NOG-Derived Peptides Can Restore Neuritogenesis on a CRASH Syndrome Cell Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Structural Modeling and Molecular Dynamics
2.2. Peptide Synthesis, Purification and Characterization
2.3. Cell Culture and Peptide Treatment
2.4. L1CAM Clones and SH-SY5Y Transfection
2.5. Neuritogenesis Assays
2.6. Immunofluorescence
2.7. Statistical Analysis
3. Results and Discussion
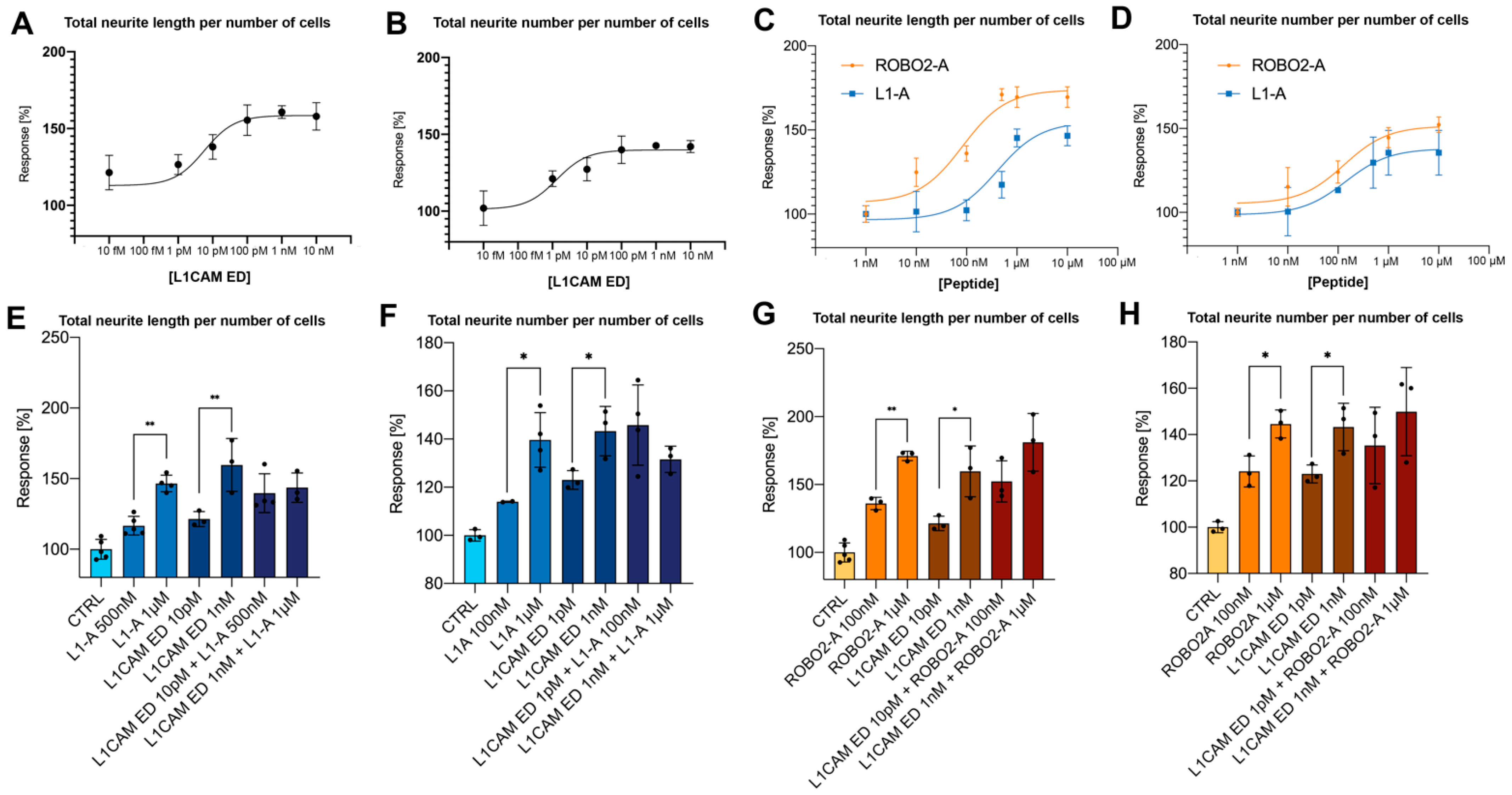
3.1. NOG-Derived Peptides and L1CAM ED Show Comparable Proneuritogenic Capacity
3.2. L1-A Peptides with Known L1CAM Mutations Show Impaired Neuritogenic Capacity
3.3. CRASH Mutations Differentially Impact on L1CAM Proneuritogenic Activity
3.4. In Silico Simulations Suggest a Rationale for CRASH Differential Severity and Translational Perspectives
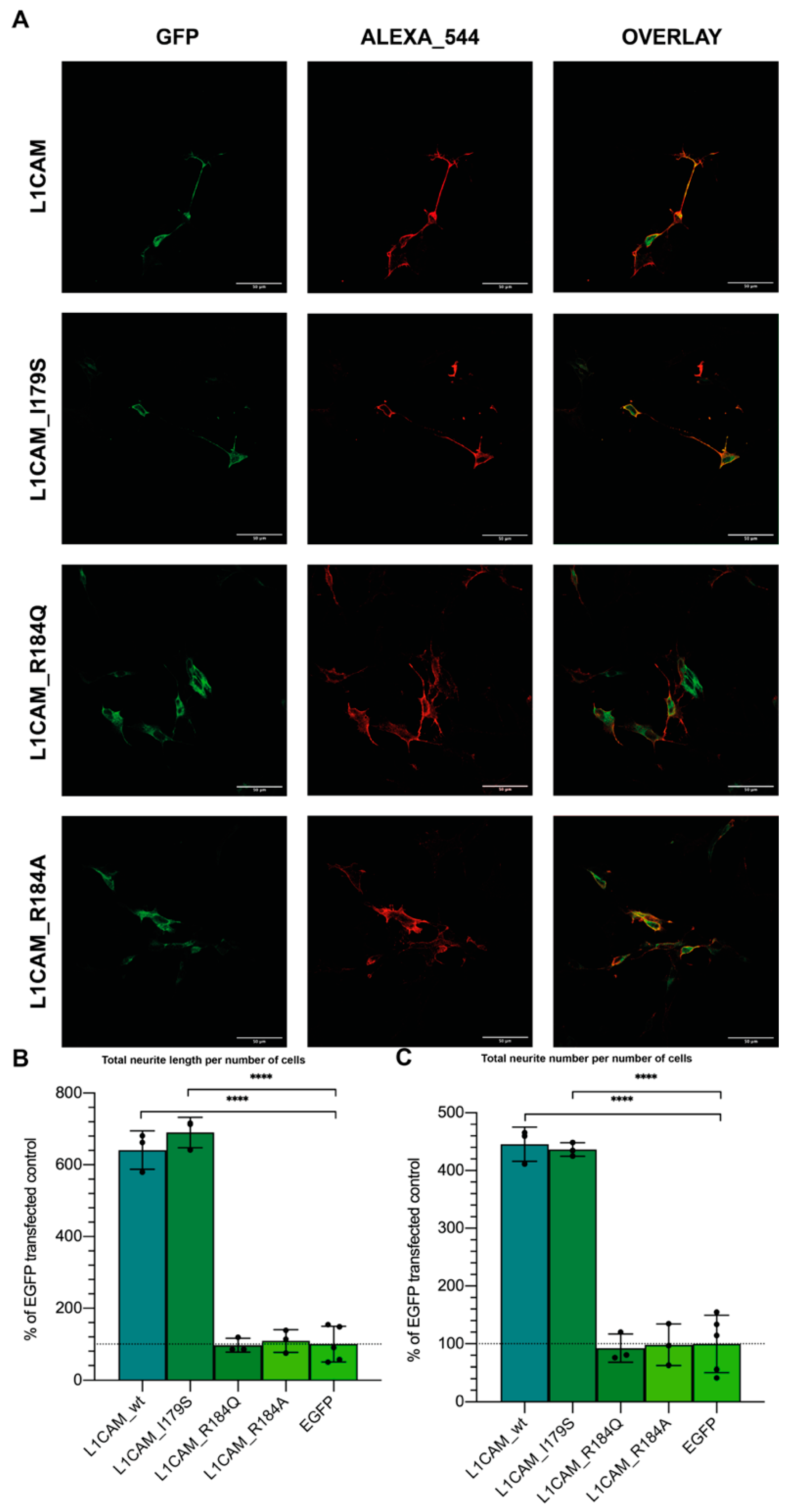
3.5. L1-A Can Restore Neuritogenesis on a CRASH Syndrome Cell Model, While L1CAM ED Is Unable
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- McFarlane, S. Attraction vs. repulsion: The growth cone decides. Biochem. Cell Biol. 2000, 78, 563–568. [Google Scholar] [CrossRef]
- Zhang, Y.; Yeh, J.; Richardson, P.M.; Bo, X. Cell adhesion molecules of the immunoglobulin superfamily in axonal regeneration and neural repair. Restor. Neurol. Neurosci. 2008, 26, 81–96. [Google Scholar]
- Hortsch, M.; Nagaraj, K.; Mualla, R. The L1 family of cell adhesion molecules: A sickening number of mutations and protein functions. Adv. Neurobiol. 2014, 8, 195–229. [Google Scholar] [CrossRef]
- Abe, K.; Katsuno, H.; Toriyama, M.; Baba, K.; Mori, T.; Hakoshima, T.; Kanemura, Y.; Watanabe, R.; Inagaki, N. Grip and slip of L1-CAM on adhesive substrates direct growth cone haptotaxis. Proc. Natl. Acad. Sci. USA 2018, 115, 2764–2769. [Google Scholar] [CrossRef]
- Duncan, B.W.; Murphy, K.E.; Maness, P.F. Molecular Mechanisms of L1 and NCAM Adhesion Molecules in Synaptic Pruning, Plasticity, and Stabilization. Front. Cell Dev. Biol. 2021, 9, 625340. [Google Scholar] [CrossRef]
- Liu, H.; Focia, P.J.; He, X. Homophilic adhesion mechanism of neurofascin, a member of the L1 family of neural cell adhesion molecules. J. Biol. Chem. 2011, 286, 797–805. [Google Scholar] [CrossRef]
- He, Y.; Jensen, G.J.; Bjorkman, P.J. Cryo-electron tomography of homophilic adhesion mediated by the neural cell adhesion molecule L1. Structure 2009, 17, 460–471. [Google Scholar] [CrossRef][Green Version]
