al. [26] | |
| Nucleotide excision repair (NER) | ERCC1 | Excision repair cross-complementation, group 1 | Chebouti et al. [58] |
| DDB2 | Damage-specific DNA binding protein 2 | Cui et al. [46] | |
| XPA | Xeroderma pigmentosum, complementation group A | Kang et al. [53]; Sancar et al. [54] | |
| XPB/ERCC3 | Xeroderma pigmentosum, complementation group B | Reed et al. [52]; Damia et al. [18] | |
| XPC | Xeroderma pigmentosum, complementation group C | Wang et al. [45]; Fleming et al. [44] | |
| XPD/ERCC2 | Xeroderma pigmentosum, complementation group D | Michalska et al. [49]; Kang et al. [50] | |
| XPG/ERCC5 | Xeroderma pigmentosum, complementation group G | Walsh et al. [59] | |
| Mismatch repair (MMR) | MLH1 | MutL homolog 1, colon cancer, nonpolyposis type 2 | Gras et al. [66]; Kawashima et al. [67] |
| MLH3 | MutL homolog 3 | Zhao et al. [60] | |
| MSH2 | MutS homolog 2, colon cancer, nonpolyposis Type 1 | Pabla et al. [64] | |
| MSH3 | MutS homolog 3 | Zhao et al. [60] | |
| MSH6 | MutS homolog 6 | Zhao et al. [60] | |
| PMS1 | PMS1 post meiotic segregation increased 1 | Zhao et al. [60] | |
| PMS2 | PMS2 post meiotic segregation increased 2 | Zhao et al. [60] | |
| Non-homologous end-joining (NHEJ) | XRCC4 | X-ray repair cross complementing 4 | Liu et al. [69] |
| Base excision repair (BER) | APE1 | Apurinic/apyrimidinic endo deoxyribonuclease 1 | Kothandapani et al. [70,71] |
| Polβ | DNA polymerase beta subunit | Kothandapani et al. [70,71] | |
| UNG | Uracil-DNA glycosylase | Kothandapani et al. [70,71] | |
| XRCC1 | X-ray repair cross complementing 1 | Abdel-Fatah et al. [72] |
4. New Therapeutic Perspectives in Epithelial Ovarian Cancer
4.1. PARP Inhibition in Epithelial Ovarian Cancer
4.2. CHK1/2, ATR Inhibitors
4.3. Wee1 Inhibitors
4.4. Immunotherapy
5. Discussion
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Stewart, C.; Ralyea, C.; Lockwood, S. Ovarian Cancer: An Integrated Review. Semin. Oncol. Nurs. 2019, 35, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.R. Ovarian cancer update: Lessons from morphology, molecules, and mice. Arch. Pathol. Lab. Med. 2009, 133, 1775–1781. [Google Scholar] [CrossRef]
- Cree, I.A.; White, V.A.; Indave, B.I.; Lokuhetty, D. Revising the WHO classification: Female genital tract tumours. Histopathology 2020, 76, 151–156. [Google Scholar] [CrossRef]
- Lynch, H.T.; Casey, M.J.; Snyder, C.L.; Bewtra, C.; Lynch, J.F.; Butts, M.; Godwin, A.K. Hereditary ovarian carcinoma: Heterogeneity, molecular genetics, pathology, and management. Mol. Oncol. 2009, 3, 97–137. [Google Scholar] [CrossRef] [PubMed]
- Kieffer, Y.; Bonneau, C.; Popova, T.; Rouzier, R.; Stern, M.-H.; Mechta-Grigoriou, F. Clinical Interest of Combining Transcriptomic and Genomic Signatures in High-Grade Serous Ovarian Cancer. Front. Genet. 2020, 11, 219. [Google Scholar] [CrossRef] [PubMed]
- Pietragalla, A.; Arcieri, M.; Marchetti, C.; Scambia, G.; Fagotti, A. Ovarian cancer predisposition beyond BRCA1 and BRCA2 genes. Int. J. Gynecol. Cancer 2020, 30, 1803–1810. [Google Scholar] [CrossRef]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer Genome Landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef]
- McConechy, M.K.; Ding, J.; Senz, J.; Yang, W.; Melnyk, N.; Tone, A.A.; Prentice, L.M.; Wiegand, K.C.; McAlpine, J.N.; Shah, S.P.; et al. Ovarian and endometrial endometrioid carcinomas have distinct CTNNB1 and PTEN mutation profiles. Mod. Pathol. 2014, 27, 128–134. [Google Scholar] [CrossRef]
- Brown, J.; Frumovitz, M. Mucinous Tumors of the Ovary: Current Thoughts on Diagnosis and Management. Curr. Oncol. Rep. 2014, 16, 389. [Google Scholar] [CrossRef]
- Prat, J. FIGO’s staging classification for cancer of the ovary, fallopian tube, and peritoneum: Abridged republication. J. Gynecol. Oncol. 2015, 26, 87–89. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.K.; Pujade-Lauraine, E.; Aoki, D.; Mirza, M.R.; Lorusso, D.; Oza, A.; du Bois, A.; Vergote, I.; Reuss, A.; Bacon, M.; et al. Fifth Ovarian Cancer Consensus Conference of the Gynecologic Cancer InterGroup: Recurrent disease. Ann. Oncol. 2016, 28, 727–732. [Google Scholar] [CrossRef] [PubMed]
