Molecular Imprinted Based Quartz Crystal Microbalance Sensors for Bacteria and Spores †
Abstract
1. Introduction
2. Materials and Methods
2.1. Surface Imprinting of Microorganisms
2.2. Measurements
3. Results and Discussion
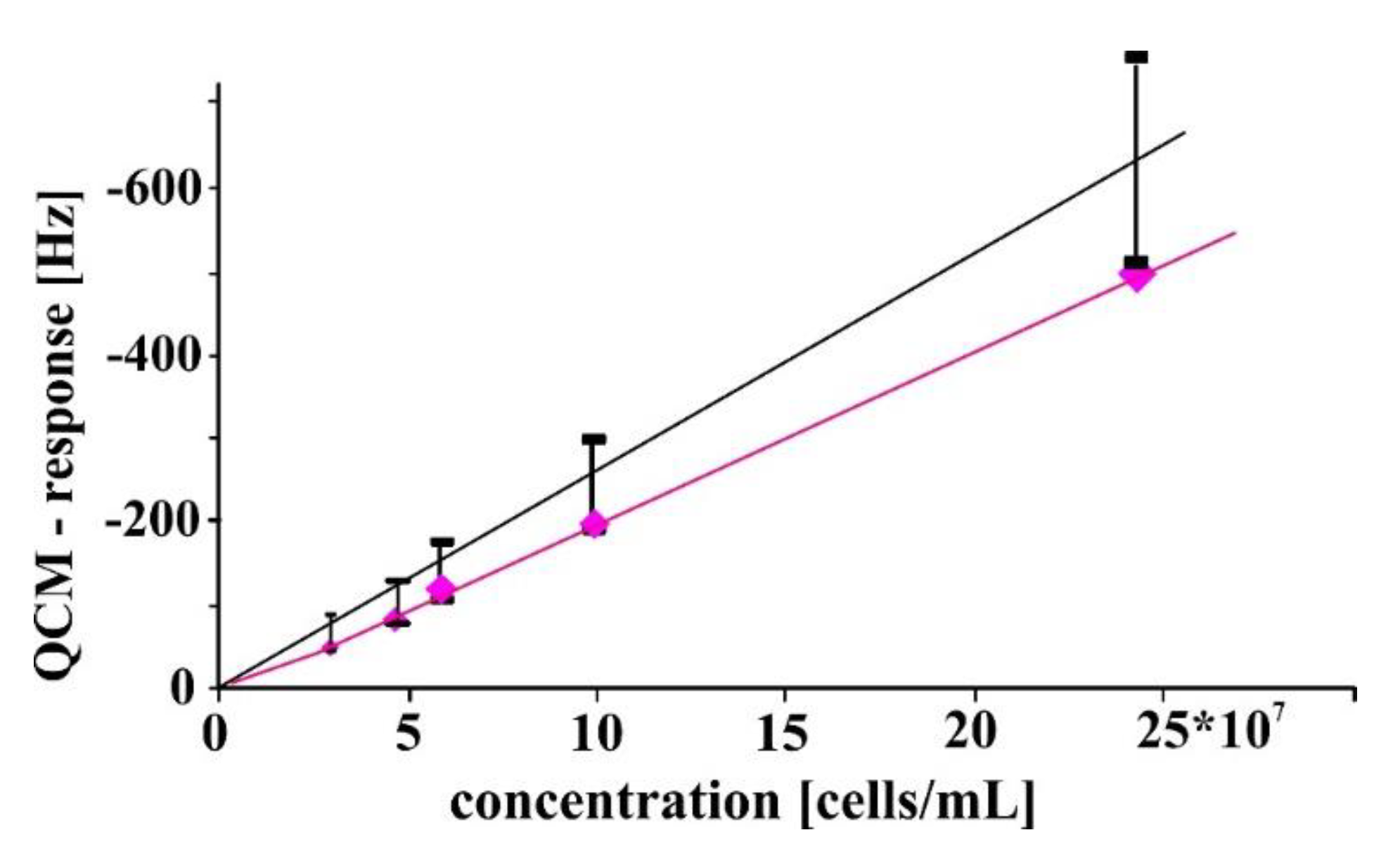
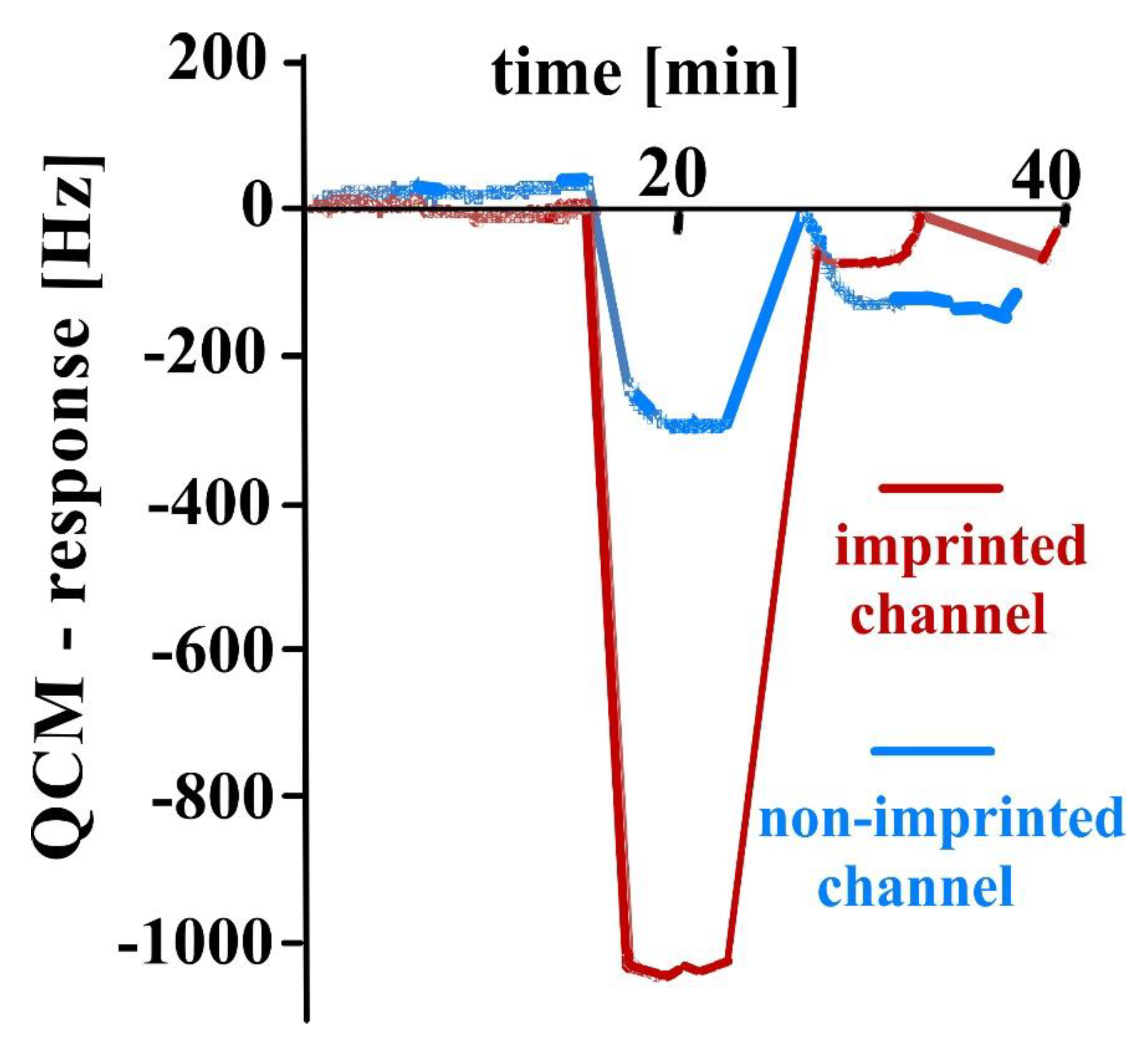
3.1. Sensor Characteristics—Comparison of QCM with AFM Measurements
3.2. Detection of Spores
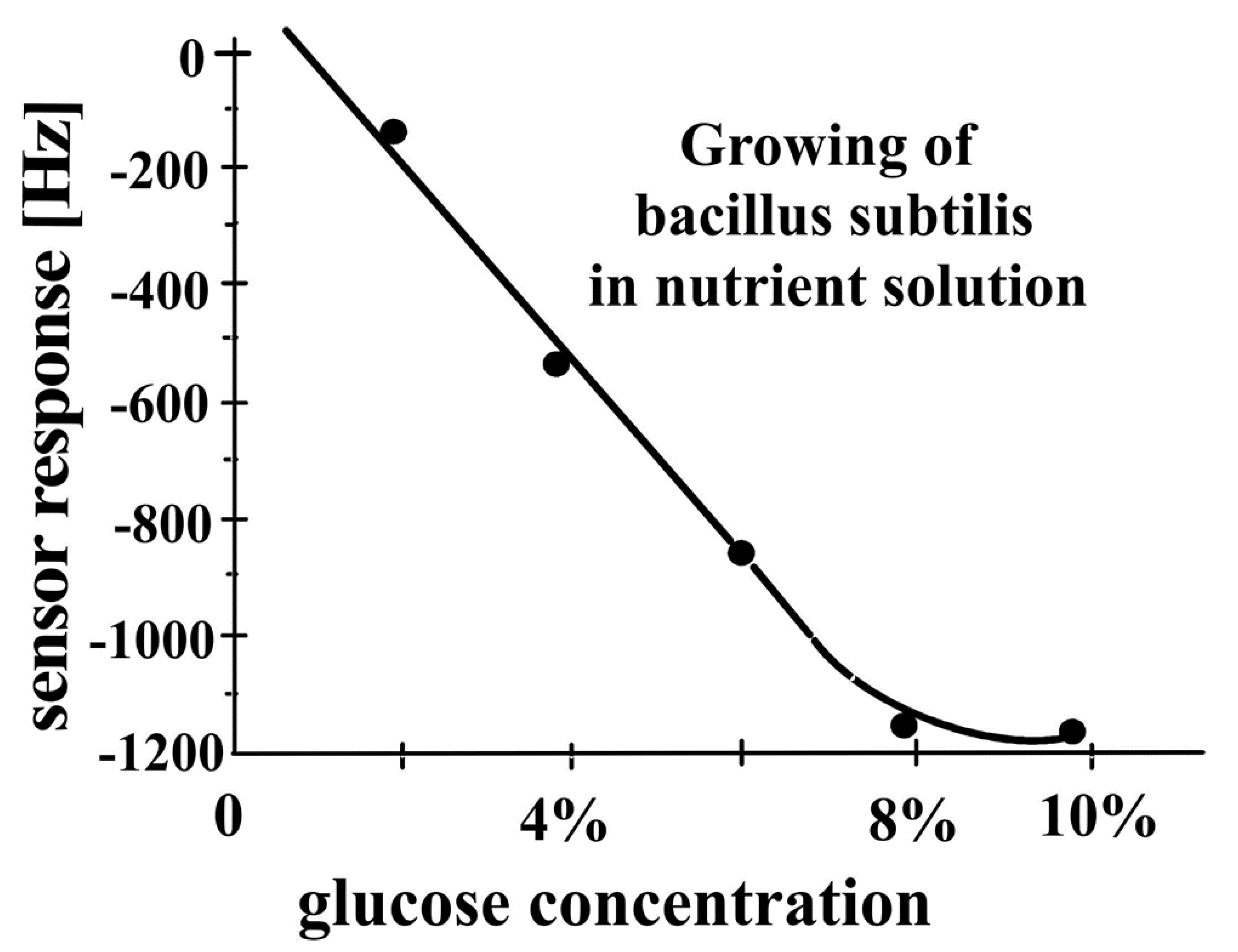
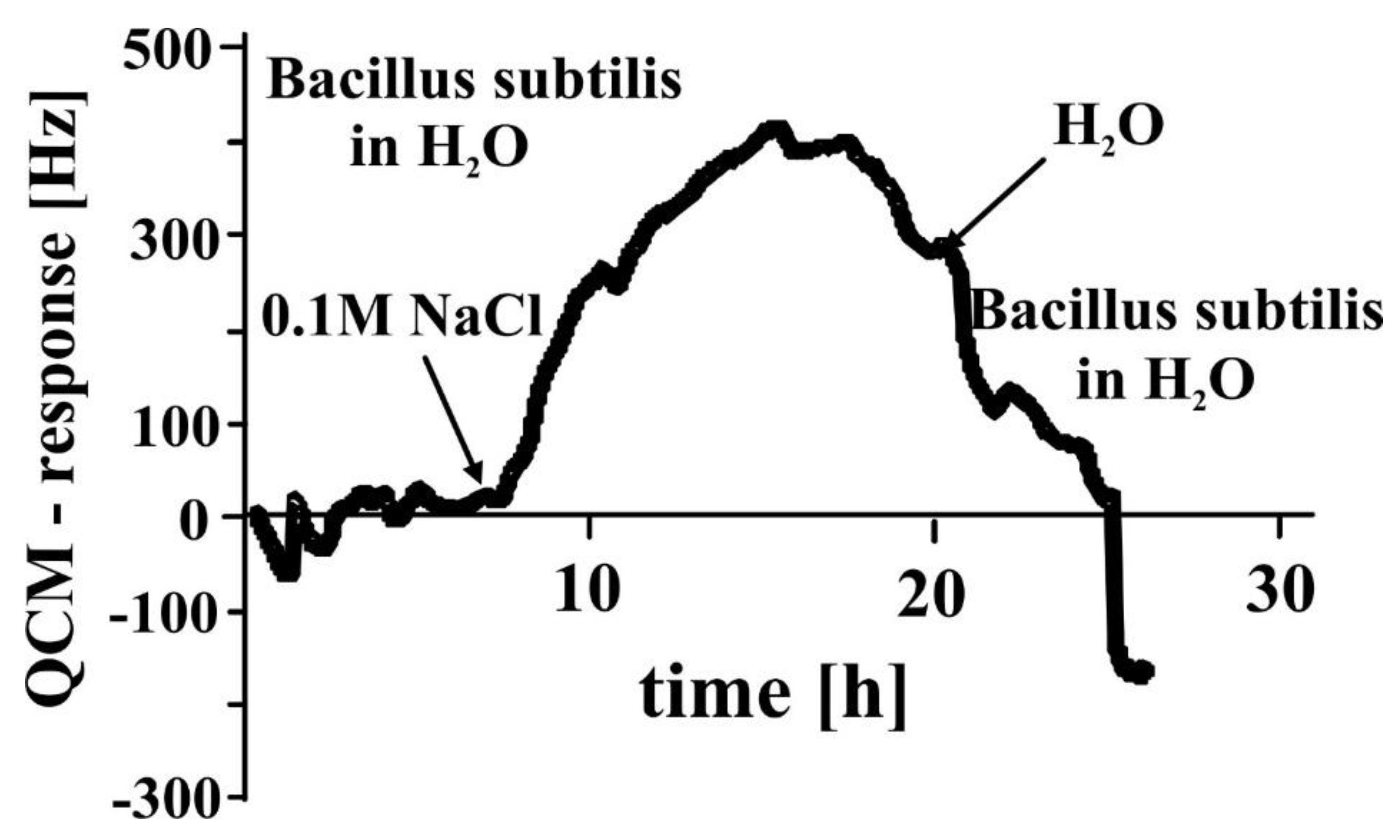
3.3. Biotechnology—Monitoring Bacterial Growth in Nutrient Solutions by QCM Sensors
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Dantism, S.; Röhlen, D.; Wagner, T.; Wagner, P.; Schöning, M.J. A laps-based differential sensor for parallelized metabolism monitoring of various bacteria. Sensors 2019, 19, 4692. [Google Scholar] [CrossRef] [PubMed]
- Dechtrirat, D.; Gajovic-Eichelmann, N.; Wojcik, F.; Hartmann, L.; Bier, F.F.; Scheller, F.W. Electrochemical displacement sensor based on ferrocene boronic acid tracer and immobilized glycan for saccharide binding proteins and e. Coli. Biosens. Bioelectron. 2014, 58, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Jun, S. Evaluation of a microwire sensor functionalized to detect escherichia coli bacterial cells. Biosens. Bioelectron. 2012, 36, 257–261. [Google Scholar] [CrossRef]