- Gouveia, R.M.; Gomes, C.M.; Sousa, M.; Alves, P.M.; Costa, J. Kinetic analysis of L1 homophilic interaction: Role of the first four immunoglobulin domains and implications on binding mechanism. J. Biol. Chem. 2008, 283, 28038–28047. [Google Scholar] [CrossRef]
- Itoh, K.; Fushiki, S. The role of L1cam in murine corticogenesis, and the pathogenesis of hydrocephalus. Pathol. Int. 2015, 65, 58–66. [Google Scholar] [CrossRef]
- Zhang, L. CRASH syndrome: Does it teach us about neurotrophic functions of cell adhesion molecules? Neurosci. Rev. J. Bringing Neurobiol. Neurol. Psychiatry 2010, 16, 470–474. [Google Scholar] [CrossRef]
- Vos, Y.J.; Hofstra, R.M.W. An updated and upgraded L1CAM mutation database. Hum. Mutat. 2010, 31, E1102–E1109. [Google Scholar] [CrossRef]
- Christaller, W.A.A.; Vos, Y.; Gebre-Medhin, S.; Hofstra, R.M.W.; Schäfer, M.K.E. L1 syndrome diagnosis complemented with functional analysis of L1CAM variants located to the two N-terminal Ig-like domains. Clin. Genet. 2017, 91, 115–120. [Google Scholar] [CrossRef]
- Vos, Y.J.; de Walle, H.E.K.; Bos, K.K.; Stegeman, J.A.; Ten Berge, A.M.; Bruining, M.; van Maarle, M.C.; Elting, M.W.; den Hollander, N.S.; Hamel, B.; et al. Genotype-phenotype correlations in L1 syndrome: A guide for genetic counselling and mutation analysis. J. Med. Genet. 2010, 47, 169–175. [Google Scholar] [CrossRef]
- Gu, S.M.; Orth, U.; Veske, A.; Enders, H.; Klunder, K.; Schlosser, M.; Engel, W.; Schwinger, E.; Gal, A. Five novel mutations in the L1CAM gene in families with X linked hydrocephalus. J. Med. Genet. 1996, 33, 103–106. [Google Scholar] [CrossRef][Green Version]
- Jouet, M.; Rosenthal, A.; Armstrong, G.; MacFarlane, J.; Stevenson, R.; Paterson, J.; Metzenberg, A.; Ionasescu, V.; Temple, K.; Kenwrick, S. X-linked spastic paraplegia (SPG1), MASA syndrome and X-linked hydrocephalus result from mutations in the L1 gene. Nat. Genet. 1994, 7, 402–407. [Google Scholar] [CrossRef]
- De Angelis, E.; MacFarlane, J.; Du, J.-S.; Yeo, G.; Hicks, R.; Rathjen, F.G.; Kenwrick, S.; Brümmendorf, T. Pathological missense mutations of neural cell adhesion molecule L1 affect homophilic and heterophilic binding activities. EMBO J. 1999, 18, 4744–4753. [Google Scholar] [CrossRef]
