An overview of Structured Biosensors for Metal Ions Determination
Abstract
:1. Introduction
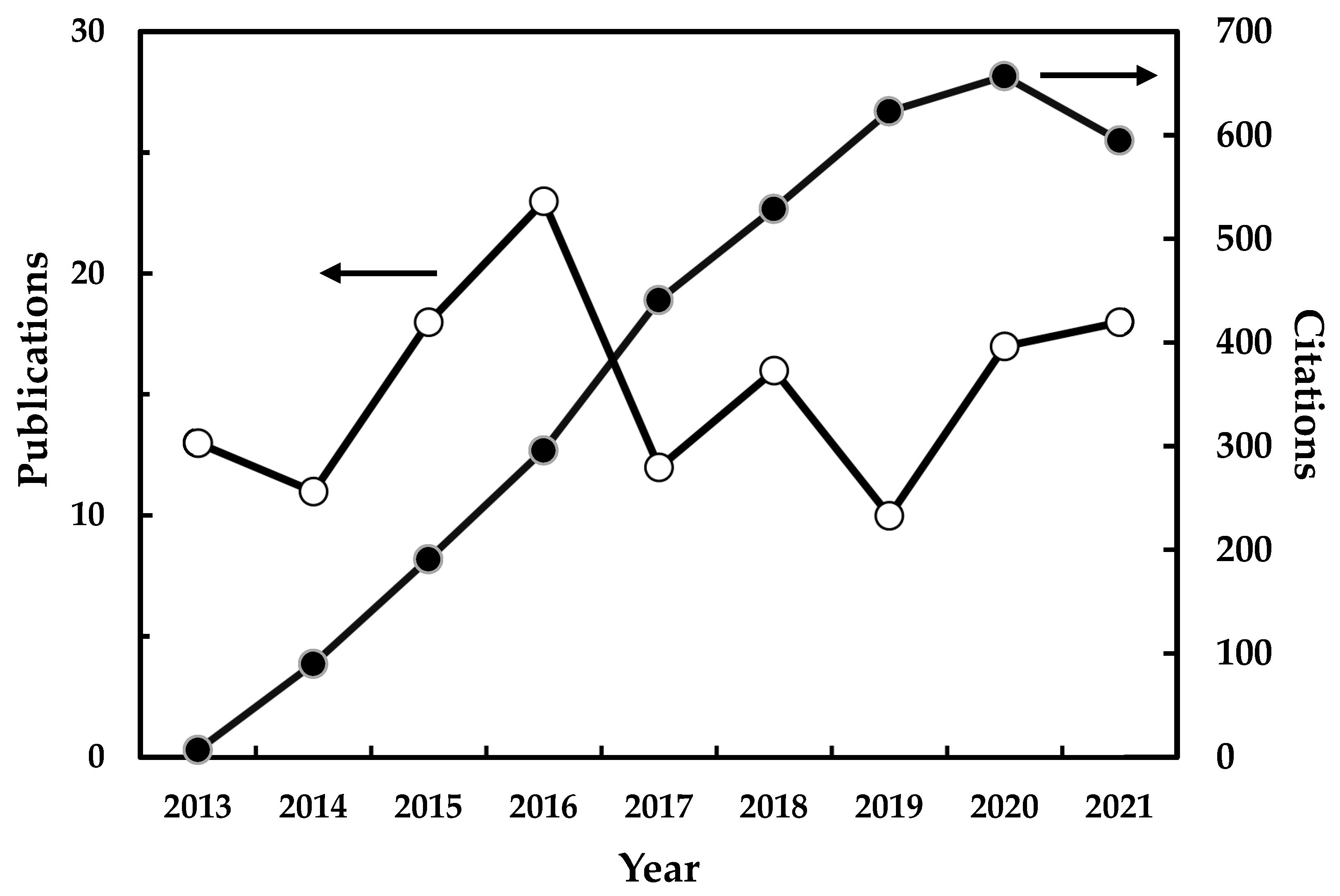
2. General Aspects and Statistics
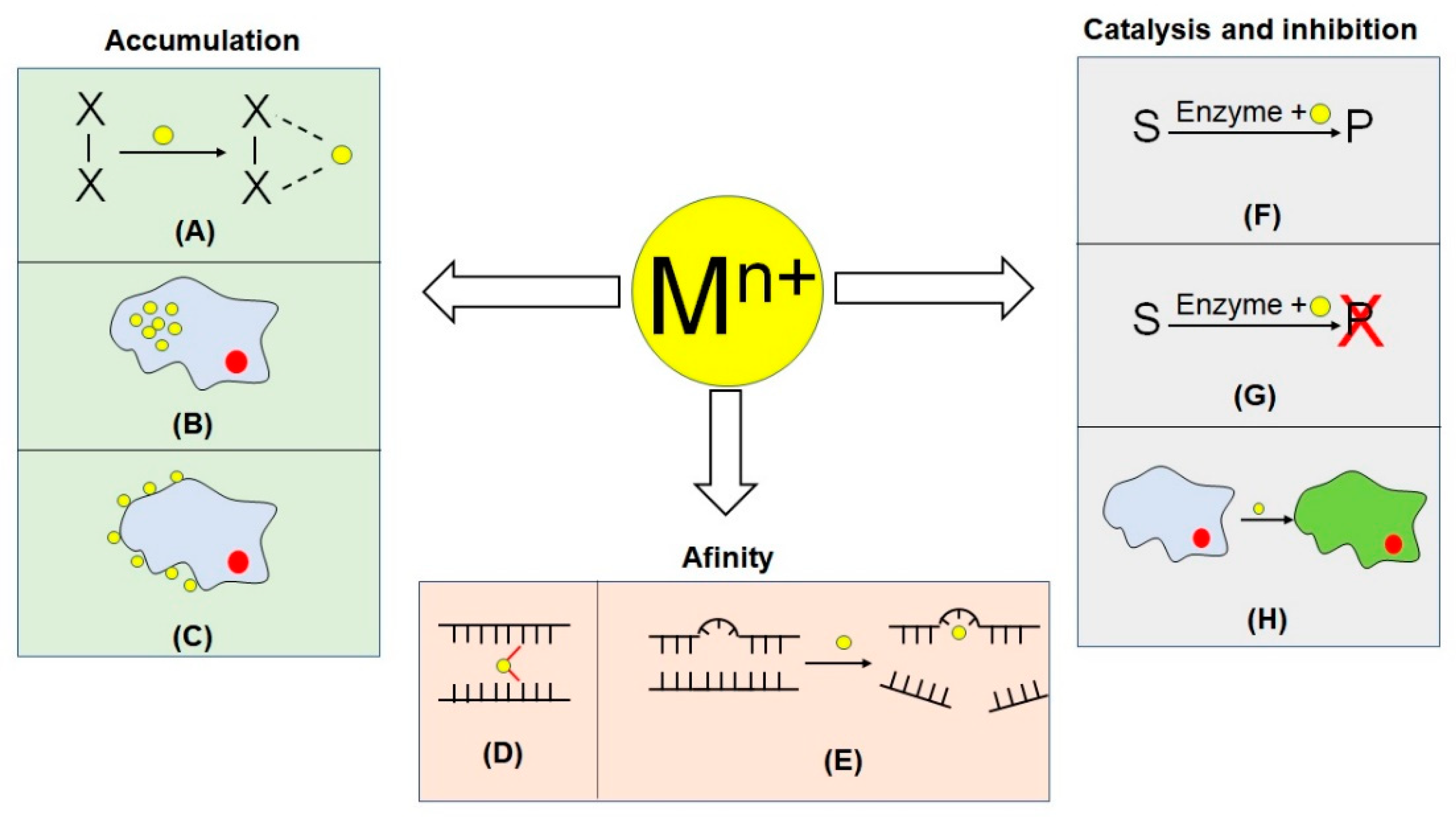
3. Biorecognition Mechanisms and Features
3.1. Metal Ion Accumulation
3.2. Catalysis and Inhibition
3.3. Affinity
4. Biosensors Platforms
4.1. Electrochemical Sensors
4.2. Optical and Piezoelectric Sensors
5. Nanomaterials in Biosensing Detection
6. Multielement Analysis
7. Conclusions and Trends
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABTS | 2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) |
| ACV | alternate current voltammetry |
| AFM | atomic force microscopy |
| AP | amperometry |
| APM | amino polymerization method |
| APTS | 3-aminopropyltriethoxysilane |
| ASV | anodic stripping voltammetry |
| BSA | bovine serum albumine |
| CA | chronoamperometry |
| CC | chronocoulometry |
| CD | conductometry |
| CH | cobalt(II) hexacyanoferrate |
| CNT | carbon nanotubes |
| CPE | carbon paste electrode |
| CSV | cathodic stripping voltammetry |
| CV | cyclic voltammetry |
| DI | digital imaging |
| DL | detection limit |
| DNA | desoxyribonucleic acid |
| DPV | differential pulse voltammetry |
| ECL | electrochemiluminescence |
| EDC | N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide |
| EDTA | ethylenediaminotetracetic acid |
| EG | ethyl green |
| EIS | electrochemical impedance spectroscopy |
| FET | field-effect transistor |
| FL | fluorescence |
| GCE | glassy carbon electrode |
| GE | gold electrode |
| GNP | gold nanoparticles |
| HRP | horseradish peroxidase |
| IRS | infrared reflectance spectrometry |
| ITCBE | 1-(4-Isothiocyanobenzyl)ethylenediamine-N,N,N′,N′-tetraacetic acid |
| ITO | indium tin oxide |
| LSV | linear sweep voltammetry |
| MA | monoclonal antibody |
| MAS | molecular absorption spectrometry |
| MB | methylene blue |
| MFC | microbial fuel cell |
| MG | methyl green |
| MOF | metal-organic frameworks |
| NADH | reduced nicotinamide adenine dinucleotide |
| NHS | N-Hydroxysuccinimide |
| NPOE | nitrophenyloctylether |
| NR | neutral red |
| PDDA | polydiallyldimethylammonium |
| PEC | photoelectrochemical detection |
| PGE | pencil graphite electrode |
| PT | potentiometry |
| PTFE | polytetrafluorethylene |
| PVC | polyvinyl chloride |
| QM | quartz microbalance |
| SERS | surface-enhanced Raman spectrometry |
| SPDP | succinimydil-3-(2-pyridyldithiol) propionate |
| SPE | screen-printed electrode |
| SPR | surface plasmon resonance |
| SWV | square wave voltammetry |
References
- McNaught, A.D.; Wilkinson, A. IUPAC Compendium of Chemical Terminology, 2nd ed.; The “Gold Book”; Blackwell Scientific Publications: Oxford, UK, 1997; ISBN 0-9678550-9-8. [Google Scholar]
- Nagel, B.; Dellweg, H.; Gierasch, L.M. IUPAC Glossary for chemist for terms used in biotechnology. Compend. Chem. Terminol. 1997, 64, 143–168. [Google Scholar] [CrossRef]
- Cui, L.; Wu, J.; Ju, H. Electrochemical sensing of heavy metal ions with in-organic, organic and biomaterials. Biosens. Bioelectron. 2015, 63, 276–286. [Google Scholar] [CrossRef]
- Chambers, J.P.; Arulanandam, B.P.; Matta, L.L.; Weis, A.; Valdes, J.J. Biosensor recognition elements. Curr. Issues Mol. Biol. 2002, 10, 1–12. [Google Scholar]
- Calvo-Pérez, A.; Domínguez-Renedo, O.; Alonso-Lomillo, M.; Arcos-Martínez, M. Speciation of chromium using chronoamperometric biosensors based on screen-printed electrodes. Anal. Chim. Acta 2014, 833, 15–21. [Google Scholar] [CrossRef]
- Prabhakaran, D.C.; Ramamurthy, P.C.; Sivry, Y.; Subramanian, S. Electro-chemical detection of Cr(VI) and Cr(III) ions present in aqueous solutions using bio-modified carbon paste electrode: A voltammetric study. Int. J. Environ. Anal. Chem. 2020, 1–21. [Google Scholar] [CrossRef]
- Tadi, K.K.; Alshanski, I.; Mervinetsky, E.; Marx, G.; Petrou, P.; Dimitrios, K.M.; Gilon, C.; Hurevich, M.; Yitzchaik, S. Oxytocin-Monolayer-Based Impedimetric Biosensor for Zinc and Copper Ions. ACS Omega 2017, 2, 8770–8778. [Google Scholar] [CrossRef] [Green Version]
- Ghica, M.E.; Carvalho, R.C.; Amine, A.; Brett, C.M.A. Glucose oxidase enzyme inhibition sensors for heavy metals at carbon film electrodes modified with cobalt or copper hexacyanoferrate. Sens. Actuators B Chem. 2013, 178, 270–278. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, S.; Ye, Z.; Peng, D.; He, L.; Yan, F.; Yang, Y.; Zhang, H.; Zhang, Z. A gold electrode modified with amino-modified reduced graphene oxide, ion specific DNA and DNAzyme for dual electrochemical determination of Pb(II) and Hg(II). Microchim. Acta 2015, 182, 2251–2258. [Google Scholar] [CrossRef]
- Do, J.S.; Lin, K.H. Kinetics of urease inhibition-based amperometric biosensors for mercury and lead ions detection. J. Taiwan Inst. Chem. Eng. 2016, 63, 25–32. [Google Scholar] [CrossRef]
- da Silva, W.; Ghica, M.E.; Brett, C.M.A. Biotoxic trace metal ion detection by enzymatic inhibition of a glucose biosensor based on a poly(brilliant green)–deep eutectic solvent/carbon nanotube modified electrode. Talanta 2020, 208, 120427. [Google Scholar] [CrossRef]
- Wong, L.S.; Lee, Y.H.; Surif, S. Whole cell biosensor using Anabaena torulosa with optical transduction for environmental toxicity evaluation. J. Sens. 2013, 2013, 567272. [Google Scholar] [CrossRef]
- Ayenimo, J.G.; Adeloju, S.B. Rapid amperometric detection of trace metals by inhibition of an ultrathin polypyrrole-based glucose biosensor. Talanta 2016, 148, 502–510. [Google Scholar] [CrossRef]
- Ilangovan, R.; Daniel, D.; Krastanov, A.; Zachariah, C.; Elizabeth, R. Enzyme based biosensor for heavy metal ions determination. Biotechnol. Biotechnol. Equip. 2006, 20, 184–189. [Google Scholar] [CrossRef]
- Attar, A.; Ghica, M.E.; Amine, A.; Brett, C.M.A. Comparison of Cobalt Hexacyanoferrate and Poly(Neutral Red) Modified Carbon Film Electrodes for the Amperometric Detection of Heavy Metals Based on Glucose Oxidase Enzyme Inhibition. Anal. Lett. 2015, 48, 659–671. [Google Scholar] [CrossRef]
- Silwana, B.; Van Der Horst, C.; Iwuoha, E.; Somerset, V. Amperometric determination of cadmium, lead, and mercury metal ions using a novel polymer immobilized horseradish peroxidase biosensor system. J. Environ. Sci. Health Part A Toxic/Hazard. Subst. Environ. Eng. 2014, 49, 1501–1511. [Google Scholar] [CrossRef]
- Baghayeri, M.; Amiri, A.; Maleki, B.; Alizadeh, Z.; Reiser, O. A simple approach for simultaneous detection of cadmium(II) and lead(II) based on glutathione coated magnetic nanoparticles as a highly selective electrochemical probe. Sens. Actuators B Chem. 2018, 273, 1442–1450. [Google Scholar] [CrossRef]
- Ayenimo, J.G.; Adeloju, S.B. Inhibitive potentiometric detection of trace metals with ultrathin polypyrrole glucose oxidase biosensor. Talanta 2015, 137, 62–70. [Google Scholar] [CrossRef]
- Moyo, M.; Okonkwo, J.O.; Agyei, N.M. An amperometric biosensor based on horseradish peroxidase immobilized onto maize tassel-multi-walled carbon nanotubes modified glassy carbon electrode for determination of heavy metal ions in aqueous solution. Enzym. Microb. Technol. 2014, 56, 28–34. [Google Scholar] [CrossRef]
- Kim, H.; Jang, G.; Yoon, Y. Specific heavy metal/metalloid sensors: Current state and perspectives. Appl. Microbiol. Biotechnol. 2020, 104, 907–914. [Google Scholar] [CrossRef]
- Butcher, D.J. Review: Recent advances in optical analytical atomic spectrometry. Appl. Spectrosc. Rev. 2013, 48, 261–328. [Google Scholar] [CrossRef]
- Zou, Z.; Deng, Y.; Hu, J.; Jiang, X.; Hou, X. Recent trends in atomic fluorescence spectrometry towards miniaturized instrumentation-A review. Anal. Chim. Acta 2018, 1019, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Carter, S.; Clough, R.; Fisher, A.; Gibson, B.; Russell, B.; Waack, J. Atomic spectrometry update: Review of advances in the analysis of metals, chemicals and materials. J. Anal. At. Spectrom. 2020, 35, 2410–2474. [Google Scholar] [CrossRef]
- Wang, L.; Peng, X.; Fu, H.; Huang, C.; Li, Y.; Liu, Z. Recent advances in the development of electrochemical aptasensors for detection of heavy metals in food. Biosens. Bioelectron. 2020, 147, 111777. [Google Scholar] [CrossRef]
- Domínguez-Renedo, O.; Alonso-Lomillo, M.A.; Arcos-Martínez, M.J. Determination of metals based on electrochemical biosensors. Crit. Rev. Environ. Sci. Technol. 2013, 43, 1042–1073. [Google Scholar] [CrossRef]
- Ma, S.; Zhang, Q.; Wu, D.; Hu, Y.; Hu, D.; Guo, Z.; Wang, S.; Liu, Q.; Peng, J. Unique G4-nanowires-mediated switch-modulated electrochemical biosensing for sensitive detection of nickel ion and histidine. J. Electroanal. Chem. 2019, 847, 113144. [Google Scholar] [CrossRef]
- Rapini, R.; Canfarotta, F.; Mazzotta, E.; Malitesta, C.; Marrazza, G.; Piletsky, S.; Piletska, E. NanoMIP-based approach for the suppression of interference signals in electrochemical sensors. Analyst 2019, 144, 7290–7295. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Li, F.Y.; Dong, X.; Zhang, J.; Xiong, Q.; Chen, P. The Electrical Detection of Lead Ions Using Gold-Nanoparticle- and DNAzyme-Functionalized Graphene Device. Adv. Healthc. Mater. 2013, 2, 271–274. [Google Scholar] [CrossRef]
- Grygo-Szymanko, E.; Tobiasz, A.; Walas, S. Speciation analysis and fractionation of manganese: A review. TrAC Trends Anal. Chem. 2016, 80, 112–124. [Google Scholar] [CrossRef]
- Cámara-Martos, F.; Da Costa, J.; Justino, C.I.L.; Cardoso, S.; Duarte, A.C.; Rocha-Santos, T. Disposable biosensor for detection of Iron(III) in wines. Talanta 2016, 154, 80–84. [Google Scholar] [CrossRef]
- Punekar, N.S. Enzymes: Catalysis, Kinetics and Mechanisms; Springer Nature: Singapore, 2018. [Google Scholar]
- EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA). Scientific Opinion on Dietary Reference Values for Calcium. EFSA J. 2015, 13, 4101. [Google Scholar] [CrossRef] [Green Version]
- Bieleski, R.L. Phosphate Pools, Phosphate Transport, and Phosphate Availability. Annu. Rev. Plant Physiol. 1973, 24, 225–252. [Google Scholar] [CrossRef]
- EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA). Scientific opinion on dietary reference values for chromium. EFSA J. 2014, 12, 3845. [Google Scholar] [CrossRef] [Green Version]
- WHO. WHO Guidelines for Drinking Water Quality, 4th ed.; World Health Organization: Geneva, Switzerland, 2011. [Google Scholar]
- Taghdisi, S.M.; Danesh, N.M.; Lavaee, P.; Ramezani, M.; Abnous, K. An electrochemical aptasensor based on gold nanoparticles, thionine and hairpin structure of complementary strand of aptamer for ultrasensitive detection of lead. Sens. Actuators B Chem. 2016, 234, 462–469. [Google Scholar] [CrossRef]
- Ma, R.N.; Wang, L.L.; Zhang, M.; Jia, L.P.; Zhang, W.; Shang, L.; Jia, W.L.; Wang, H.S. A novel one-step triggered “signal-on/off” electrochemical sensing platform for lead based on the dual-signal ratiometric output and electrode-bound DNAzyme assembly. Sens. Actuators B Chem. 2018, 257, 678–684. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, P.; Zhu, X.; Peng, Q.; Zhou, Y.; Yin, T.; Liang, Y.; Yin, X. Combined determination of copper ions and β-amyloid peptide by a single ratiometric electrochemical biosensor. Analyst 2018, 143, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Wustoni, S.; Hideshima, S.; Kuroiwa, S.; Nakanishi, T.; Mori, Y.; Osaka, T. Label-free detection of Cu(II) in a human serum sample by using a prion protein-immobilized FET sensor. Analyst 2015, 140, 6485–6488. [Google Scholar] [CrossRef]
- Shi, L.; Wang, Y.; Ding, S.; Chu, Z.; Yin, Y.; Jiang, D.; Luo, J.; Jin, W. A facile and green strategy for preparing newly-designed 3D graphene/gold film and its application in highly efficient electrochemical mercury assay. Biosens. Bioelectron. 2017, 89, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Xiong, E.; Wu, L.; Zhou, J.; Yu, P.; Zhang, X.; Chen, J. A ratiometric electrochemical biosensor for sensitive detection of Hg2+ based on thymine-Hg2+-thymine structure. Anal. Chim. Acta 2015, 853, 242–248. [Google Scholar] [CrossRef]
- Gao, X.; Huang, H.; Niu, S.; Ye, H.; Lin, Z.; Qiu, B.; Chen, G. Determination of magnesium ion in serum samples by a DNAzyme-based electrochemical biosensor. Anal. Methods 2012, 4, 947–952. [Google Scholar] [CrossRef]
- Nie, J.; He, B.; Zang, Y.; Yin, W.; Han, L.; Li, W.; Hou, C.; Huo, D.; Yang, M.; Fa, H. A multi-functional minimally-disruptive portable electrochemical system based on yeast/Co3O4/Au/SPEs for blood lead(II) measurement. Bioelectrochemistry 2019, 126, 156–162. [Google Scholar] [CrossRef]
- Taghdisi, S.M.; Danesh, N.M.; Ramezani, M.; Alibolandi, M.; Abnous, K. Voltammetric determination of lead(II) by using exonuclease III and gold nanoparticles, and by exploiting the conformational change of the complementary strand of an aptamer. Microchim. Acta 2017, 184, 2783–2790. [Google Scholar] [CrossRef]
- Liao, X.; Luo, J.; Wu, J.; Fan, T.; Yao, Y.; Gao, F.; Qian, Y. A sensitive DNAzyme-based electrochemical sensor for Pb2+ detection with platinum nanoparticles decorated TiO2/α-Fe2O3 nanocomposite as signal labels. J. Electroanal. Chem. 2018, 829, 129–137. [Google Scholar] [CrossRef]
- Wang, X.; Gao, W.; Yan, W.; Li, P.; Zou, H.; Wei, Z.; Guan, W.; Ma, Y.; Wu, S.; Yu, Y.; et al. A Novel Aptasensor Based on Graphene/Graphite Carbon Nitride Nanocomposites for Cadmium Detection with High Selectivity and Sensitivity. ACS Appl. Nano Mater. 2018, 1, 2341–2346. [Google Scholar] [CrossRef]
- Wang, N.; Dai, H.; Wang, D.; Ma, H.; Lin, M. Determination of copper ions using a phytic acid/polypyrrole nanowires modified glassy carbon electrode. Mater. Sci. Eng. C 2017, 76, 139–143. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, H.; Jiang, X.; Sun, B.; Song, B.; Su, Y.; He, Y. Ultrasensitive, Specific, Recyclable, and Reproducible Detection of Lead Ions in Real Systems through a Polyadenine-Assisted, Surface-Enhanced Raman Scattering Silicon Chip. Anal. Chem. 2016, 88, 3723–3729. [Google Scholar] [CrossRef]
- Chay, T.C.; Surif, S.; Heng, L.Y. A copper toxicity biosensor using immobilized cyanobacteria, Anabaena torulosa. Sens. Lett. 2005, 3, 49–54. [Google Scholar] [CrossRef]
- Adekunle, A.; Rickwood, C.; Tartakovsky, B. Online monitoring of heavy metal–related toxicity using flow-through and floating microbial fuel cell biosensors. Environ. Monit. Assess. 2020, 192. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, X.; Lan, L.; Pan, Y.; Zhu, G.; Miao, P. Gly-Gly-His tripeptide- and silver nanoparticle-assisted electrochemical evaluation of copper(II) ions in aqueous environment. New J. Chem. 2018, 42, 14733–14737. [Google Scholar] [CrossRef]
- Wang, G.H.; Cheng, C.Y.; Liu, M.H.; Chen, T.Y.; Hsieh, M.C.; Chung, Y.C. Utility of Ochrobactrum anthropi yc152 in a microbial fuel cell as an early warning device for hexavalent chromium determination. Sensors 2016, 16, 1272. [Google Scholar] [CrossRef] [Green Version]
- Nepomuscene, N.J.; Daniel, D.; Krastanov, A. Biosensor to detect chromium in wastewater. Biotechnol. Biotechnol. Equip. 2007, 21, 377–381. [Google Scholar] [CrossRef] [Green Version]
- Mandl, M.; Macholán, L. Membrane biosensor for the determination of iron(II, III) based on immobilized cells of Thiobacillus ferrooxidans. Folia Microbiol. 1990, 35, 363–367. [Google Scholar] [CrossRef]
- Singh, J.; Mittal, S.K. A new assembly for biosensing ultra-trace levels of mercury in a continuous flow system. Anal. Methods 2014, 6, 5741–5745. [Google Scholar] [CrossRef]
- Verma, N.; Singh, M. A Bacillus sphaericus based biosensor for monitoring nickel ions in industrial effluents and foods. J. Autom. Methods Manag. Chem. 2006, 2006, 083427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocha, D.L.; Batista, A.D.; Rocha, F.R.P.; Donati, G.L.; Nóbrega, J.A. Greening sample preparation in inorganic analysis. TrAC Trends Anal. Chem. 2013, 45, 79–92. [Google Scholar] [CrossRef]
- Isarankura-Na-Ayudhya, C.; Tantimongcolwat, T.; Galla, H.J.; Prachayasit-tikul, V. Fluorescent protein-based optical biosensor for copper ion quantitation. Biol. Trace Elem. Res. 2010, 134, 352–363. [Google Scholar] [CrossRef]
- Fang, X.; Zhao, Q.; Cao, H.; Liu, J.; Guan, M.; Kong, J. Rapid detection of Cu2+ by a paper-based microfluidic device coated with bovine serum albumin (BSA)—Au nanoclusters. Analyst 2015, 140, 7823–7826. [Google Scholar] [CrossRef]
- Tantimongcolwat, T.; Isarankura-Na-Ayudhya, C.; Srisarin, A.; Galla, H.J.; Prachayasittikul, V. Polyacrylamide hydrogel encapsulated E. coli expressing metal-sensing green fluorescent protein as a potential tool for copper ion determination. EXCLI J. 2014, 13, 401–415. [Google Scholar] [CrossRef]
- Lam, C.K.S.C.C.; Jickells, T.D.; Richardson, D.J.; Russell, D.A. Fluorescence-based siderophore biosensor for the determination of bioavailable iron in oceanic waters. Anal. Chem. 2006, 78, 5040–5045. [Google Scholar] [CrossRef]
- Marieeswaran, M.; Panneerselvam, P. Fluorescent Polyaniline Nanoclips (PANCs): A Highly Sensitive and Selective Chemical Sensor for the Detection of Hg(II) Ions in Aqueous Media. ChemistrySelect 2020, 5, 4481–4487. [Google Scholar] [CrossRef]
- Teh, H.B.; Li, H.; Yau Li, S.F. Highly sensitive and selective detection of Pb2+ ions using a novel and simple DNAzyme-based quartz crystal microbalance with dissipation biosensor. Analyst 2014, 139, 5170–5175. [Google Scholar] [CrossRef]
- May, L.M.; Russell, D.A. Novel determination of cadmium ions using an enzyme self-assembled monolayer with surface plasmon resonance. Anal. Chim. Acta 2003, 500, 119–125. [Google Scholar] [CrossRef]
- Cennamo, N.; Alberti, G.; Pesavento, M.; D’Agostino, G.; Quattrini, F.; Biesuz, R.; Zeni, L. A simple small size and low cost sensor based on Surface Plasmon Resonance for selective detection of Fe(III). Sensors 2014, 14, 4657–4671. [Google Scholar] [CrossRef] [PubMed]
- Shervedani, R.K.; Akrami, Z. Gold-deferrioxamine nanometric interface for selective recognition of Fe(III) using square wave voltammetry and electro-chemical impedance spectroscopy methods. Biosens. Bioelectron. 2013, 39, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Stoytcheva, M.; Zlatev, R.; Magnin, J.P.; Ovalle, M.; Valdez, B. Leptospirillum ferrooxidans based Fe2+ sensor. Biosens. Bioelectron. 2009, 25, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Zlatev, R.; Magnin, J.P.; Ozil, P.; Stoytcheva, M. Bacterial sensors based on Acidithiobacillus ferrooxidans: Part I. Fe2+ and S2O32− determination. Biosens. Bioelectron. 2006, 21, 1493–1500. [Google Scholar] [CrossRef] [PubMed]
- Torrinha, Á.; Montenegro, M.C.B.S.M.; Araújo, A.N. Microfluidic Platform with an Embedded Pencil Graphite Electrode Biosensor for the Detection of Glucose and Cadmium. J. Electrochem. Soc. 2019, 166, B155–B160. [Google Scholar] [CrossRef]
- Lin, M.; Hu, X.; Ma, Z.; Chen, L. Functionalized polypyrrole nanotube arrays as electrochemical biosensor for the determination of copper ions. Anal. Chim. Acta 2012, 746, 63–69. [Google Scholar] [CrossRef]
- Gao, F.; Gao, C.; He, S.; Wang, Q.; Wu, A. Label-free electrochemical lead(II) aptasensor using thionine as the signaling molecule and graphene as signal-enhancing platform. Biosens. Bioelectron. 2016, 81, 15–22. [Google Scholar] [CrossRef]
- Liu, S.; Kang, M.; Yan, F.; Peng, D.; Yang, Y.; He, L.; Wang, M.; Fang, S.; Zhang, Z. Electrochemical DNA biosensor based on microspheres of cuprous oxide and nano-chitosan for Hg(II) detection. Electrochim. Acta 2015, 160, 64–73. [Google Scholar] [CrossRef]