- Pateras, I.S.; Havaki, S.; Nikitopoulou, X.; Vougas, K.; Townsend, P.A.; Panayiotidis, M.I.; Georgakilas, A.G.; Gorgoulis, V.G. The DNA damage response and immune signaling alliance: Is it good or bad? Nature decides when and where. Pharmacol. Ther. 2015, 154, 36–56. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Chen, X. Drug resistance and combating drug resistance in cancer. Cancer Drug Resist. 2019, 2, 141–160. [Google Scholar] [CrossRef] [PubMed]
- Woźniak, K.; Walter, Z. Induction of DNA-Protein Cross-Links by Platinum Compounds. Z. Für Nat. C 2000, 55, 731–736. [Google Scholar] [CrossRef]
- Chválová, K.; Brabec, V.; Kašpárková, J. Mechanism of the formation of DNA–protein cross-links by antitumor cisplatin. Nucleic Acids Res. 2007, 35, 1812–1821. [Google Scholar] [CrossRef]
- Ming, X.; Groehler, I.A.; Michaelson-Richie, E.D.; Villalta, P.W.; Campbell, C.; Tretyakova, N.Y. Mass Spectrometry Based Proteomics Study of Cisplatin-Induced DNA–Protein Cross-Linking in Human Fibrosarcoma (HT1080) Cells. Chem. Res. Toxicol. 2017, 30, 980–995. [Google Scholar] [CrossRef] [PubMed]
- Kubelac, P.; Genestie, C.; Auguste, A.; Mesnage, S.; Le Formal, A.; Pautier, P.; Gouy, S.; Morice, P.; Bentivegna, E.; Maulard, A.; et al. Changes in DNA Damage Response Markers with Treatment in Advanced Ovarian Cancer. Cancers 2020, 12, 707. [Google Scholar] [CrossRef] [PubMed]
- Damia, G.; Broggini, M. Platinum Resistance in Ovarian Cancer: Role of DNA Repair. Cancers 2019, 11, 119. [Google Scholar] [CrossRef]
- Helleday, T. Homologous recombination in cancer development, treatment and development of drug resistance. Carcinogenesis 2010, 31, 955–960. [Google Scholar] [CrossRef]
- West, S.C. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003, 4, 435–445. [Google Scholar] [CrossRef]
- Yang, D.; Khan, S.; Sun, Y.; Hess, K.; Shmulevich, I.; Sood, A.K.; Zhang, W. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. JAMA 2011, 306, 1557–1565. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.S.; Rothermundt, C.; Thomas, K.; Bancroft, E.; Eeles, R.; Shanley, S.; Ardern-Jones, A.; Norman, A.; Kaye, S.B.; Gore, M.E. “BRCAness” syndrome in ovarian cancer: A case-control study describing the clinical features and outcome of patients with epithelial ovarian cancer associated with BRCA1 and BRCA2 mutations. J. Clin. Oncol. 2008, 26, 5530–5536. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef]
- Hughes-Davies, L.; Huntsman, D.; Ruas, M.; Fuks, F.; Bye, J.; Chin, S.-F.; Milner, J.; Brown, L.; Hsu, F.; Gilks, B.; et al. EMSY Links the BRCA2 Pathway to Sporadic Breast and Ovarian Cancer. Cell 2003, 115, 523–535. [Google Scholar] [CrossRef]
- Joshi, P.M.; Sutor, S.L.; Huntoon, C.J.; Karnitz, L.M. Ovarian cancer-associated mutations disable catalytic activity of CDK12, a kinase that promotes homologous recombination repair and resistance to cisplatin and poly(ADP-ribose) polymerase inhibitors. J. Biol. Chem. 2014, 289, 9247–9253. [Google Scholar] [CrossRef]
- Hurley, R.M.; McGehee, C.D.; Nesic, K.; Correia, C.; Weiskittel, T.M.; Kelly, R.L.; Venkatachalam, A.; Hou, X.; Pathoulas, N.M.; Meng, X.W.; et al. Characterization of a RAD51C-silenced high-grade serous ovarian cancer model during development of PARP inhibitor resistance. NAR Cancer 2021, 3, zcab028. [Google Scholar] [CrossRef]
- Frey, M.K.; Pothuri, B. Homologous recombination deficiency (HRD) testing in ovarian cancer clinical practice: A review of the literature. Gynecol. Oncol. Res. Pract. 2017, 4, 1–11. [Google Scholar] [CrossRef]
- Hoppe, M.M.; Sundar, R.; Tan, D.S.P.; Jeyasekharan, A.D. Biomarkers for Homologous Recombination Deficiency in Cancer. JNCI J. Natl. Cancer Inst. 2018, 110, 704–713. [Google Scholar] [CrossRef]