- Wang, Y.; Ping, J.; Ye, Z.; Wu, J.; Ying, Y. Impedimetric immunosensor based on gold nanoparticles modified graphene paper for label-free detection of escherichia coli o157:H7. Biosens. Bioelectron. 2013, 49, 492–498. [Google Scholar] [CrossRef]
- Pierucci, O. Dimensions of escherichia coli at various growth rates: Model for envelope growth. J. Bacteriol. 1978, 135, 559–574. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Chen, F.; Wang, R.; Li, Y. Whole-bacterium selex of DNA aptamers for rapid detection of e.Coli o157:H7 using a qcm sensor. J. Biotechnol. 2018, 266, 39–49. [Google Scholar] [CrossRef]
- Basu, P.K.; Indukuri, D.; Keshavan, S.; Navratna, V.; Vanjari, S.R.K.; Raghavan, S.; Bhat, N. Graphene based e. Coli sensor on flexible acetate sheet. Sens Actuators B Chem. 2014, 190, 342–347. [Google Scholar] [CrossRef]
- Levine, M.M. Escherichia coli that cause diarrhea: Enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic, and enteroadherent. J. Infect. Dis. 1987, 155, 377–389. [Google Scholar] [CrossRef]
- Horner, S.R.; Mace, C.R.; Rothberg, L.J.; Miller, B.L. A proteomic biosensor for enteropathogenic e. Coli. Biosens. Bioelectron. 2006, 21, 1659–1663. [Google Scholar] [CrossRef]
- Li, Y.; Afrasiabi, R.; Fathi, F.; Wang, N.; Xiang, C.; Love, R.; She, Z.; Kraatz, H.-B. Impedance based detection of pathogenic e. Coli o157:H7 using a ferrocene-antimicrobial peptide modified biosensor. Biosens. Bioelectron. 2014, 58, 193–199. [Google Scholar] [CrossRef]
- Zhang, W.; Luo, C.; Zhong, L.; Nie, S.; Cheng, W.; Zhao, D.; Ding, S. Sensitive detection of enteropathogenic e. Coli using a bfpa gene-based electrochemical sensor. Microchim. Acta 2013, 180, 1233–1240. [Google Scholar] [CrossRef]
- Tharad, S.; Promdonkoy, B.; Toca-Herrera, J.L. Lipid phase influences the binding of bacillus thuringiensis cyt2aa2 toxin on model lipid membranes. Biochem. Biophys. Res. Commun. 2019, 511, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, B.; Rippere, K.E.; Yousten, A.A.; Priest, F.G. Transfer of bacillus lentimorbus and bacillus popilliae to the genus paenibacillus with emended descriptions of paenibacillus lentimorbus comb. Nov. And paenibacillus popilliae comb. Nov. Int. J. Syst. Bacteriol. 