- Tang, L.; Xie, X.; Zhou, Y.; Zeng, G.; Tang, J.; Wu, Y.; Long, B.; Peng, B.; Zhu, J. A reusable electrochemical biosensor for highly sensitive detection of mercury ions with an anionic intercalator supported on ordered mesoporous carbon/self-doped polyaniline nanofibers platform. Biochem. Eng. J. 2017, 117, 7–14. [Google Scholar] [CrossRef]
- Kaleli-Can, G.; Ozlu, B.; Özgüzar, H.F.; Onal-Ulusoy, B.; Kabay, G.; Eom, T.; Shim, B.S.; Mutlu, M. Natural Melanin Nanoparticle-decorated Screen-printed Carbon Electrode: Performance Test for Amperometric Determination of Hexavalent Chromium as Model Trace. Electroanalysis 2020, 32, 1696–1706. [Google Scholar] [CrossRef]
- Norocel, L.; Gutt, G. Screen-printed voltammetric biosensors for the determination of copper in wine. Sensors 2019, 19, 4618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liana, D.D.; Raguse, B.; Wieczorek, L.; Baxter, G.R.; Chuah, K.; Gooding, J.J.; Chow, E. Sintered gold nanoparticles as an electrode material for paper-based electrochemical sensors. RSC Adv. 2013, 3, 8683–8691. [Google Scholar] [CrossRef]
- Norocel, L.; Gutt, G. Method and electrochemical biosensor for detection of copper in wine. Rev. Chim. 2018, 69, 3010–3012. [Google Scholar] [CrossRef]
- Zhou, J.; Huang, H.; Chen, S.; Wang, M.; Zhao, D.; Yu, J.; Jin, S.; Zhong, Y.; Chen, X.; Yu, X.; et al. A high sensitive chemiresistive-biosensor based on self-assembly grown GaN porous layer. Sens. Actuators B Chem. 2021, 345, 130360. [Google Scholar] [CrossRef]
- Yang, S.; Liu, P.; Wang, Y.; Guo, Z.; Tan, R.; Qu, L. Electrochemical sensor using poly-(l-cysteine) functionalized CuO nanoneedles/N-doped reduced graphene oxide for detection of lead ions. RSC Adv. 2020, 10, 18526–18532. [Google Scholar] [CrossRef]
- Wu, X.; Liu, W.; Dai, H.; Chen, G. A novel sensitive biosensor for Ca2+ based on electropolymerized melatonin modified electrode. Electrochem. Commun. 2009, 11, 393–396. [Google Scholar] [CrossRef]
- Akbari Hasanjani, H.R.; Zarei, K. An electrochemical sensor for attomolar determination of mercury(II) using DNA/poly-L-methionine-gold nanoparticles/pencil graphite electrode. Biosens. Bioelectron. 2019, 128, 1–8. [Google Scholar] [CrossRef]
- Chen, G.; Bai, W.; Jin, Y.; Zheng, J. Fluorescence and electrochemical assay for bimodal detection of lead ions based on Metal—Organic framework nanosheets. Talanta 2021, 232, 122405. [Google Scholar] [CrossRef]
- Weng, C.; Li, X.; Lu, Q.; Yang, W.; Wang, J.; Yan, X.; Li, B.; Sakran, M.; Hong, J.; Zhu, W.; et al. A label-free electrochemical biosensor based on magnetic biocomposites with DNAzyme and hybridization chain reaction dual signal amplification for the determination of Pb2+. Microchim. Acta 2020, 187, 575. [Google Scholar] [CrossRef]
- Bi, X.; Wong, W.L.; Ji, W.; Agarwal, A.; Balasubramanian, N.; Yang, K.L. Development of electrochemical calcium sensors by using silicon nanowires modified with phosphotyrosine. Biosens. Bioelectron. 2008, 23, 1442–1448. [Google Scholar] [CrossRef] [PubMed]
- Rabai, S.; Benounis, M.; Catanante, G.; Baraket, A.; Errachid, A.; Jaffrezic Renault, N.; Marty, J.L.; Rhouati, A. Development of a label-free electrochemical aptasensor based on diazonium electrodeposition: Application to cadmium detection in water. Anal. Biochem. 2021, 612, 113956. [Google Scholar] [CrossRef]
- Niu, Y.; Chen, Y.; Zhang, X.; Xie, H.; Luo, G.; Sun, W. Target-enhanced photoelectrochemical aptasensor for Cd(II) detection using graphite-like carbon nitride as sensitizer with high sensitivity. Microchem. J. 2021, 168, 106394. [Google Scholar] [CrossRef]
- Yu, Y.; Yu, C.; Yin, T.; Ou, S.; Sun, X.; Wen, X.; Zhang, L.; Tang, D.; Yin, X. Functionalized poly(ionic liquid) as the support to construct a ratiometric electrochemical biosensor for the selective determination of copper ions in AD rats. Biosens. Bioelectron. 2017, 87, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Oztekin, Y.; Yazicigil, Z.; Ramanaviciene, A.; Ramanavicius, A. Polyphenol-modified glassy carbon electrodes for copper detection. Sens. Actuators B Chem. 2011, 152, 37–48. [Google Scholar] [CrossRef]
- Yüce, M.; Nazir, H.; Dönmez, G. A voltammetric Rhodotorula mucilaginosa modified microbial biosensor for Cu(II) determination. Bioelectrochemistry 2010, 79, 66–70. [Google Scholar] [CrossRef]
- Alpat, S.K.; Alpat, Ş.; Kutlu, B.; Özbayrak, Ö.; Büyükişik, H.B. Development of biosorption-based algal biosensor for Cu(II) using Tetraselmis chuii. Sens. Actuators B Chem. 2007, 128, 273–278. [Google Scholar] [CrossRef]
- Gu, H.; Hou, Q.; Liu, Y.; Cai, Y.; Guo, Y.; Xiang, H.; Chen, S. On-line regeneration of electrochemical biosensor for in vivo repetitive measurements of striatum Cu2+ under global cerebral ischemia/reperfusion events. Biosens. Bioelectron. 2019, 135, 111–119. [Google Scholar] [CrossRef]
- Atapour, M.; Amoabediny, G.; Ahmadzadeh-Raji, M. Integrated optical and electrochemical detection of Cu2+ ions in water using a sandwich amino acid-gold nanoparticle-based nano-biosensor consisting of a transparent-conductive platform. RSC Adv. 2019, 9, 8882–8893. [Google Scholar] [CrossRef] [Green Version]
- Wawrzyniak, U.E.; Ciosek, P.; Zaborowski, M.; Liu, G.; Gooding, J.J. Gly-Gly-His immobilized on monolayer modified back-side contact miniaturized sensors for complexation of copper ions. Electroanalysis 2013, 25, 1461–1471. [Google Scholar] [CrossRef]
- Amin, N.U.; Kun-Lin, Y.; Majeed, N.; Siddiqi, H.M. Fabrication of a Fluorophore/Liquid-Crystal-Based Oligopeptide Biosensor for the Detection of Cu(II) Ions. ChemistrySelect 2021, 6, 6607–6618. [Google Scholar] [CrossRef]
- Singh, V.K.; Kushwaha, C.S.; Shukla, S.K. Potentiometric detection of copper ion using chitin grafted polyaniline electrode. Int. J. Biol. Macromol. 2020, 147, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Ezhil-Vilian, A.T.; Shahzad, A.; Chung, J.; Choe, S.R.; Kim, W.S.; Huh, Y.S.; Yu, T.; Han, Y.K. Square voltammetric sensing of mercury at very low working potential by using oligomer-functionalized Ag@Au core-shell nanoparticles. Microchim. Acta 2017, 184, 3547–3556. [Google Scholar] [CrossRef]
- Bala, A.; Górski, Ł. Determination of mercury cation using electrode modified with phosphorothioate oligonucleotide. Sens. Actuators B Chem. 2016, 230, 731–735. [Google Scholar] [CrossRef]
- Wang, N.; Lin, M.; Dai, H.; Ma, H. Functionalized gold nanoparticles/reduced graphene oxide nanocomposites for ultrasensitive electrochemical sensing of mercury ions based on thymine-mercury-thymine structure. Biosens. Bioelectron. 2016, 79, 320–326. [Google Scholar] [CrossRef]
- An, J.H.; Park, S.J.; Kwon, O.S.; Bae, J.; Jang, J. High-performance flexible graphene aptasensor for mercury detection in mussels. ACS Nano 2013, 7, 10563–10571. [Google Scholar] [CrossRef]
- Lü, H.; Zhao, Y.; Ma, J.; Li, J.; Wang, H.; Lu, Z. Electrochemical detection of magnesium ions using PVC membrane trapped chlorophyll A molecules. Mol. Cryst. Liq. Cryst. Sci. Technol. Sect. A Mol. Cryst. Liq. Cryst. 2001, 371, 391–396. [Google Scholar] [CrossRef]
- Liu, H.; Wang, J.; Jin, H.; Wei, M.; Ren, W.; Zhang, Y.; Wu, L.; He, B. Electrochemical biosensor for sensitive detection of Hg2+ baesd on clustered peonylike copper-based metal-organic frameworks and DNAzyme-driven DNA Walker dual amplification signal strategy. Sens. Actuators B Chem. 2021, 329, 129215. [Google Scholar] [CrossRef]
- Ding, J.; Liu, Y.; Zhang, D.; Yu, M.; Zhan, X.; Zhang, D.; Zhou, P. An electrochemical aptasensor based on gold@polypyrrole composites for detection of lead ions. Microchim. Acta 2018, 185, 545. [Google Scholar] [CrossRef]
- Gil, R.L.; Amorim, C.G.; Montenegro, M.C.B.S.M.; Araújo, A.N. Potentiometric detection in liquid chromatographic systems: An overview. J. Chromatogr. A 2019, 1602, 326–340. [Google Scholar] [CrossRef]
- Chandra, S.; Dhawangale, A.; Mukherji, S. Hand-held optical sensor using denatured antibody coated electroactive polymer for ultra-trace detection of copper in blood serum and environmental samples. Biosens. Bioelectron. 2018, 110, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chang, H.; Hirata, A.; Wu, H.; Xue, Q.K.; Chen, M. Nanoporous gold based optical sensor for sub-ppt detection of mercury ions. ACS Nano 2013, 7, 4595–4600. [Google Scholar] [CrossRef] [PubMed]
- Akyilmaz, E.; Kozgus, O. Determination of calcium in milk and water samples by using catalase enzyme electrode. Food Chem. 2009, 115, 347–351. [Google Scholar] [CrossRef]
- Moyo, M.; Okonkwo, J.O. Horseradish peroxidase biosensor based on maize tassel-MWCNTs composite for cadmium detection. Sens. Actuators B Chem. 2014, 193, 515–521. [Google Scholar] [CrossRef]
- Shtenberg, G.; Massad-Ivanir, N.; Segal, E. Detection of trace heavy metal ions in water by nanostructured porous Si biosensors. Analyst 2015, 140, 4507–4514. [Google Scholar] [CrossRef]
- Vopálenská, I.; Váchová, L.; Palková, Z. New biosensor for detection of copper ions in water based on immobilized genetically modified yeast cells. Biosens. Bioelectron. 2015, 72, 160–167. [Google Scholar] [CrossRef]
- Ballen, S.C.; Ostrowski, G.M.; Steffens, J.; Steffens, C. Graphene Oxide/Urease Nanobiosensor Applied for Cadmium Detection in River Water. IEEE Sens. J. 2021, 21, 9626–9633. [Google Scholar] [CrossRef]
- Swain, K.K.; Bhand, S. A colorimetric paper-based ATONP-ALP nanobiosensor for selective detection of Cd2+ ions in clams and mussels. Anal. Bioanal. Chem. 2021, 413, 1715–1727. [Google Scholar] [CrossRef]
- Dabhade, A.; Jayaraman, S.; Paramasivan, B. Development of glucose oxidase-chitosan immobilized paper biosensor using screen-printed electrode for amperometric detection of Cr(VI) in water. 3 Biotech 2021, 11, 183. [Google Scholar] [CrossRef]