- Mukhopadhyay, A.; Elattar, A.; Cerbinskaite, A.; Wilkinson, S.J.; Drew, Y.; Kyle, S.; Los, G.; Hostomsky, Z.; Edmondson, R.J.; Curtin, N.J. Development of a functional assay for homologous recombination status in primary cultures of epithelial ovarian tumor and correlation with sensitivity to poly(ADP-ribose) polymerase inhibitors. Clin. Cancer Res. 2010, 16, 2344–2351. [Google Scholar] [CrossRef]
- Cruz, C.; Castroviejo-Bermejo, M.; Gutiérrez-Enríquez, S.; Llop-Guevara, A.; Ibrahim, Y.; Oliver, A.G.; Bonache, S.; Morancho, B.; Bruna, A.; Rueda, O.; et al. RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCA-mutated breast cancer. Ann. Oncol. 2018, 29, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Stronach, E.A.; Paul, J.; Timms, K.M.; Hughes, E.; Brown, K.; Neff, C.; Perry, M.; Gutin, A.; El-Bahrawy, M.; Steel, J.H.; et al. Biomarker Assessment of HR Deficiency, Tumor BRCA1/2 Mutations, and CCNE1 Copy Number in Ovarian Cancer: Associations with Clinical Outcome Following Platinum Monotherapy. Mol. Cancer Res. 2018, 16, 1103–1111. [Google Scholar] [CrossRef]
- Jóhannsson, O.T.; Ranstam, J.; Borg, A.; Olsson, H. Survival of BRCA1 breast and ovarian cancer patients: A population-based study from southern Sweden. J. Clin. Oncol. 1998, 16, 397–404. [Google Scholar] [CrossRef]
- Pharoah, P.D.; Easton, D.F.; Stockton, D.L.; Gayther, S.; Ponder, B.A. Survival in familial, BRCA1-associated, and BRCA2-associated epithelial ovarian cancer. Cancer Res. 1999, 59, 868–871. [Google Scholar] [PubMed]
- Bolton, K.L.; Chenevix-Trench, G.; Goh, C.; Sadetzki, S.; Ramus, S.J.; Karlan, B.Y.; Lambrechts, D.; Despierre, E.; Barrow-dale, D.; McGuffog, L.; et al. Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA 2012, 307, 382–390. [Google Scholar] [CrossRef]
- Weigelt, B.; Comino-Méndez, I.; de Bruijn, I.; Tian, L.; Meisel, J.L.; García-Murillas, I.; Fribbens, C.; Cutts, R.; Martelotto, L.G.; Ng, C.K.Y.; et al. Diverse. Clin. Cancer Res. 2017, 23, 6708–6720. [Google Scholar] [CrossRef]
- Patch, A.M.; Christie, E.L.; Etemadmoghadam, D.; Garsed, D.W.; George, J.; Fereday, S.; Nones, K.; Cowin, P.; Alsop, K.; Bailey, P.J.; et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature 2015, 521, 489–494. [Google Scholar] [CrossRef]
- Dhillon, K.K.; Swisher, E.M.; Taniguchi, T. Secondary mutations of BRCA1/2 and drug resistance. Cancer Sci. 2011, 102, 663–669. [Google Scholar] [CrossRef]
- Kondrashova, O.; Nguyen, M.; Shield-Artin, K.; Tinker, A.V.; Teng, N.N.; Harrell, M.I.; Kuiper, M.J.; Ho, G.-Y.; Barker, H.; Jasin, M.; et al. Secondary Somatic Mutations Restoring RAD51C and RAD51D Associated with Acquired Resistance to the PARP Inhibitor Rucaparib in High-Grade Ovarian Carcinoma. Cancer Discov. 2017, 7, 984–998. [Google Scholar] [CrossRef] [PubMed]
- Johnson, N.; Johnson, S.F.; Yao, W.; Li, Y.-C.; Choi, Y.-E.; Bernhardy, A.J.; Wang, Y.; Capelletti, M.; Sarosiek, K.A.; Moreau, L.A.; et al. Stabilization of mutant BRCA1 protein confers PARP inhibitor and platinum resistance. Proc. Natl. Acad. Sci. USA 2013, 110, 17041–17046. [Google Scholar] [CrossRef] [PubMed]
- Shuck, S.C.; Short, E.A.; Turchi, J.J. Eukaryotic nucleotide excision repair: From understanding mechanisms to influencing biology. Cell Res. 2008, 18, 64–72. [Google Scholar] [CrossRef]
- Mouw, K.W.; D’Andrea, A.D.; Konstantinopoulos, P.A. Nucleotide excision repair (NER) alterations as evolving biomarkers and therapeutic targets in epithelial cancers. Oncoscience 2005, 2, 942–943. [Google Scholar] [CrossRef]
- Rubatt, J.M.; Darcy, K.M.; Tian, C.; Muggia, F.; Dhir, R.; Armstrong, D.K.; Bookman, M.A.; Niedernhofer, L.J.; Deloia, J.; Birrer, M.; et al. Pre-treatment tumor expression of ERCC1 in women with advanced stage epithelial ovarian cancer is not predictive of clinical outcomes: A gynecologic oncology group study. Gynecol. Oncol. 2012, 125, 421–426. [Google Scholar] [CrossRef]