1999, 49, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-B.; Tian, B.; Zhang, Z.-P.; Deng, J.-Y.; Cui, Z.-Q.; Yang, R.-F.; Wang, X.-Y.; Wei, H.-P.; Zhang, X.-E. Rapid detection of bacillus anthracis spores using a super-paramagnetic lateral-flow immunological detectionsystem. Biosens. Bioelectron. 2013, 42, 661–667. [Google Scholar] [CrossRef] [PubMed]
- Knurr, J.; Benedek, O.; Heslop, J.; Vinson, R.B.; Boydston, J.A.; McAndrew, J.; Kearney, J.F.; Turnbough, C.L. Peptide ligands that bind selectively to spores of bacillus subtilis and closely related species. Appl. Environ. Microbiol. 2003, 69, 6841–6847. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, W.L.; Munakata, N.; Horneck, G.; Melosh, H.J.; Setlow, P. Resistance of bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 2000, 64, 548–572. [Google Scholar] [CrossRef]
- Cheng, H.-W.; Chen, Y.-Y.; Lin, X.-X.; Huan, S.-Y.; Wu, H.-L.; Shen, G.-L.; Yu, R.-Q. Surface-enhanced raman spectroscopic detection of bacillus subtilis spores using gold nanoparticle based substrates. Anal. Chim. Acta 2011, 707, 155–163. [Google Scholar] [CrossRef]
- Sengupta, A.; Shende, C.; Farquharson, S.; Inscore, F. Detection of bacillus anthracis spores using peptide functionalized sers-active substrates. Int. J. Spectrosc. 2012, 2012, 6. [Google Scholar] [CrossRef]