- Attar, A.; Emilia Ghica, M.; Amine, A.; Brett, C.M.A. Poly(neutral red) based hydrogen peroxide biosensor for chromium determination by inhibition measurements. J. Hazard. Mater. 2014, 279, 348–355. [Google Scholar] [CrossRef]
- Kaur, G.; Verma, N. Colorimetric determination of Cu2+ ions in water and milk by apotyrosinase disc. Sens. Actuators B Chem. 2018, 263, 524–532. [Google Scholar] [CrossRef]
- Elsebai, B.; Ghica, M.E.; Abbas, M.N.; Brett, C.M.A. Catalase based hydrogen peroxide biosensor for mercury determination by inhibition measurements. J. Hazard. Mater. 2017, 340, 344–350. [Google Scholar] [CrossRef]
- Smit, M.H.; Rechnitz, G.A. Reagentless Enzyme Electrode for the Determination of Manganese through Biocatalytic Enhancement. Anal. Chem. 1992, 64, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Magar, H.S.; Ghica, M.E.; Abbas, M.N.; Brett, C.M.A. Highly Sensitive Choline Oxidase Enzyme Inhibition Biosensor for Lead Ions Based on Multiwalled Carbon Nanotube Modified Glassy Carbon Electrodes. Electroanalysis 2017, 29, 1741–1748. [Google Scholar] [CrossRef]
- Sabir, S.; Akash, M.S.H.; Fiayyaz, F.; Saleem, U.; Mehmood, M.H.; Rehman, K. Role of cadmium and arsenic as endocrine disruptors in the metabolism of carbohydrates: Inserting the association into perspectives. Biomed. Pharmacother. 2019, 114, 108802. [Google Scholar] [CrossRef]
- Liu, S.; Wei, W.; Sun, X.; Wang, L. Ultrasensitive electrochemical DNAzyme sensor for lead ion based on cleavage-induced template-independent polymerization and alkaline phosphatase amplification. Biosens. Bioelectron. 2016, 83, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.C.; Kharkwal, S.; Chew, K.K.W.; Alwi, R.; Mak, S.F.W.; Ng, H.Y. Enhancing the robustness of microbial fuel cell sensor for continuous copper(II) detection against organic strength fluctuations by acetate and glucose addition. Bioresour. Technol. 2018, 259, 357–364. [Google Scholar] [CrossRef]
- Yu, D.; Bai, L.; Zhai, J.; Wang, Y.; Dong, S. Toxicity detection in water containing heavy metal ions with a self-powered microbial fuel cell-based biosensor. Talanta 2017, 168, 210–216. [Google Scholar] [CrossRef]
- Wang, D.; Liang, P.; Jiang, Y.; Liu, P.; Miao, B.; Hao, W.; Huang, X. Open external circuit for microbial fuel cell sensor to monitor the nitrate in aquatic environment. Biosens. Bioelectron. 2018, 111, 97–101. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Liu, B.; Dong, Q.; Lei, Y.; Li, Y.; Ren, J.; McCutcheon, J.; Li, B. Flat microliter membrane-based microbial fuel cell as “on-line sticker sensor” for self-supported in situ monitoring of wastewater shocks. Bioresour. Technol. 2015, 197, 244–251. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, P.; Yan, P.; Wang, M.; Tang, Q.; Deng, A.; Li, J. Quantum Dots Based Electrochemiluminescent Immunosensor for Ultrasensitive and Specific Determination of Mercury(II) Ions Using Gold Nanoparticles and a Monoclonal Antibody. J. Electrochem. Soc. 2015, 162, B22–B26. [Google Scholar] [CrossRef]
- Wang, H.; Luo, Q.; Zhao, Y.; Nan, X.; Zhang, F.; Wang, Y.; Wang, Y.; Hua, D.; Zheng, S.; Jiang, L.; et al. Electrochemical device based on nonspecific DNAzyme for the high-accuracy determination of Ca2+ with Pb2+ interference. Bioelectrochemistry 2021, 140, 107732. [Google Scholar] [CrossRef]
- Ebrahimi, M.; Raoof, J.B.; Ojani, R. Design of an electrochemical DNA-based biosensor for selective determination of cadmium ions using a DNA hybridization indicator. Int. J. Biol. Macromol. 2018, 108, 1237–1241. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Wu, L.; Liu, H.; Li, J.; Lv, H.; Fu, X.; Song, Y. A novel electrochemical biosensor based on DNA for rapid and selective detection of cadmium. Int. J. Electrochem. Sci. 2015, 10, 4020–4028. [Google Scholar]
- Sreekanth, S.P.; Alodhayb, A.; Assaifan, A.K.; Alzahrani, K.E.; Muthuramamoorthy, M.; Alkhammash, H.I.; Pandiaraj, S.; Alswieleh, A.M.; Van Le, Q.; Mangaiyarkarasi, R.; et al. Multi-walled carbon nanotube-based nanobiosensor for the detection of cadmium in water. Environ. Res. 2021, 197, 111148. [Google Scholar] [CrossRef]
- Wang, H.; Zheng, S.; Nan, X.; Zhao, Y.; Wang, Y.; Zhang, F.; Yang, L.; Lixing, X.; Xiong, B. Non-specific DNAzyme-based biosensor with interfering ions for the Cd2+ determination in feed. Sens. Actuators B Chem. 2021, 329, 129139. [Google Scholar] [CrossRef]
- Ocaña, C.; Malashikhina, N.; Del Valle, M.; Pavlov, V. Label-free selective impedimetric detection of Cu2+ ions using catalytic DNA. Analyst 2013, 138, 1995–1999. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Dai, B.; Xu, J.; Jiang, L.; Huang, H. An Electrochemical Sensor for the Detection of Cu2+ Based on Gold Nanoflowers-modifed Electrode and DNAzyme Functionalized Au@MIL-101 (Fe). Electroanalysis 2019, 31, 2330–2338. [Google Scholar] [CrossRef]
- Tian, R.; Chen, X.; Liu, D.; Yao, C. A Sensitive Biosensor for Determination of Cu2+ by One-step Electrodeposition. Electroanalysis 2016, 28, 1617–1624. [Google Scholar] [CrossRef]
- Hu, W.; Min, X.; Li, X.; Yang, S.; Yi, L.; Chai, L. DNAzyme catalytic beacons-based a label-free biosensor for copper using electrochemical impedance spectroscopy. RSC Adv. 2016, 6, 6679–6685. [Google Scholar] [CrossRef]
- Tang, D.; Zhang, J.; Tang, Y.; Teng, L.; Xia, B.; Tang, D. Hairpin DNA-Dependent Click Conjugation of Oligonucleotides for Electrochemical Monitoring of Copper(II). Electroanalysis 2015, 27, 2513–2517. [Google Scholar] [CrossRef]
- Zhang, Y.; Chu, G.; Guo, Y.; Zhao, W.; Yang, Q.; Sun, X. An electrochemical biosensor based on Au nanoparticles decorated reduced graphene oxide for sensitively detecting of Hg2+. J. Electroanal. Chem. 2018, 824, 201–206. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, C.; Ma, R.; Du, X.; Dong, W.; Chen, Y.; Chen, Q. An ultra-sensitive Au nanoparticles functionalized DNA biosensor for electrochemical sensing of mercury ions. Mater. Sci. Eng. C 2017, 75, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Huang, Y.; Zhang, C.; Liu, H.; Tang, D. DNA-based electrochemical determination of mercury(II) by exploiting the catalytic formation of gold amalgam and of silver nanoparticles. Microchim. Acta 2016, 183, 1805–1812. [Google Scholar] [CrossRef]
- Ebrahimi, M.; Raoof, J.B.; Ojani, R.; Bagheryan, Z. A novel electrochemical biosensor for selective determination of mercury ions based on DNA hybridization. Anal. Biochem. 2015, 488, 12–13. [Google Scholar] [CrossRef] [PubMed]
- He, L.L.; Cheng, L.; Lin, Y.; Cui, H.F.; Hong, N.; Peng, H.; Kong, D.R.; Chen, C.D.; Zhang, J.; Wei, G.; et al. A sensitive biosensor for mercury ions detection based on hairpin hindrance by thymine-Hg(II)-thymine structure. J. Electroanal. Chem. 2018, 814, 161–167. [Google Scholar] [CrossRef]
- Ziółkowski, R.; Jarczewska, M.; Górski, Ł.; Malinowska, E. Oligonucleotide-Based Electrochemical Biosensor for Hg2+ Using Methylene Blue as a Redox Indicator. J. Electrochem. Soc. 2013, 160, B152–B155. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, H.; Wu, Z.; Xue, Y.; Zhang, X.; He, Y.; Li, X.; Yuan, Z. A novel graphene-DNA biosensor for selective detection of mercury ions. Biosens. Bioelectron. 2013, 48, 180–187. [Google Scholar] [CrossRef]
- Qiu, Z.; Tang, D.; Shu, J.; Chen, G.; Tang, D. Enzyme-triggered formation of enzyme-tyramine concatamers on nanogold-functionalized dendrimer for impedimetric detection of Hg(II) with sensitivity enhancement. Biosens. Bioelectron. 2016, 75, 108–115. [Google Scholar] [CrossRef]
- Huang, Y.L.; Gao, Z.F.; Jia, J.; Luo, H.Q.; Li, N.B. A label-free electrochemical sensor for detection of Mercury(II) ions based on the direct growth of guanine nanowire. J. Hazard. Mater. 2016, 308, 173–178. [Google Scholar] [CrossRef]
- Tortolini, C.; Bollella, P.; Antonelli, M.L.; Antiochia, R.; Mazzei, F.; Favero, G. DNA-based biosensors for Hg2+ determination by polythymine–methylene blue modified electrodes. Biosens. Bioelectron. 2015, 67, 524–531. [Google Scholar] [CrossRef]
- He, Z.J.; Kang, T.F.; Lu, L.P.; Cheng, S.Y. An electrochemiluminescence sensor based on CdSe@CdS-functionalized MoS2 and a GOD-labeled DNA probe for the sensitive detection of Hg(II). Anal. Methods 2020, 12, 491–498. [Google Scholar] [CrossRef]
- Fan, X.; Wang, S.; Li, Z.; Wang, Y.; Fan, X.; Yu, L. An electrochemiluminescence biosensor for the determination of mercury ion via dual-amplification strategy. J. Braz. Chem. Soc. 2020, 31, 2620–2627. [Google Scholar] [CrossRef]
- Duan, Y.; Wang, F.; Zhang, X.; Liu, Q.; Lu, M.; Ji, W.; Zhang, Y.; Jing, Z.; Peng, W. TFBG-SPR DNA-Biosensor for Renewable Ultra-Trace Detection of Mercury Ions. J. Lightwave Technol. 2021, 39, 3903–3910. [Google Scholar] [CrossRef]
- He, W.; Qiao, B.; Li, F.; Pan, L.; Chen, D.; Cao, Y.; Tu, J.; Wang, X.; Lv, C.; Wu, Q. A novel electrochemical biosensor for ultrasensitive Hg2+ detection: Via a triple signal amplification strategy. Chem. Commun. 2021, 57, 619–622. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chen, B.; Luo, L.; Liu, X.; Bi, X.; You, T. Sensitive and selective detection of Hg2+ in tap and canal water via self-enhanced ECL aptasensor based on NH2–Ru@SiO2-NGQDs. Talanta 2021, 222, 121579. [Google Scholar] [CrossRef]
- De Acha, N.; Elosúa, C.; Arregui, F.J. Development of an aptamer based luminescent optical fiber sensor for the continuous monitoring of Hg2+ in Aqueous media. Sensors 2020, 20, 2372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qing, M.; Chen, S.; Xie, S.; Tang, Y.; Zhang, J.; Yuan, R. Encapsulation and Release of Recognition Probes Based on a Rigid Three-Dimensional DNA “nanosafe-box” for Construction of a Electrochemical Biosensor. Anal. Chem. 2020, 92, 1811–1817. [Google Scholar] [CrossRef]