- Zhao, M.; Li, S.; Zhou, L.; Shen, Q.; Zhu, H.; Zhu, X. Prognostic values of excision repair cross-complementing genes mRNA expression in ovarian cancer patients. Life Sci. 2018, 194, 34–39. [Google Scholar] [CrossRef]
- Fleming, N.D.; Agadjanian, H.; Nassanian, H.; Miller, C.W.; Orsulic, S.; Karlan, B.Y.; Walsh, C.S. Xeroderma pigmentosum complementation group C single-nucleotide polymorphisms in the nucleotide excision repair pathway correlate with prolonged progression-free survival in advanced ovarian cancer. Cancer 2012, 118, 689–697. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Chuang, L.; Zhang, X.; Colton, S.; Dombkowski, A.; Reiners, J.; Diakiw, A.; Xu, X.S. The initiative role of XPC protein in cisplatin DNA damaging treatment-mediated cell cycle regulation. Nucleic Acids Res. 2004, 32, 2231–2240. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cui, T.; Srivastava, A.K.; Han, C.; Wu, D.; Wani, N.; Liu, L.; Gao, Z.; Qu, M.; Zou, N.; Zhang, X.; et al. DDB2 represses ovarian cancer cell dedifferentiation by suppressing ALDH1A1. Cell Death Dis. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Barakat, B.M.; Wang, Q.E.; Han, C.; Milum, K.; Yin, D.T.; Zhao, Q.; Wani, G.; Arafa, E.-S.; El-Mahdy, M.A.; Wani, A.A. Overexpression of DDB2 enhances the sensitivity of human ovarian cancer cells to cisplatin by augmenting cellular apoptosis. Int. J. Cancer 2010, 127, 977–988. [Google Scholar] [PubMed]
- Furuta, T.; Ueda, T.; Aune, G.; Sarasin, A.; Kraemer, K.H.; Pommier, Y. Transcription-coupled nucleotide excision repair as a determinant of cisplatin sensitivity of human cells. Cancer Res. 2002, 62, 4899–4902. [Google Scholar] [PubMed]
- Michalska, M.M.; Samulak, D.; Romanowicz, H.; Sobkowski, M.; Smolarz, B. An Association between Single Nucleotide Polymorphisms of Lys751GlnERCC2Gene and Ovarian Cancer in Polish Women. Adv. Med. 2015, 1–6. [Google Scholar] [CrossRef]
- Kang, S.; Sun, H.-Y.; Zhou, R.-M.; Wang, N.; Hu, P.; Li, Y. DNA Repair Gene Associated with Clinical Outcome of Epithelial Ovarian Cancer Treated with Platinum-based Chemotherapy. Asian Pac. J. Cancer Prev. 2013, 14, 941–946. [Google Scholar] [CrossRef]
- Khrunin, A.V.; Moisseev, A.; Gorbunova, V.; Limborska, S. Genetic polymorphisms and the efficacy and toxicity of cisplatin-based chemotherapy in ovarian cancer patients. Pharm. J. 2010, 10, 54–61. [Google Scholar]
- Reed, E.; Yu, J.J.; Davies, A.; Gannon, J.; Armentrout, S.L. Clear cell tumors have higher mRNA levels of ERCC1 and XPB than other histological types of epithelial ovarian cancer. Clin. Cancer Res. 2003, 9, 5299–5305. [Google Scholar]
- Kang, T.-H.; Reardon, J.T.; Kemp, M.; Sancar, A. Circadian oscillation of nucleotide excision repair in mammalian brain. Proc. Natl. Acad. Sci. USA 2009, 106, 2864–2867. [Google Scholar] [CrossRef]
- Sancar, A.; Lindsey-Boltz, L.A.; Gaddameedhi, S.; Selby, C.P.; Ye, R.; Chiou, Y.Y.; Kemp, M.G.; Hu, J.; Lee, J.H.; Ozturk, N. Circadian clock, cancer, and chemotherapy. Biochemistry 2015, 54, 110–123. [Google Scholar] [CrossRef]
- Kuo, M.-S.; Adam, J.; Dorvault, N.; Robin, A.; Friboulet, L.; Soria, J.-C.; Olaussen, K.A. A novel antibody-based approach to detect the functional ERCC1-202 isoform. DNA Repair 2018, 64, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Ferry, K.V.; Hamilton, T.C.; Johnson, S.W. Increased nucleotide excision repair in cisplatin-resistant ovarian cancer cells: Role of ercc1–xpf. Biochem. Pharmacol. 2000, 60, 1305–1313. [Google Scholar] [CrossRef]