- Dalton, R. Genetic sleuths rush to identify anthrax strains in mail attacks. Nature 2001, 413, 657–658. [Google Scholar] [CrossRef]
- Skottrup, P.D.; Nicolaisen, M.; Justesen, A.F. Towards on-site pathogen detection using antibody-based sensors. Biosens. Bioelectron. 2008, 24, 339–348. [Google Scholar] [CrossRef]
- Chenau, J.; Fenaille, F.; Ezan, E.; Morel, N.; Lamourette, P.; Goossens, P.L.; Becher, F. Sensitive detection of bacillus anthracis spores by immunocapture and liquid chromatography–tandem mass spectrometry. Anal. Chem. 2011, 83, 8675–8682. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Rands, A.D.; Losee, S.C.; Holt, B.C.; Williams, J.R.; Lammert, S.A.; Robison, R.A.; Tolley, H.D.; Lee, M.L. Automated thermochemolysis reactor for detection of bacillus anthracis endospores by gas chromatography–mass spectrometry. Anal. Chim. Acta 2013, 775, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Weerasekara, M.L.M.A.W.; Ryuda, N.; Miyamoto, H.; Okumura, T.; Ueno, D.; Inoue, K.; Someya, T. Double-color fluorescence in situ hybridization (fish) for the detection of bacillus anthracis