- Cao, S.P.; Hu, H.M.; Liang, R.P.; Qiu, J.D. An ultrasensitive electrochemiluminescence resonance energy transfer biosensor for divalent mercury monitoring. J. Electroanal. Chem. 2020, 856, 113494. [Google Scholar] [CrossRef]
- Jarczewska, M.; Górski, Ł.; Malinowska, E. Application of DNA aptamers as sensing layers for electrochemical detection of potassium ions. Sens. Actuators B Chem. 2016, 226, 37–43. [Google Scholar] [CrossRef]
- Li, L.D.; Huang, X.Q.; Guo, L. Electrochemical potassium ion sensor based on DNA G-quadruplex conformation and gold nanoparticle amplification. Rare Met. 2013, 32, 369–374. [Google Scholar] [CrossRef]
- Chen, Z.; Zhou, T.; Zhang, C.; Ma, H.; Lin, Y.; Li, K. Aptasensor for label-free square-wave voltammetry detection of potassium ions based on gold nanoparticle amplification. RSC Adv. 2014, 4, 48671–48675. [Google Scholar] [CrossRef]
- Chen, Z.; Chen, L.; Ma, H.; Zhou, T.; Li, X. Aptamer biosensor for label-free impedance spectroscopy detection of potassium ion based on DNA G-quadruplex conformation. Biosens. Bioelectron. 2013, 48, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yuan, Z.; Liu, X.P.; Liu, Q.; Mao, C.J.; Niu, H.L.; Jin, B.K.; Zhang, S.Y. Electrochemical biosensor for Ni2+ detection based on a DNAzyme-CdSe nanocomposite. Biosens. Bioelectron. 2016, 77, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yu, C.; Yu, Y.; Chen, J.; Gao, R.; He, J. DNAzyme assisted recycling amplification method for ultrasensitive amperometric determination of lead(II) based on the use of a hairpin assembly on a composite prepared from nitrogen doped graphene, perylenetetracarboxylic anhydride, thionine and gold nanoparticles. Microchim. Acta 2019, 186, 677. [Google Scholar] [CrossRef]
- Xie, X.; Chai, Y.; Yuan, Y.; Yuan, R. Dual triggers induced disassembly of DNA polymer decorated silver nanoparticle for ultrasensitive electrochemical Pb2+ detection. Anal. Chim. Acta 2018, 1034, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Feng, J.; Li, J.; Feng, M.; Huang, S. A label-free and ultrasensitive electrochemical aptasensor for lead(II) using a N,P dual-doped carbon dot-chitosan composite as a signal-enhancing platform and thionine as a signaling molecule. Analyst 2018, 143, 4764–4773. [Google Scholar] [CrossRef]
- Yu, Y.; Yu, C.; Niu, Y.; Chen, J.; Zhao, Y.; Zhang, Y.; Gao, R.; He, J. Target triggered cleavage effect of DNAzyme: Relying on Pd-Pt alloys functionalized Fe-MOFs for amplified detection of Pb2+. Biosens. Bioelectron. 2018, 101, 297–303. [Google Scholar] [CrossRef]
- Ebrahimi, M.; Raoof, J.B.; Ojani, R. Design of a novel electrochemical biosensor based on intramolecular G-quadruplex DNA for selective determination of lead(II) ions. Anal. Bioanal. Chem. 2017, 409, 4729–4739. [Google Scholar] [CrossRef]
- Yuan, M.; Song, Z.; Fei, J.; Wang, X.; Xu, F.; Cao, H.; Yu, J. Aptasensor for lead(II) based on the use of a quartz crystal microbalance modified with gold nanoparticles. Microchim. Acta 2017, 184, 1397–1403. [Google Scholar] [CrossRef]
- Bala, A.; Pietrzak, M.; Górski, L.; Malinowska, E. Electrochemical determination of lead ion with DNA oligonucleotide-based biosensor using anionic redox marker. Electrochim. Acta 2015, 180, 763–769. [Google Scholar] [CrossRef]
- Jarczewska, M.; Kierzkowska, E.; Ziółkowski, R.; Górski, Ł.; Malinowska, E. Electrochemical oligonucleotide-based biosensor for the determination of lead ion. Bioelectrochemistry 2015, 101, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.F.; Cai, Z.M.; Wu, G.H.; Rong, M.C.; Jiang, Y.Q.; Yang, C.Y.J.; Chen, X. A novel signal-on DNAzyme-based electrochemiluminescence sensor for Pb2+. Sens. Actuators B Chem. 2014, 191, 60–66. [Google Scholar] [CrossRef]
- Lai, C.; Zhang, Y.; Liu, X.; Liu, S.; Li, B.; Zhang, M.; Qin, L.; Yi, H.; Li, M.; Li, L.; et al. Electrochemical biosensor for amplified detection of Pb2+ based on perfect match of reduced graphene oxide–gold nanoparticles and single-stranded DNAzyme. Anal. Bioanal. Chem. 2019, 411, 7499–7509. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Lin, Y.; Lin, Y.; Wei, Q.; Chen, G.; Tang, D. Highly sensitive electrochemical sensing platform for lead ion based on synergetic catalysis of DNAzyme and Au-Pd porous bimetallic nanostructures. Biosens. Bioelectron. 2016, 78, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Jing, P.; Xu, W. Hemin on graphene nanosheets functionalized with flower-like MnO2 and hollow AuPd for the electrochemical sensing lead ion based on the specific DNAzyme. Biosens. Bioelectron. 2016, 86, 958–965. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wen, Y.; Li, L.; Yang, X.; Jia, N.; Li, W.; Meng, J.; Duan, M.; Sun, X.; Liu, G. Sensitive and label-free electrochemical lead ion biosensor based on a DNAzyme triggered G-quadruplex/hemin conformation. Biosens. Bioelectron. 2018, 115, 91–96. [Google Scholar] [CrossRef]
- Kuang, H.; Xing, C.; Hao, C.; Liu, L.; Wang, L.; Xu, C. Rapid and highly sensitive detection of lead ions in drinking water based on a strip immunosensor. Sensors 2013, 13, 4214–4224. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Lai, C.; Zeng, G.; Huang, D.; Tang, L.; Yang, C.; Zhou, Y.; Qin, L.; Cheng, M. Nanoporous Au-based chronocoulometric aptasensor for amplified detection of Pb2+ using DNAzyme modified with Au nanoparticles. Biosens. Bioelectron. 2016, 81, 61–67. [Google Scholar] [CrossRef]
- Tang, S.; Tong, P.; You, X.; Lu, W.; Chen, J.; Li, G.; Zhang, L. Label free electrochemical sensor for Pb2+ based on graphene oxide mediated deposition of silver nanoparticles. Electrochim. Acta 2016, 187, 286–292. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, S.; Li, H.; Liu, H.; Pang, P.; Wang, H.; Wu, Z.; Yang, W. A Pb2+-ion electrochemical biosensor based on single-stranded DNAzyme catalytic beacon. Sens. Actuators B Chem. 2016, 222, 1083–1089. [Google Scholar] [CrossRef]
- Lei, Y.M.; Huang, W.X.; Zhao, M.; Chai, Y.Q.; Yuan, R.; Zhuo, Y. Electrochemiluminescence Resonance Energy Transfer System: Mechanism and Application in Ratiometric Aptasensor for Lead Ion. Anal. Chem. 2015, 87, 7787–7794. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Tang, L.; Zeng, G.; Zhang, C.; Xie, X.; Liu, Y.; Wang, J.; Tang, J.; Zhang, Y.; Deng, Y. Label free detection of lead using impedimetric sensor based on ordered mesoporous carbon-gold nanoparticles and DNAzyme catalytic beacons. Talanta 2016, 146, 641–647. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Wu, J.; Li, J.; Ju, H. Electrochemical Sensor for Lead Cation Sensitized with a DNA Functionalized Porphyrinic Metal-Organic Framework. Anal. Chem. 2015, 87, 10635–10641. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Huang, S.; Yang, X.; Yuan, R.; Chai, Y. A SERS biosensor constructed by calcined ZnO substrate with high-efficiency charge transfer for sensitive detection of Pb2+. Sens. Actuators B Chem. 2021, 343, 130142. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, Y.; Lu, M.; Du, Y.; Chen, M.; Meng, S.; Ji, W.; Sun, C.; Peng, W. Near-infrared band Gold nanoparticles-Au film “hot spot” model based label-free ultratrace lead(II) ions detection via fiber SPR DNAzyme biosensor. Sens. Actuators B Chem. 2021, 337, 129816. [Google Scholar] [CrossRef]
- Meng, J.; Huang, J.; Oueslati, R.; Jiang, Y.; Chen, J.; Li, S.; Dai, S.; He, Q.; Wu, J. A single-step DNAzyme sensor for ultra-sensitive and rapid detection of Pb2+ ions. Electrochim. Acta 2021, 368, 137551. [Google Scholar] [CrossRef]
- Zhao, G.; Li, C.; Wang, X.; Liu, G.; Thuy, N.T.D. A Reusable Electrochemical Aptasensor for the Sensitive Detection of Pb(II) with an Electrodeposited AuNP-Modified Electrode based on the Formation of a Target-Induced G-Quadruplex. Int. J. Electrochem. Sci. 2021, 16, 150956. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, G.; Zhang, G.; Zhang, Y.; Wang, H.; Cao, W.; Li, T.; Wei, Q. An electrochemical aptasensor based on gold-modified MoS2/rGO nanocomposite and gold-palladium-modified Fe-MOFs for sensitive detection of lead ions. Sens. Actuators B Chem. 2020, 319, 128313. [Google Scholar] [CrossRef]
- Wu, H.; Wang, S.; Li, S.F.Y.; Bao, Q.; Xu, Q. A label-free lead(II) ion sensor based on surface plasmon resonance and DNAzyme-gold nanoparticle conjugates. Anal. Bioanal. Chem. 2020, 412, 7525–7533. [Google Scholar] [CrossRef]
- He, Y.; Hu, X.; Gong, Z.; Chen, S.; Yuan, R. A novel electrochemiluminescence biosensor based on the self-ECL emission of conjugated polymer dots for lead ion detection. Microchim. Acta 2020, 187, 237–245. [Google Scholar] [CrossRef]
- Jin, H.; Zhang, D.; Liu, Y.; Wei, M. An electrochemical aptasensor for lead ion detection based on catalytic hairpin assembly and porous carbon supported platinum as signal amplification. RSC Adv. 2020, 10, 6647–6653. [Google Scholar] [CrossRef]
- Feng, D.; Li, P.; Tan, X.; Wu, Y.; Wei, F.; Du, F.; Ai, C.; Luo, Y.; Chen, Q.; Han, H. Electrochemiluminescence aptasensor for multiple determination of Hg2+ and Pb2+ ions by using the MIL-53 (Al)@CdTe-PEI modified electrode. Anal. Chim. Acta 2020, 1100, 232–239. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, J.; Guo, Y.; Li, J.; Fu, F.; Yang, H.H.; Chen, G. An ultrasensitive electrochemical biosensor for detection of DNA species related to oral cancer based on nuclease-assisted target recycling and amplification of DNAzyme. Chem. Commun. 2011, 47, 8004–8006. [Google Scholar] [CrossRef]
- Chen, D.; Li, B.; Jiang, L.; Duan, D.; Li, Y.; Wang, J.; He, J.; Zeng, Y. Highly efficient colorimetric detection of cancer cells utilizing Fe-MIL-101 with intrinsic peroxidase-like catalytic activity over a broad pH range. RSC Adv. 2015, 5, 97910–97917. [Google Scholar] [CrossRef]