- Mesquita, K.A.; Alabdullah, M.; Griffin, M.; Toss, M.S.; Fatah, T.M.A.; Alblihy, A.; Moseley, P.; Chan, S.; Rakha, E.A.; Madhusudan, S. ERCC1-XPF deficiency is a predictor of olaparib induced synthetic lethality and platinum sensitivity in epithelial ovarian cancers. Gynecol. Oncol. 2019, 153, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Chebouti, I.; Kuhlmann, J.D.; Buderath, P.; Weber, S.; Wimberger, P.; Bokeloh, Y.; Hauch, S.; Kimmig, R.; Kasimir-Bauer, S. ERCC1-expressing circulating tumor cells as a potential diagnostic tool for monitoring response to platinum-based chemotherapy and for predicting post-therapeutic outcome of ovarian cancer. Oncotarget 2016, 8, 24303–24313. [Google Scholar] [CrossRef]
- Walsh, C.S.; Ogawa, S.; Karahashi, H.; Scoles, D.R.; Pavelka, J.C.; Tran, H.; Miller, C.W.; Kawamata, N.; Ginther, C.; Dering, J.; et al. ERCC5Is a Novel Biomarker of Ovarian Cancer Prognosis. J. Clin. Oncol. 2008, 26, 2952–2958. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Li, S.; Zhao, M.; Zhu, H.; Zhu, X. Prognostic values of DNA mismatch repair genes in ovarian cancer patients treated with platinum-based chemotherapy. Arch. Gynecol. Obstet. 2018, 297, 153–159. [Google Scholar] [CrossRef]
- Xiao, X.; Melton, D.W.; Gourley, C. Mismatch repair deficiency in ovarian cancer—Molecular characteristics and clinical implications. Gynecol. Oncol. 2014, 132, 506–512. [Google Scholar] [CrossRef] [PubMed]
- Jiricny, J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006, 7, 335–346. [Google Scholar] [CrossRef]
- Shaheen, M.; Allen, C.; Nickoloff, J.A.; Hromas, R. Synthetic lethality: Exploiting the addiction of cancer to DNA repair. Blood 2011, 117, 6074–6082. [Google Scholar] [CrossRef] [PubMed]
- Pabla, N.; Ma, Z.; McIlhatton, M.A.; Fishel, R.; Dong, Z. hMSH2 Recruits ATR to DNA Damage Sites for Activation during DNA Damage-induced Apoptosis. J. Biol. Chem. 2011, 286, 10411–10418. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Yan, L.; Xiao-Fei, L.; Hai-Yan, S.; Juan, C.; Shan, K. Hypermethylation of mismatch repair gene hMSH2 associates with platinum-resistant disease in epithelial ovarian cancer. Clin. Epigenetics 2019, 11, 153. [Google Scholar] [CrossRef]
- Gras, E.; Catasus, L.; Argüelles, R.; Moreno-Bueno, G.; Palacios, J.; Gamallo, C.; Matias-Guiu, X.; Prat, J. Microsatellite instability, MLH-1 promoter hypermethylation, and frameshift mutations at coding mononucleotide repeat microsatellites in ovarian tumors. Cancer 2001, 92, 2829–2836. [Google Scholar] [CrossRef]
- Kawashima, N.; Yoshida, H.; Miwa, M.; Fujiwara, K. MLH1 Is a Prognostic Biomarker for Serous Ovarian Cancer Treated With Platinum- and Taxane-based Chemotherapy. Anticancer Res. 2019, 39, 5505–5513. [Google Scholar] [CrossRef]
- Davis, A.J.; Chen, D.J. DNA double strand break repair via non-homologous end-joining. Transl. Cancer Res. 2013, 2, 130–143. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, Y.; Jiang, M.; Chen, W.; Zhu, X. Significant value of XRCC2 and XRCC9 expression in the prognosis of human ovarian carcinoma. J. Cancer 2021, 12, 6254–6264. [Google Scholar] [CrossRef]
- Kothandapani, A.; Dangeti, V.S.M.N.; Brown, A.R.; Banze, L.A.; Wang, X.-H.; Sobol, R.; Patrick, S.M. Novel Role of Base Excision Repair in Mediating Cisplatin Cytotoxicity. J. Biol. Chem. 2011, 286, 14564–14574. [Google Scholar] [CrossRef]
- Kothandapani, A.; Patrick, S.M. Evidence for base excision repair processing of DNA interstrand crosslinks. Mutat. Res. Mol. Mech. Mutagen. 2012, 743–744, 44–52. [Google Scholar] [CrossRef]
- Abdel-Fatah, T.; Sultana, R.; Abbotts, R.; Hawkes, C.; Seedhouse, C.; Chan, S.; Madhusudan, S. Clinicopathological and functional significance of XRCC1 expression in ovarian cancer. Int. J. Cancer 2012, 132, 2778–2786. [Google Scholar] [CrossRef]
- Lord, C.J.; Ashworth, A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017, 355, 1152–1158. [Google Scholar] [CrossRef]
- Ledermann, J.A.; Drew, Y.; Kristeleit, R.S. Homologous recombination deficiency and ovarian cancer. Eur. J. Cancer 2016, 60, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Coleman, R.L.; Oza, A.M.; Lorusso, D.; Aghajanian, C.; Oaknin, A.; Dean, A.; Colombo, N.; Weberpals, J.I.; Clamp, A.; Scambia, G.; et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017, 390, 1949–1961, Erratum in Lancet 2017, 390, 1948. [Google Scholar] [CrossRef]