- Zhao, X.; Yip, P.M.; Siu, C.H. Identification of a homophilic binding site in immunoglobulin-like domain 2 of the cell adhesion molecule L1. J. Neurochem. 1998, 71, 960–971. [Google Scholar] [CrossRef]
- Fryns, J.P.; Spaepen, A.; Cassiman, J.J.; van den Berghe, H. X linked complicated spastic paraplegia, MASA syndrome, and X linked hydrocephalus owing to congenital stenosis of the aqueduct of Sylvius: Variable expression of the same mutation at Xq28. J. Med. Genet. 1991, 28, 429–431. [Google Scholar] [CrossRef][Green Version]
- Ruiz, J.C.; Cuppens, H.; Legius, E.; Fryns, J.P.; Glover, T.; Marynen, P.; Cassiman, J.J. Mutations in L1-CAM in two families with X linked complicated spastic paraplegia, MASA syndrome, and HSAS. J. Med. Genet. 1995, 32, 549–552. [Google Scholar] [CrossRef]
- Michelson, P.; Hartwig, C.; Schachner, M.; Gal, A.; Veske, A.; Finckh, U. Missense mutations in the extracellular domain of the human neural cell adhesion molecule L1 reduce neurite outgrowth of murine cerebellar neurons. Hum. Mutat. 2002, 20, 481–482. [Google Scholar] [CrossRef]
- Scapin, G.; Salice, P.; Tescari, S.; Menna, E.; De Filippis, V.; Filippini, F. Enhanced neuronal cell differentiation combining biomimetic peptides and a carbon nanotube-polymer scaffold. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 621–632. [Google Scholar] [CrossRef]
- Scapin, G.; Gasparotto, M.; Peterle, D.; Tescari, S.; Porcellato, E.; Piovesan, A.; Righetto, I.; Acquasaliente, L.; De Filippis, V.; Filippini, F. A conserved Neurite Outgrowth and Guidance motif with biomimetic potential in neuronal Cell Adhesion Molecules. Comput. Struct. Biotechnol. J. 2021, 19, 5622–5636. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2005, 22, 195–201. [Google Scholar] [CrossRef]
- Kiefer, F.; Arnold, K.; Künzli, M.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2008, 37, D387–D392. [Google Scholar] [CrossRef]
- Wang, Q.; Canutescu, A.A.; Dunbrack, R.L. SCWRL and MolIDE: Computer programs for side-chain conformation prediction and homology modeling. Nat. Protoc. 2008, 3, 1832–1847. [Google Scholar] [CrossRef]
- Canutescu, A.A.; Shelenkov, A.A.; Dunbrack, R.L. A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003, 12, 2001–2014. [Google Scholar] [CrossRef]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2010, 27, 343–350. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindah, E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T.; et al. A Point-Charge Force Field for Molecular Mechanics Simulations of Proteins Based on Condensed-Phase Quantum Mechanical Calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef]