- Xue, Y.; Li, X.; Li, H.; Zhang, W. Quantifying thiol-gold interactions towards the efficient strength control. Nat. Commun. 2014, 5, 4348. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Mohanty, S.K.; Mahendra, S. Nanomaterial-Supported Enzymes for Water Purification and Monitoring in Point-of-Use Water Supply Systems. Acc. Chem. Res. 2019, 52, 876–885. [Google Scholar] [CrossRef]
- Chey, C.O.; Ibupoto, Z.H.; Khun, K.; Nur, O.; Willander, M. Indirect determination of mercury ion by inhibition of a glucose biosensor based on ZnO nanorods. Sensors 2012, 12, 15063–15077. [Google Scholar] [CrossRef] [PubMed]
- Santos, E.; Montenegro, M.C.B.S.M.; Couto, C.; Araújo, A.N.; Pimentel, M.F.; Da Silva, V.L. Sequential injection analysis of chloride and nitrate in waters with improved accuracy using potentiometric detection. Talanta 2004, 63, 721–727. [Google Scholar] [CrossRef]



| Analyte | Principle | Remarks | Sample | Technique | DL (µg L−1) | Response Time (min) | Ref. |
|---|---|---|---|---|---|---|---|
| Ca(II) | Bonding with polymerized melatonin | GCE coated with polymerized melatonin after electrochemical deposition | Cerebrospinal fluid | DPV | 18 | NI | [80] |
| Bonding with immobilized tyrosine | Modified FET with amino groups and crosslink with tyrosinase | Intracellular fluids | CD | NI | NI | [84] | |
| Cd(II) | Complexation with aptamer | Aptamer immobilized on GCE modified with carbon nitride and reduced graphene oxide. | Tap, natural, and waste waters | ASV | 0.038 | NI | [46] |
| Bonding with immobilized aptamer | Immobilization of aptamer on a GE modified with a reduced diazonium salt | River waters | EIS | 0.030 | 30 | [85] | |
| Complexation with aptamer | Aptamer immobilized on ITO electrode modified with gold layer | Lake waters | PEC | 0.0012 | 60 | [86] | |
| Cu(II) | Complexation with BSA | Immobilization of BSA with benzophenone mediated using UV radiation on a SPE | Wines | AP | 173 | 180 | [75] |
| Complexation with neurokinin B | Immobilization of neurokinin B and ABTS (mediator) on GCE modified with CNT | Animal plasma and tissues | DPV | 2.5 | NI | [38] | |
| Complexation with phytate | Phytic acid and polypirrole nanowires sealed with Nafion® on GCE | Wastewater | ASV | 3.3 | 300 | [47] | |
| Complexation with neurokinin B | Neurokinin B and ABTS sealed with polymeric membrane of ionic liquid on GCE | Animal cerebrospinal fluid and tissues | DPV | 15 | NI | [87] | |
| Complexation with tripeptide | Tripeptide immobilized on polypirrole and ZnO nanoparticles on ITO electrode. | Drinking water | SWV | 3.0 | 11 | [70] | |
| Complexation with polymerized polyphenols | Electropolymerization of luteolin and kaempferon on GCE | NA | DPV | 0.00064 | 60 | [88] | |
| Preconcentration by passive biosorption | Addition of lyophilized biomass of Rhodotorula mucilaginosa to a CPE | NA | CSV | NI | 15 | [89] | |
| Preconcentration by passive biosorption | Addition of lyophilized biomass of Tetraselmis chuii to a CPE | Multivitamin tablets | CSV | 0.029 | 30 | [90] | |
| Complexation with modified cysteine | Immobilization of modified cysteine on GCE by APM | Animal tissue | AP | 1.0 | 15 | [91] | |
| Preconcentration on GNP modified with cysteine | Cysteine immobilization on GNP via thiol groups and electrodeposition on ITO electrode | Tap water | SWV | 0.30 | 12 | [92] | |
| Complexation with immobilized glycine | Carbon SPE modified with glycine using agarose layer | Wine | CV | 40 | 0.5 | [77] | |
| Complexation with immobilized synthetic tripeptide | Deposition of SNP modified with tripeptide on a GCE coated with Poly-diallyldimethylammonium | Tap, natural, and waste waters | EIS | 0.54 | 15 | [51] | |
| Complexation and quenching of BSA/gold fluorescence | BSA/gold nanoclusters adsorbed on paper platform | NA | FL | 317 | NI | [59] | |
| Complexation with immobilized prion protein | Protein bonding with adsorbed APTS on FET silicon gate surface | Human serum | PT | 0.63 | 30 | [39] | |
| Complexation with immobilized cysteine | Cysteine immobilization on nail polisher material coated with GNP | NA | CV | 50 | 10 | [76] | |
| Complexation with immobilized synthetic tripeptide | Immobilized tripeptide glycine-glycine-histidine on GE. | NA | SWV | 6.3 | 15 | [93] | |
| Complexation with cysteine | Immobilization of cysteine on semiconductor modified with GNP | NA | LSV | 6.4 × 10−8 | 1.5 | [78] | |
| Complexation with fluorescent oligopeptide | Oligopeptide immobilized on liquid crystal surface | NA | FL | 6.4 | 180 | [94] | |
| Complexation with chitin | Chitin-polyaniline film deposited on ITO electrode | Ground and waste waters | PT | 13000 | 4 | [95] | |
| Cr(VI) | Complexation with melanin followed by a reduction in the analyte | Melanin adsorbed on SPE coated with polyvinyl alcohol | Tap and river waters | AP | 1.6 | 1.5 | [74] |
| Cr(VI), Cr(III) | Bioaccumulation in bacterial cells | Drop coating of CPE with a suspension of Sphingopyxis acrogoltabida | River water | CSV | Cr(VI): 0.052 Cr(III): 5.2 | 5 | [6] |
| Fe(III) | Complexation with immobilized deferoxamine | Immobilization of deferoxamine by APM on gold disk coupled an optical fiber | NA | SPR | 111 | 5 | [65] |
| Quenching of parabactin fluorescence | Silica wall of fluorescence flow cell coated with parabactin sealed with sol-gel matrix | Seawater | FL | 0.0022 | 10 | [61] | |
| Complexation with immobilized deferoxamine | Immobilization of deferoxamine by APM on GE | Plants, alloys, and pharmaceuticals | EIS | 0.0011 | 20 | [66] | |
| Hg(II) | Bonding with DNA strands via thymine base | Immobilization of DNA on PGE modified with GNP and methionine | Seawaters and seafood | ASV | 8.0 × 10−13 | 4 | [81] |
| Bonding with DNA strands | SPE coated with DNA strands bonded with of GNP | Tap and river waters | SWV | 0.0012 | 80 | [96] | |
| Complexation with immobilized oligonucleotide | Phosphorothioate oligonucleotide adsorbed on GCE. | NA | SWV | 0.0047 | 60 | [97] | |
| Bonding with DNA strands | Cysteamine reaction with thymin-1-ylacetic acid and immobilization on GCE via APM | Tap waters | DPV | 0.0015 | 15 | [98] | |
| Complexation with synthetic aptamer | Immobilization of aptamers and diaminonaphthalene on FET modified with graphene | Mussel digests | AP | 0.0020 | 0.02 | [99] | |
| Complexation with chlorophyll | Chlorophyll entrapped in a mixture of PVC and NPOE inside a glass disk. | NA | PT | 78 | 2 | [100] | |
| Bonding with immobilized DNA via thymine base | Immobilization on GE | Milk powder | SWV | 0.00010 | 120 | [101] | |
| Pb(II) | Biosorption by yeast cells | SPE coated with lyophilized yeast modified with Co3O4 | Human blood and serum | SWV | 3.4 × 10−9 | 2.5 | [43] |
| Complexation with immobilized aptamer | Immobilization of aptamers on SPE modified with gold and polypyrrol nanoparticles | Biological material and soils | DPV | 0.074 | 30 | [102] | |
| Complexation with immobilized DNA strand | Immobilization of DNA strands on GCE modified with MOF | Tap waters and fertilizers | EIS | 0.0018 | NI | [82] | |
| Bonding with immobilized DNA | Immobilization of DNA on magnetic particles coated with gold layer and retention under a GCE with a magnet | Tap and lake waters, and sediments | DPV | 0.0030 | 5 | [83] | |
| Complexation with immobilized cysteine polymer | Immobilization of cysteine by electropolymerization on CPE | Natural and wastewaters | EIS | 0.000016 | 4 | [79] | |
| Cu(II), Zn(II) | Bonding with oxytocin | Immobilized oxytocin on GCE by APM | Human serum | EIS | NI | 5 | [7] |
| Cd(II), Pb(II) | Complexation with glutathione | Adsorption of glutathione on magnetic solid entrapped over GCE surface using a magnet | Sea, Tap, and mineral waters | ASV | Cd: 0.17 Pb: 0.18 | 3.5 | [17] |
| Analyte | Principle | Remarks | Sample | Technique | DL (µg L−1) | Response Time (min) | Ref. |
|---|---|---|---|---|---|---|---|
| Ca(II) | Enhancement of catalase activity increased O2 generation | Gold cathode of a commercial oximeter coated with a gelatin layer to entrap catalase | Milk | AP | 40 | 60 | [106] |
| Cd(II) | Inhibition of urease activity | Modification of urease with SPDP on GE | NA | SPR | NI | 30 | [64] |
| Inhibition of glucose oxidase activity | Immobilization of enzyme onto PGE modified with carboxylated CNT | NI | DPV | 1600 | <1 | [69] | |
| Inhibition of HRP activity | HRP linked to maize tassel/CNT composite and sealed with Nafion® on GCE | Natural waters | CV | 0.51 | 20 | [107] | |
| Inhibition of photosynthetic O2 release | Anabaena torulosa sealed on the cathode of oximeter with poly(2-hydroxyl ethyl methacrylate) | Wastewaters | AP | NI | <5 | [49] | |
| Inhibition of fluorescence of green fluorescent protein | Encapsulation of the protein using a tetramethoxysilane sol-gel on optical fibers | NA | FL | 32 | NI | [58] | |
| Inhibition of organic matter decomposition | MFC with Shewanella Putrefaciens biofilm. | Wastewaters | PT | 40 | <1 | [50] | |
| Inhibition of HRP activity | Enzyme immobilization on a porous SiO2 surface modified with APTS | Tap, drain and irrigation waters | IRS | 80 | 40 | [108] | |
| Fluorescence quenching of genetically modified E. coli | Modified E. coli encapsulated in polyacrylamide hydrogel platform | NA | FL | 317 | 30 | [60] | |
| Alteration of S. cerevisiae metabolism | Yeast sealed in calcium alginate beads | Well water | DI | 13 | 15 | [109] | |
| Inhibition of urease activity | Immobilization of urease on a silicon surface modified with graphene oxide and gold layers | Rain and river waters | AFM | 0.018 | 15 | [110] | |
| Inhibition of phosphatase activity | Immobilization of phosphatase on antimony tin oxide nanoparticles deposited on a paper support | Seafood | MAS | 0.006 | NI | [111] | |
| Cr(VI) | Potential drop of MFC due to anthropic side reaction with Ochrobactrum | MFC with a polymeric biofilm | Drinking, natural and waste waters | PT | 12 | 45 | [52] |