- Wu, X.H.; Zhu, J.Q.; Yin, R.T.; Yang, J.X.; Liu, J.H.; Wang, J.; Wu, L.Y.; Liu, Z.L.; Gao, Y.N.; Wang, D.B.; et al. Niraparib maintenance therapy in patients with platinum-sensitive recurrent ovarian cancer using an individualized starting dose (NORA): A randomized, double-blind, placebo-controlled phase III trial(☆). Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2021, 32, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Ledermann, J.; Harter, P.; Gourley, C.; Friedlander, M.; Vergote, I.; Rustin, G.; Scott, C.; Meier, W.; Shapira-Frommer, R.; Safra, T.; et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N. Engl. J. Med. 2012, 366, 1382–1392. [Google Scholar] [CrossRef] [PubMed]
- Gibson, B.A.; Kraus, W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012, 13, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Topatana, W.; Juengpanich, S.; Li, S.; Cao, J.; Hu, J.; Lee, J.; Suliyanto, K.; Ma, D.; Zhang, B.; Chen, M.; et al. Advances in synthetic lethality for cancer therapy: Cellular mechanism and clinical translation. J. Hematol. Oncol. 2020, 13, 1–22. [Google Scholar] [CrossRef]
- Konstantinopoulos, P.A.; Ceccaldi, R.; Shapiro, G.I.; D’Andrea, A.D. Homologous Recombination Deficiency: Exploiting the Fundamental Vulnerability of Ovarian Cancer. Cancer Discov. 2015, 5, 1137–1154. [Google Scholar] [CrossRef]
- Banerjee, S.; Kaye, S.B.; Ashworth, A. Making the best of PARP inhibitors in ovarian cancer. Nat. Rev. Clin. Oncol. 2010, 7, 508–519. [Google Scholar] [CrossRef]
- Murai, J.; NHuang, S.; Brata Das, B.; Renaud, A.; Zhang, Y.; Doroshow, J.H.; Ji, J.; Takeda, S.; Pommier, Y. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012, 72, 5588–5599. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Kang, Y.; Chen, L.; Wang, H.; Liu, J.; Zeng, S.; Yu, L. The Drug-Resistance Mechanisms of Five Platinum-Based Antitumor Agents. Front. Pharmacol. 2020, 11, 343. [Google Scholar] [CrossRef]
- Edwards, S.L.; Brough, R.; Lord, C.J.; Natrajan, R.; Vatcheva, R.; Levine, D.A.; Boyd, J.; Reis-Filho, J.S.; Ashworth, A. Resistance to therapy caused by intragenic deletion in BRCA2. Nat. Cell Biol. 2008, 451, 1111–1115. [Google Scholar] [CrossRef]
- Lord, C.J.; Ashworth, A. BRCAness revisited. Nat. Rev. Cancer 2016, 16, 110–120. [Google Scholar] [CrossRef]
- Pujade-Lauraine, E.; Ledermann, J.; Selle, F.; Gebski, V.; Penson, R.; Oza, A.M.; Korach, J.; Huzarski, T.; Poveda, A.; Pignata, S.; et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SO-LO2/ENGOT-Ov21): A double-blind, randomised, placebo-controlled, phase 3 trial. Lancet 2017, 18, 1274–1284. [Google Scholar] [CrossRef]
- Frenel, J.S.; Kim, J.W.; Berton-Rigaud, D.; Asher, R.; Vidal, L.; Pautier, P.; Ledermann, J.A.; Penson, R.T.; Oza, A.M.; Korach, J.; et al. Efficacy of subsequent chemotherapy for patients with BRCA1/2 mutated platinum-sensitive recurrent epithelial ovarian cancer (EOC) progressing on olaparib vs placebo: The SOLO2/ENGOT Ov-21 trial. Ann. Oncol. 2020, 31 (Suppl. 4), S615. [Google Scholar] [CrossRef]
- Halazonetis, T.D.; Gorgoulis, V.G.; Bartek, J. An Oncogene-Induced DNA Damage Model for Cancer Development. Science 2008, 319, 1352–1355. [Google Scholar] [CrossRef] [PubMed]
- Parmar, K.; Kochupurakkal, B.S.; Lazaro, J.B.; Wang, Z.C.; Palakurthi, S.; Kirschmeier, P.T.; Yang, C.; Sambel, L.A.; Färkkilä, A.; Reznichenko, E.; et al. The CHK1 Inhibitor Prexasertib Exhibits Monotherapy Activity in High-Grade Serous Ovarian Cancer Models and Sensitizes to PARP Inhibition. Clin. Cancer Res. 2019, 25, 6127–6140. [Google Scholar] [CrossRef]