- Berendsen, H.; Postma, J.P.M.; van Gunsteren, W.; DiNola, A.D.; Haak, J.R. Molecular-Dynamics with Coupling to An External Bath. J. Chem. Phys. 1984, 81, 3684. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Huang, C.C.; Meng, E.C.; Morris, J.H.; Pettersen, E.F.; Ferrin, T.E. Enhancing UCSF Chimera through web services. Nucleic Acids Res. 2014, 42, W478–W484. [Google Scholar] [CrossRef]
- Lavi, A.; Ngan, C.H.; Movshovitz-Attias, D.; Bohnuud, T.; Yueh, C.; Beglov, D.; Schueler-Furman, O.; Kozakov, D. Detection of peptide-binding sites on protein surfaces: The first step toward the modeling and targeting of peptide-mediated interactions. Proteins Struct. Funct. Bioinform. 2013, 81, 2096–2105. [Google Scholar] [CrossRef]
- Brenke, R.; Kozakov, D.; Chuang, G.-Y.; Beglov, D.; Hall, D.; Landon, M.R.; Mattos, C.; Vajda, S. Fragment-based identification of druggable “hot spots” of proteins using Fourier domain correlation techniques. Bioinformatics 2009, 25, 621–627. [Google Scholar] [CrossRef]
- Schrödinger, LLC. The {PyMOL} Molecular Graphics System, Version 1.8; Schrödinger, LLC: New York, NY, USA, 2015. [Google Scholar]
- Atherton, E. Solid Phase Peptide Synthesis: A Practical Approach; IRL Press: Oxford, UK, 1989. [Google Scholar]
- De Filippis, V.; Quarzago, D.; Vindigni, A.; Di Cera, E.; Fontana, A. Synthesis and characterization of more potent analogues of hirudin fragment 1-47 containing non-natural amino acids. Biochemistry 1998, 37, 13507–13515. [Google Scholar] [CrossRef]
- Ross, R.A.; Spengler, B.A.; Biedler, J.L. Coordinate Morphological and Biochemical Interconversion of Human Neuroblastoma Cells2. JNCI J. Natl. Cancer Inst. 1983, 71, 741–747. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Hu, H.; Ni, Y.; Montana, V.; Haddon, R.C.; Parpura, V. Chemically Functionalized Carbon Nanotubes as Substrates for Neuronal Growth. Nano Lett. 2004, 4, 507–511. [Google Scholar] [CrossRef]
- Munnamalai, V.; Weaver, C.J.; Weisheit, C.E.; Venkatraman, P.; Agim, Z.S.; Quinn, M.T.; Suter, D.M. Bidirectional interactions between NOX2-type NADPH oxidase and the F-actin cytoskeleton in neuronal growth cones. J. Neurochem. 2014, 130, 526–540. [Google Scholar] [CrossRef]