| Inhibition of catalytic activity of urease | Crude extract containing urease sealed with tetramethyl orthosilicate sol-gel on GE | Wastewater | AP | NI | 25 | [53] | |
| Inhibition of glucose oxidase activity | Entrapment of glucose oxidase with chitosan on paper device | NA | AP | 50 | 5 | [112] | |
| Cr(III) | Inhibition of catalytic activity of HRP | Crosslinked HRP/BSA with poly(neutral red) layer on carbon film electrode. | NA | AP | 1.5 | 1.3 | [113] |
| Cr(III), Cr(VI) | Inhibition of tyrosinase and glucose oxidase activities | Tyrosinase, glucose oxidase, and mediators immobilized on separate SPE | Tap and waste waters | CA | Cr(III): 104 Cr(VI): 4.7 | 17 | [5] |
| Cu(II) | Activation of tyrosinase and oxidation of dopamine | Entrapment of enzyme on polyacrylamide sol-gel after removal of prosthetic Cu(II) ions | Drinking water and milk | MAS | 0.010 | 15 | [114] |
| Fe(II), Fe(III) | Fe(II) oxidation catalyzed by Thiobacillus ferrooxidans | Thiobacillus ferrooxidans/jarosite suspension adsorbed on cellulose and fixed on the cathode of an oximeter using a membrane | Mine waste and mineral extracts | AP | 3300 | <5 | [54] |
| Fe(II), Cr(VI) | Biocatalytic oxidation by Leptospirillum ferrooxidans | Leptospirillum ferrooxidans cells adsorbed on cellulose assembled on cathode of oximeter sealed by a plastic membrane | NA | AP | Fe: 134 Cr: 22 | 0.3 | [67] |
| Fe(II) | Biocatalytic oxidation by Acidithiobacillus ferrooxidans | Cathode of oximeter coated with Acidithiobacillus ferrooxidans sealed by cellulose membrane | NA | AP | 50 | 1.4 | [68] |
| Hg(II) | Inhibition of catalase activity | Catalase/BSA immobilized on GCE via crosslink with glutaraldehyde | Drinking and natural waters | AP | 0.0036 | 10 | [115] |
| Inhibition of Chlorella sp. metabolism | Adsorption of Chlorella sp. on GCE | Wastewaters | AP | 0.014 | 5 | [55] | |
| Mn(II) | Enhancement of HRP activity | Mixing of HRP with carbon paste | NA | AP | 28 | 1 | [116] |
| Ni(II) | Inhibition of Bacillus sphaericus metabolism | Adsorption of Bacillus sphaericus onto cellulose membrane followed by fixation on potentiometric electrode | Wastewaters and food digests | PT | 0.0018 | 1.5 | [56] |
| Pb(II) | Inhibition of choline oxidase | Immobilization of choline oxidase on GCE modified with CNT | Tap waters | AP | 0.0083 | 5 | [117] |
| Cd(II), Co(II), Cu(II), Ni(II) | Inhibition of glucose oxidase activity | Carbon film electrodes modified with Cu or Co hexacyanoferrate and dip-coated with glucose oxidase. | NA | EIS | Cd:135 Co:53 Cu:13 Ni:282 | 5 | [8] |
| Cu(II), Pb(II), Cd(II) | Hindrance of metabolism of Anabaena torulosa | Adsorption of whole cells on cellulose membrane after filtration. | NA | EIS | Cd: 0.027 Cu: 1.2 Pb: 0.10 | 60 | [12] |
| Cd(II), Cu(II), Hg(II), Pb(II) | Inhibition of glucose oxidase activity | Electrodeposition of polypyrrole/glucose oxidase mixture on Pt electrode. | Tap water | AP | Cd:450 Cu:95 Hg: 96 Pb: 332 | 0.3 | [13] |
| Cd(II), Cu(II), Pb(II) | Inhibition of urease activity | Immobilization of urease SPE sealed with tetramethoxysilicate sol-gel | NA | CD | NI | 10 | [14] |
| Cd(II), Co(II), Cu(II) | Inhibition of glucose oxidase activity | Glucose oxidase immobilized on carbon film electrode coated with cobalt hexacyanoferrate (CH) or poly-neutral red (NR) | Tap water | AP | Cd:34 (CH), 888 (NR); Co: 100 (CH), 1100 (NR); Cu: 5.7 (CH), 76 (NR) | NI | [15] |
| Cd(II), Hg(II), Pb(II) | Inhibition of peroxidase activity | Electrodeposition of HRP on Pt disk coated with polyaniline and copolymer poly(2,2′-dithiodianiline) | Tap and river waters | AP | Cd: 8.0 × 10−4 Hg: 7.9 × 10−4 Pb: 9.4 × 10−4 | NI | [16] |
| Cd(II), Cu(II), Hg(II), Pb(II) | Inhibition of glucose oxidase activity | Electrodeposition of a polypyrrole/glucose oxidase film on Pt disk | Tap water | PT | NI | 1.6 | [18] |
| Cu(II), Pb(II) | Inhibition of peroxidase activity | Immobilized HRP onto maize tassel/CNT composite sealed by Nafion® on GCE | Tap water | AP | Cu: 4.2 Pb: 2.5 | 20 | [19] |
| Hg(II), Pb(II) | Inhibition of urease activity | Urease sealed with Nafion® on alumina coated with Au/polyaniline | NA | AP | Hg: 10 Pb: 100 | NI | [10] |
| Hg(II), Cd(II), Pb(II), Cr(VI) | Inhibition of glucose oxidase | Glucose oxidase and brilliant green polymer immobilized on GCE modified with CNT/chitosan | Milk | AP | Hg: 0.46 Cd: 0.20 Pb: 0.50 Cr: 0.12 | NI | [11] |
| Analyte | Principle | Remarks | Sample | Technique | DL (µg L−1) | Response Time (min) | Ref. |
|---|---|---|---|---|---|---|---|
| Ca(II) | Cleavage of immobilized DNAzyme | Immobilization of DNAzyme on a FET modified with CNT | Milk | LSV | 220 | 15 | [125] |
| Cd(II) | Cleavage of DNA followed by hybridization with a single strand labeled with EG | Electrochemical deposition of double stranded DNA on CPE | Tap and sea waters | AP | 1.0 × 10−5 | NI | [126] |
| Complexation with single stranded DNA labeled with MB | Dip coating of GE with thiolated DNA | NA | CV | 0.30 | 15 | [127] | |
| Competition with EG by immobilized double stranded DNA | DNA immobilization on a GCE modified with CNT | NA | DPV | 0.22 | 10 | [128] | |
| Cleavage of immobilized DNAzyme | Immobilization of DNAzyme on a FET modified with CNT | Foods | LSV | 0.0038 | 5 | [129] | |
| Cu(II) | Bonding with immobilized DNA followed by poisoning of the electrode with dehydroascorbic acid | Immobilization of DNA modified with biotin on a carbon-avidin epoxy resin | NA | EIS | 400 | 30 | [130] |
| Bonding with immobilized DNA strand labeled with MOF | Immobilization of DNA on GNP electrodeposited on ITO electrode | Tap and natural waters, and soils | DPV | 0.029 | 80 min | [131] | |
| Spectral shift of polyaniline platform due complexation with immunoglobulin | Immobilization of denatured immunoglobulin on optical fiber coated with polyaniline | Natural waters, soils, and blood | MAS | 0.063 | 25 | [104] | |
| Hybridization of immobilized DNA strand with aptamer labeled with glucose oxidase | Immobilization of DNA strands on a GCE modified with 6-mercaptohexanol and Prussian blue (mediator) | Natural waters | DPV | 6.3 × 10−12 | 30 | [132] | |
| Cleavage of immobilized DNA | Immobilization of thiolated DNA strands on CGE modified with GNP | Natural waters | EIS | 0.0046 | 50 | [133] | |
| Bonding and conformation shift of immobilized DNA labeled with MB | Immobilized thiolated DNA on GE. | Tap water | SWV | 0.078 | 100 | [134] | |
| Fe(III) | Redox immunoreaction of Fe(III) with transferrin | Immobilization of transferrin on the surface of a FET coated with CNT functionalized with anionic surfactants. | Wine | AP | 0.050 | 15 | [30] |
| Hg(II) | GNP release due to bonding with DNA | Thiolated DNA strands immobilized on GE coated with GNP and reduced graphene oxide. | Tap and natural waters | CV | 0.0080 | <0.5 | [135] |
| Inhibition of hybridization of DNA strands labeled with MB | Thiolated DNA strands immobilized on GE modified with GNP | Natural waters | CV | 0.010 | 120 | [136] | |
| Bonding with immobilized oligomer followed by deposition of Ag | Oligonucleotide immobilized on GCE via APM | River waters | DPV | 4.0 × 10−4 | 50 | [137] | |
| Hybridization of DNA strands labeled with MG | Immobilized DNA on CPE modified with SNP. | Tap waters | DPV | 0.0062 | 10 | [138] | |
| Bonding with DNA strands labeled with recognition protein | Immobilized avidin on GNP deposited on GCE using EPM | Herb digests | DPV | 4.2 × 10−5 | 60 | [139] | |
| Bonding with MA labeled with quantum dots | Immobilization of conjugate CH3Hg-MA-ovoalbumin on GCE using GNP | Cosmetic digests | ECL | 0.0026 | 60 | [124] | |
| Hybridization of DNA strands labeled with MB | Immobilization of thiolated single-stranded DNA on GE | Natural waters | SWV | 0.93 | 30 | [140] | |
| Complexation with DNA strands followed by cleavage by exonuclease III | DNA strands immobilized on GE modified with three-dimensional graphene structure | Tap and lake waters, and human serum | SWV | 1.0 × 10−8 | 240 | [40] | |
| Hybridization of labeled DNA strands with MB and ferrocene | Immobilized single-stranded DNA labeled with MB on GE. | Tap and river waters, and human serum | DPV | 0.016 | 120 | [41] | |
| Hybridization of DNA single strands labeled with [Ru(NH3)6]3+ | Immobilized thymine-rich single-stranded DNA on GCE coated with polydopamine-capped graphene oxide. | River water | DPV | 1.0 | 30 | [141] | |
| Hybridization of DNA strands labeled with HRP | Immobilization of thiolated thymine-rich single-stranded DNA on GE. | Drinking water | EIS | 8.0 × 10−5 | 90 | [142] | |
| Hybridization of DNA strands labeled with hemin | Immobilization of thiolated single-stranded DNA on GE | Tap water | AP | 0.0066 | 120 | [143] | |
| Complexation with DNA labeled with MB | Immobilization of DNA labeled with MB on GE. | Tap, river, and drinking waters | SWV | 0.020 | 60 | [144] | |
| Complexation and conformational shift of immobilized DNA | Single-stranded DNA immobilized on GE modified with chitosan and Cu2O nanospheres | River water | EIS | 0.030 | NI | [72] | |
| Complexation and conformational shift of immobilized DNA | Immobilized DNA on GCE coated with polyaniline nanofiber, ordered mesoporous carbon, and GNP | Lake and tap waters | DPV | 1.2 × 10−7 | 870 | [73] | |
| Complexation and conformational shift of immobilized DNA | DNA strands labeled with cysteine immobilized on nanoporous gold surface | NA | SERS | 0.0002 | 30 | [105] | |
| Hybridization of DNA strands | DNA immobilized on GCE modified with MoS2, PDDA and quantum dots | River water, soil, and milk | ECL | 2.0 × 10−5 | 120 | [145] | |
| Hybridization of DNA strands labeled with fluorophore | DNA single strands immobilized on polyaniline nanoclips support | Natural waters | FL | 0.80 | 60 | [62] | |