- Lee, J.-M.; Nair, J.; Zimmer, A.; Lipkowitz, S.; Annunziata, C.M.; Merino, M.J.; Swisher, E.M.; I Harrell, M.; Trepel, J.B.; Lee, M.-J.; et al. Prexasertib, a cell cycle checkpoint kinase 1 and 2 inhibitor, in BRCA wild-type recurrent high-grade serous ovarian cancer: A first-in-class proof-of-concept phase 2 study. Lancet Oncol. 2018, 19, 207–215. [Google Scholar] [CrossRef]
- Angius, G.; Tomao, S.; Stati, V.; Vici, P.; Bianco, V.; Tomao, F. Prexasertib, a checkpoint kinase inhibitor: From preclinical data to clinical development. Cancer Chemother. Pharmacol. 2019, 85, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, A.; Hall, S.; Curtin, N.; Drew, Y. Targeting ATR as Cancer Therapy: A new era for synthetic lethality and synergistic combinations? Pharmacol. Ther. 2020, 207, 107450. [Google Scholar] [CrossRef]
- Konstantinopoulos, P.A.; Cheng, S.-C.; Hendrickson, A.E.W.; Penson, R.T.; Schumer, S.T.; Doyle, L.A.; Lee, E.K.; Kohn, E.C.; Duska, L.R.; Crispens, M.A.; et al. Berzosertib plus gemcitabine versus gemcitabine alone in platinum-resistant high-grade serous ovarian cancer: A multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 2020, 21, 957–968. [Google Scholar] [CrossRef]
- Konstantinopoulos, P.A.; da Costa, A.A.B.A.; Gulhan, D.; Lee, E.K.; Cheng, S.C.; Hendrickson, A.E.W.; Kochupurakkal, B.; Kolin, D.L.; Kohn, E.C.; Liu, J.F.; et al. A Replication stress biomarker is associated with response to gemcitabine versus combined gemcitabine and ATR in-hibitor therapy in ovarian cancer. Nat. Commun. 2021, 22, 5574. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.D.; Wethington, S.L.; Pagan, C.; Latif, N.; Tanyi, J.; Martin, L.P.; Morgan, M.; Burger, R.A.; Haggerty, A.; Zarrin, H.; et al. Combination ATR and PARP Inhibitor (CAPRI): A phase 2 study of ceralasertib plus olaparib in patients with recurrent, platinum-resistant epithelial ovarian cancer. Gynecol. Oncol. 2021, 163, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Stewart, J.; Porta, N.; Toms, C.; Leary, A.; Lheureux, S.; Khalique, S.; Tai, J.; Attygalle, A.; Vroobel, K.; et al. ATARI trial: ATR inhibitor in combination with olaparib in gynecological cancers with ARID1A loss or no loss (ENGOT/GYN1/NCRI). Int. J. Gynecol. Cancer 2021, 31, 1471–1475. [Google Scholar] [CrossRef] [PubMed]
- De Witt Hamer, P.C.; Mir, S.E.; Noske, D.; Van Noorden, C.J.; Würdinger, T. WEE1 kinase targeting combined with DNA-damaging cancer therapy catalyzes mitotic catastrophe. Clin. Cancer Res. 2001, 17, 4200–4207. [Google Scholar] [CrossRef]
- Takebe, N.; Naqash, A.R.; O’Sullivan Coyne, G.; Kummar, S.; Do, K.; Bruns, A.; Juwara, L.; Zlott, J.; Rubinstein, L.; Piekarz, R.; et al. Safety, Antitumor Activity, and Biomarker Analysis in a Phase I Trial of the Once-daily Wee1 In-hibitorAdavosertib (AZD1775) in Patients with Advanced Solid Tumors. Clin. Cancer Res. 2021, 27, 3834–3844. [Google Scholar] [CrossRef]
- Moore, K.N.; Chambers, S.K.; Hamilton, E.P.; Chen, L.-M.; Oza, A.M.; Ghamande, S.A.; Konecny, G.E.; Plaxe, S.C.; Spitz, D.L.; Geenen, J.J.; et al. Adavosertib with Chemotherapy in Patients with Primary Platinum-Resistant Ovarian, Fallopian Tube, or Peritoneal Cancer: An Open-Label, Four-Arm, Phase II Study. Clin. Cancer Res. 2021, 27. [Google Scholar] [CrossRef]
- Lheureux, S.; Cristea, M.C.; Bruce, J.P.; Garg, S.; Cabanero, M.; Mantia-Smaldone, G.; Olawaiye, A.B.; Ellard, S.L.; I Weberpals, J.; Hendrickson, A.E.W.; et al. Adavosertib plus gemcitabine for platinum-resistant or platinum-refractory recurrent ovarian cancer: A double-blind, randomised, placebo-controlled, phase 2 trial. Lancet 2021, 397, 281–292. [Google Scholar] [CrossRef]
- Cristescu, R.; Mogg, R.; Ayers, M.; Albright, A.; Murphy, E.; Yearley, J.; Sher, X.; Liu, X.Q.; Lu, H.; Nebozhyn, M.; et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade–based immunotherapy. Science 2018, 362, 6411. [Google Scholar] [CrossRef] [PubMed]