- Yamasaki, M.; Thompson, P.; Lemmon, V. CRASH Syndrome: Mutations in L1CAM Correlate with Severity of the Disease. Neuropediatrics 1997, 28, 175–178. [Google Scholar] [CrossRef]
- Romano, N.H.; Madl, C.M.; Heilshorn, S.C. Matrix RGD ligand density and L1CAM-mediated Schwann cell interactions synergistically enhance neurite outgrowth. Acta Biomater. 2015, 11, 48–57. [Google Scholar] [CrossRef]
- Bellet, P.; Gasparotto, M.; Pressi, S.; Fortunato, A.; Scapin, G.; Mba, M.; Menna, E.; Filippini, F. Graphene-based scaffolds for regenerative medicine. Nanomaterials 2021, 11, 404. [Google Scholar] [CrossRef]
- De Angelis, E.; Watkins, A.; Schäfer, M.; Brümmendorf, T.; Kenwrick, S. Disease-associated mutations in L1 CAM interfere with ligand interactions and cell-surface expression. Hum. Mol. Genet. 2002, 11, 1–12. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gasparotto, M.; Hernandez Gomez, Y.S.; Peterle, D.; Grinzato, A.; Zen, F.; Pontarollo, G.; Acquasaliente, L.; Scapin, G.; Bergantino, E.; De Filippis, V.; et al. NOG-Derived Peptides Can Restore Neuritogenesis on a CRASH Syndrome Cell Model. Biomedicines 2022, 10, 102. https://doi.org/10.3390/biomedicines10010102
Gasparotto M, Hernandez Gomez YS, Peterle D, Grinzato A, Zen F, Pontarollo G, Acquasaliente L, Scapin G, Bergantino E, De Filippis V, et al. NOG-Derived Peptides Can Restore Neuritogenesis on a CRASH Syndrome Cell Model. Biomedicines. 2022; 10(1):102. https://doi.org/10.3390/biomedicines10010102
Chicago/Turabian StyleGasparotto, Matteo, Yuriko Suemi Hernandez Gomez, Daniele Peterle, Alessandro Grinzato, Federica Zen, Giulia Pontarollo, Laura Acquasaliente, Giorgia Scapin, Elisabetta Bergantino, Vincenzo De Filippis, and et al. 2022. "NOG-Derived Peptides Can Restore Neuritogenesis on a CRASH Syndrome Cell Model" Biomedicines 10, no. 1: 102. https://doi.org/10.3390/biomedicines10010102
APA StyleGasparotto, M., Hernandez Gomez, Y. S., Peterle, D., Grinzato, A., Zen, F., Pontarollo, G., Acquasaliente, L., Scapin, G., Bergantino, E., De Filippis, V., & Filippini, F. (2022). NOG-Derived Peptides Can Restore Neuritogenesis on a CRASH Syndrome Cell Model. Biomedicines, 10(1), 102. https://doi.org/10.3390/biomedicines10010102