| Hybridization of DNA strands labeled with Ru complex | Single-stranded DNA labeled with silica/Ru nanoparticles immobilized on GCE modified with GNP. | River waters | ECL | 4.0 × 10−6 | 70 | [146] | |
| Hybridization of DNA via thymine bonding | DNA strands immobilized on fiber support coated with gold film and GNP | Tap water and human serum | SPR | 0.00060 | NI | [147] | |
| DNA dual cycle triggered by exonuclease III | DNA strands immobilized on GCE modified with gold film | Drinking water | SWV | 2.4 × 10−8 | 60 | [148] | |
| Conformational shift of immobilized aptamer labeled with Ru-based composite | Immobilization of aptamer on GCE coated with GNP | Tap and river waters | ECL | 0.0060 | 90 | [149] | |
| Quenching of fluorescence of labeled aptamer after conformational shift | Immobilization on optical fibers after modification of surface with amino-terminated groups | Tap waters | FL | 9.5 × 10−5 | 25 | [150] | |
| DNA dual cycle triggered by exonuclease III | Immobilization of a 3D DNA “safebox” on GCE modified with GNP | Tap waters | DPV | 6.6 × 10−6 | 90 | [151] | |
| Hindrance of hybridization of immobilized DNA | DNA strands immobilized on GCE modified with semiconductor | Tap, river and lake waters | ECL | 0.0010 | 120 | [152] | |
| K(I) | Complexation and conformational shift of immobilized DNA | Single-stranded DNA immobilized on GE | NA | SWV | 0.083 | 5 | [153] |
| Conformational shift of guanine-rich DNA | Immobilized DNA labeled with ferrocene on GE coated with GNP | NA | SWV | NI | <0.5 | [154] | |
| Complexation and conformational shift of DNA | Immobilized DNA on GE coated with GNP | Human urine | SWV | 5.0 × 10−6 | <0.5 | [155] | |
| Complexation and conformational shift of immobilized DNA | Immobilized thiolated DNA on GE | Human urine | EIS | 0.0039 | NI | [156] | |
| Mg(II) | Cleavage of DNAzymes labeled with ferrocene | Immobilization of thiolated DNAzymes labeled with ferrocene on GE | Human serum | DPV | 1200 | NI | [42] |
| Ni(II) | Complexation and conformational shift of DNA structure with peroxidase-like activity | Immobilized tetrahedron DNA structure on GE | Human blood | CV | 0.0088 | 30 | [26] |
| Cleavage of immobilized DNA strands labeled with CdSe | Immobilized DNA strands on GE via APM | NA | DPV | 0.39 | 60 | [157] | |
| Pb(II) | Bonding and cleavage of DNA strands | Immobilization of thiolated DNA strands on GCE modified with GNP. | Natural waters | AP | 8.7 × 10−5 | 120 | [158] |
| Cleavage of DNA chain labeled with Ag nanoparticles | Immobilized labeled DNA on GCE coated with GNP | Tap waters | LSV | 5.0 × 10−5 | 200 | [159] | |
| Complexation and conformational shift of DNA labeled with carbon dots/thionine | Immobilization of thiol-modified aptamer on GCE | River, tap and mineral waters | DPV | 7.9 × 10−4 | 80 | [160] | |
| Hybridization of DNA labeled with MOF | Immobilization of DNAzymes on GNP fixed on GCE by reduced graphene oxide | Natural and tap waters | CA | 4.1 × 10−4 | 45 | [161] | |
| Competitive bonding with EG by DNA | Electrochemical deposition of DNA on bare CPE or after modification with CNT | Tap and sea waters | DPV | 0.021 (bare); 0.0055 (CNT) | 10 | [162] | |
| Competitive bonding with DNA strands labeled with GNP | Immobilization of single-stranded DNA on GE | Human serum | DPV | 0.031 | 210 | [44] | |
| Inhibition of hybridization of DNA strands | Immobilization of DNA aptamer on quartz crystal coated with gold layer | NA | QM | 0.83 | 100 | [163] | |
| Competitive bonding with anthraquinone-2-sulfonic acid by DNA strands | Immobilized thiolated single-stranded DNA on GE | NA | DPV | 0.87 | 30 | [164] | |
| Complexation and conformational shift of DNA strands | Immobilized thiolated single-stranded DNA on GE | Tap waters | EIS | 7.2 | 10 | [165] | |
| Hybridizations of DNA strands labeled with Ru(II) complex | Immobilization of DNA strands on GCE via APM | NA | ECL | 0.0013 | 60 | [166] | |
| Cleavage of DNAzyme labeled with ferrocene | Immobilization of ferrocene-labeled DNAzyme GCE modified with GNP and reduced graphene oxide | Tap and natural waters | DPV | 0.0031 | 40 | [167] | |
| Hybridization of DNA strands labeled with composite with peroxidase-like activity | Immobilized single-stranded DNA on GE | Lake water and human serum | DPV | 6.0 × 10−5 | 40 | [45] | |
| Hybridization of DNA strands labeled with hemin, assigning peroxidase-like activity | Immobilization of thiolated DNA on GE | River waters | DPV | 7.0 × 10−5 | 90 | [168] | |
| Hybridization of DNA strands labeled with hemin, assigning peroxidase-like activity | Immobilization of thiolated DNA on GCE after electrodeposition of a gold layer | Tap and lake waters | DPV | 7.0 × 10−6 | 60 | [169] | |
| Cleavage of DNAzyme | Immobilization of thiolated DNAzyme on Si support | Tap, river, and waste waters | SERS | 0.0018 | 70 | [48] | |
| Hybridization of DNA strands labeled with hemin, assigning peroxidase-like activity | Immobilization of thiolated DNA organized in tetrahedral geometry on GE | Tap and pool waters | CV | 0.0020 | 120 | [170] | |
| Hybridization of DNA strands labeled with recognizing protein | Immobilization of DNA strands on GE | Tap water | DPV | 0.0089 | 230 | [119] | |
| Hindrance of hybridization of DNA strands labeled with thionine and GNP | Immobilization of thiolated single-stranded DNA on gold SPE | Tap water and rat serum | DPV | 0.065 | 240 | [36] | |
| Cleavage of labeled DNAzyme | Immobilization of DNAzyme labeled with methylene blue and ferrocene on GE | Human serum | ACV | 0.0095 | 30 | [37] | |
| Complexation and conformational shift of DNA strands labeled with thionine | Immobilization of single-stranded DNA labeled with GNP and thionine | Tap and river waters | DPV | 6.6 × 10−6 | 70 | [71] | |
| Interaction with anti-Pb(II)-ITCBE MA | Antibody immobilized on nitrocellulose device using GNP and glass fiber | Drinking water | DI | 0.19 | 15 | [171] | |
| Cleavage of DNAzyme labeled with GNP | Immobilization of DNAzyme on gold-coated quartz crystal | Tap water | QM | 2.9 | 40 | [63] | |
| Cleavage of DNAzyme labeled with GNP | DNAzyme immobilized on GE. | Tap and river waters, and soils | CC | 0.0025 | 45 | [172] | |
| Cleavage of immobilized DNA and deposition of Ag | DNA strands immobilized on GE. | River water | SWV | 0.016 | 155 | [173] | |
| Cleavage of labeled DNAzyme | Immobilized ferrocene-labeled DNAzyme on GE | Lake water | DPV | 0.052 | 25 | [174] | |
| Cleavage of immobilized DNAzyme | DNAzyme immobilized on glass surface modified with GNP and graphene | NA | CD | 0.0041 | 20 | [28] | |
| Complexation and conformational shift of DNA strands labeled with chemiluminescent molecule | Immobilized DNA labeled with aminoperylene derivative on GCE coated with fullerene and GNP | Soil leachates | ECL | 7.2 × 10−5 | 60 | [175] | |
| Cleavage of DNA strands | Immobilized thiolated DNA on GCE coated with GNP | Tap, lake, and river waters | EIS | 0.041 | 50 | [176] | |
| Hybridization of DNA strands labeled with MOF with peroxidase-like activity | Immobilized labeled DNA on SPE coated with GNP sealed with chitosan | Soil extracts | CA | 0.0070 | 90 | [177] | |
| DNA cleavage followed by hybridization and labeling with MB | DNA strands immobilized on silicon wafer coated with GNP | Tap waters | SERS | 0.00073 | 90 | [178] | |
| Cleavage of immobilized DNAzyme | Immobilization of DNAzyme on gold surface | Tap water and human serum | SPR | 0.0018 | NI | [179] | |
| Cleavage of immobilized DNAzyme | Immobilization of DNAzyme on GE | Tap waters | EIS | 2.6 × 10−7 | 0.25 | [180] | |
| Conformational shift of immobilized DNA strands | Immobilization of DNA on GCE coated with GNP via APM | NA | EIS | 0.00095 | 40 | [181] | |
| Cleavage of immobilized DNAzyme | Immobilization of DNAzyme on GCE modified with MOF | Lake and tap waters | AP | 0.000014 | 60 | [182] | |
| Cleavage of immobilized DNAzyme labeled with GNP | Immobilization of DNAzyme on gold surface | Groundwaters | SPR | 0.016 | 25 | [183] | |
| Cleavage of immobilized DNAzymes | DNAzymes immobilized on GCE modified with quantum dots | Cell lysates | ECL | 0.000033 | 140 | [184] | |
| Bonding with DNA strands followed by hybridization of labeled DNA strands | DNA strands immobilized on GE | Tap and lake waters | DPV | 0.0037 | NI | [185] | |
| Hg(II), Pb(II) | Cleaved DNA strands (for Pb) and conformational shift (for Hg) | DNA strands immobilized on GE coated with amino-functionalized reduced graphene oxide | Human serum and tomato juice | EIS | Hg: 0.0011 Pb:0.0016 | 5 | [9] |
| Conformational shift of immobilized DNA altered ECL | DNA strands immobilized on GCE modified with quantum dots | Seafood | ECL | Hg(II): 0.00082 Pb(II): 0.0077 | 60 | [186] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rocha, D.L.; Maringolo, V.; Araújo, A.N.; Amorim, C.M.P.G.; Montenegro, M.d.C.B.S.M. An overview of Structured Biosensors for Metal Ions Determination. Chemosensors 2021, 9, 324. https://doi.org/10.3390/chemosensors9110324
Rocha DL, Maringolo V, Araújo AN, Amorim CMPG, Montenegro MdCBSM. An overview of Structured Biosensors for Metal Ions Determination. Chemosensors. 2021; 9(11):324. https://doi.org/10.3390/chemosensors9110324
Chicago/Turabian StyleRocha, Diogo L., Vivian Maringolo, Alberto N. Araújo, Célia M. P. G. Amorim, and Maria da Conceição B. S. M. Montenegro. 2021. "An overview of Structured Biosensors for Metal Ions Determination" Chemosensors 9, no. 11: 324. https://doi.org/10.3390/chemosensors9110324
APA StyleRocha, D. L., Maringolo, V., Araújo, A. N., Amorim, C. M. P. G., & Montenegro, M. d. C. B. S. M. (2021). An overview of Structured Biosensors for Metal Ions Determination. Chemosensors, 9(11), 324. https://doi.org/10.3390/chemosensors9110324