- Pujade-Lauraine, E.; Fujiwara, K.; Ledermann, J.A.; Oza, A.M.; Kristeleit, R.; Ray-Coquard, I.L.; Richardson, G.E.; Sessa, C.; Yonemori, K.; Banerjee, S.; et al. Avelumab alone or in combination with chemotherapy versus chemotherapy alone in platinum-resistant or platinum-refractory ovarian cancer (JAVELIN Ovarian 200): An open-label, three-arm, randomised, phase 3 study. Lancet Oncol. 2021, 22, 1034–1046. [Google Scholar] [CrossRef]
- Moore, K.N.; Bookman, M.; Sehouli, J.; Miller, A.; Anderson, C.; Scambia, G.; Myers, T.; Taskiran, C.; Robison, K.; Mäenpää, J.; et al. Atezolizumab, Bevacizumab, and Chemotherapy for Newly Diagnosed Stage III or IV Ovarian Cancer: Placebo-Controlled Randomized Phase III Trial (IMagyn050/GOG 3015/ENGOT-OV39). J. Clin. Oncol. 2021, 39, 1842–1855. [Google Scholar] [CrossRef]
- Reisländer, T.; Lombardi, E.P.; Groelly, F.J.; Miar, A.; Porru, M.; Di Vito, S.; Wright, B.; Lockstone, H.; Biroccio, A.; Harris, A.; et al. BRCA2 abrogation triggers innate immune responses potentiated by treatment with PARP inhibitors. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Domchek, S.M.; Postel-Vinay, S.; Im, S.-A.; Park, Y.H.; Delord, J.-P.; Italiano, A.; Alexandre, J.; You, B.; Bastian, S.; Krebs, M.G.; et al. Olaparib and durvalumab in patients with germline BRCA-mutated metastatic breast cancer (MEDIOLA): An open-label, multicentre, phase 1/2, basket study. Lancet Oncol. 2020, 21, 1155–1164. [Google Scholar] [CrossRef]
- Konstantinopoulos, P.A.; Waggoner, S.; Vidal, G.A.; Mita, M.; Moroney, J.W.; Holloway, R.; Van Le, L.; Sachdev, J.C.; Chapman-Davis, E.; Colon-Otero, G.; et al. Single-Arm Phases 1 and 2 Trial of Niraparib in Combination with Pembrolizumab in Patients with Recurrent Platinum-Resistant Ovarian Carcinoma. JAMA Oncol. 2019, 5, 1141–1149. [Google Scholar] [CrossRef]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef]
- Marabelle, A.; Le, D.T.; Ascierto, P.A.; Di Giacomo, A.M.; De Jesus-Acosta, A.; Delord, J.-P.; Geva, R.; Gottfried, M.; Penel, N.; Hansen, A.R.; et al. Efficacy of Pembrolizumab in Patients with Noncolorectal High Microsatellite Instability/Mismatch Repair–Deficient Cancer: Results from the Phase II KEYNOTE-158 Study. J. Clin. Oncol. 2020, 38, 1–10. [Google Scholar] [CrossRef]
- Watanabe, Y.; Koi, M.; Hemmi, H.; Hoshai, H.; Noda, K. A change in microsatellite instability caused by cisplatin-based chemotherapy of ovarian cancer. Br. J. Cancer 2001, 85, 1064–1069. [Google Scholar] [CrossRef]
- Varadé, J.; Magadán, S.; González-Fernández, Á. Human immunology and immunotherapy: Main achievements and challenges. Cell. Mol. Immunol. 2020, 18, 805–828. [Google Scholar] [CrossRef]
- Zhu, J.; Yan, L.; Wang, Q. Efficacy of PD-1/PD-L1 inhibitors in ovarian cancer: A single-arm meta-analysis. J. Ovarian Res. 2021, 14, 112. [Google Scholar] [CrossRef] [PubMed]
- Brandsma, I.; Fleuren, E.D.; Williamson, C.T.; Lord, C.J. Directing the use of DDR kinase inhibitors in cancer treatment. Expert Opin. Investig. Drugs 2017, 26, 1341–1355. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stefanou, D.T.; Souliotis, V.L.; Zakopoulou, R.; Liontos, M.; Bamias, A. DNA Damage Repair: Predictor of Platinum Efficacy in Ovarian Cancer? Biomedicines 2022, 10, 82. https://doi.org/10.3390/biomedicines10010082
Stefanou DT, Souliotis VL, Zakopoulou R, Liontos M, Bamias A. DNA Damage Repair: Predictor of Platinum Efficacy in Ovarian Cancer? Biomedicines. 2022; 10(1):82. https://doi.org/10.3390/biomedicines10010082
Chicago/Turabian StyleStefanou, Dimitra T., Vassilis L. Souliotis, Roubini Zakopoulou, Michalis Liontos, and Aristotelis Bamias. 2022. "DNA Damage Repair: Predictor of Platinum Efficacy in Ovarian Cancer?" Biomedicines 10, no. 1: 82. https://doi.org/10.3390/biomedicines10010082
APA StyleStefanou, D. T., Souliotis, V. L., Zakopoulou, R., Liontos, M., & Bamias, A. (2022). DNA Damage Repair: Predictor of Platinum Efficacy in Ovarian Cancer? Biomedicines, 10(1), 82. https://doi.org/10.3390/biomedicines10010082