spores in environmental samples with a novel permeabilization protocol. J. Microbiol. Methods 2013, 93, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, P.M.; Fell, N.F., Jr.; Gillespie, J.B. Enhanced spore detection using dipicolinate extraction techniques. Anal. Chim. Acta 2002, 455, 167–177. [Google Scholar] [CrossRef]
- Hathout, Y.; Setlow, B.; Cabrera-Martinez, R.-M.; Fenselau, C.; Setlow, P. Small, acid-soluble proteins as biomarkers in mass spectrometry analysis of bacillus spores. Appl. Environ. Microbiol. 2003, 69, 1100–1107. [Google Scholar] [CrossRef] [PubMed]
- Goodacre, R.; Shann, B.; Gilbert, R.J.; Timmins, É.M.; McGovern, A.C.; Alsberg, B.K.; Kell, D.B.; Logan, N.A. Detection of the dipicolinic acid biomarker in bacillus spores using curie-point pyrolysis mass spectrometry and fourier transform infrared spectroscopy. Anal. Chem. 1999, 72, 119–127. [Google Scholar] [CrossRef]
- Farquharson, S.; Grigely, L.; Khitrov, V.; Smith, W.; Sperry, J.F.; Fenerty, G. Detecting bacillus cereus spores on a mail sorting system using raman spectroscopy. J. Raman Spectrosc. 2004, 35, 82–86. [Google Scholar] [CrossRef]
- Cowcher, D.P.; Xu, Y.; Goodacre, R. Portable, quantitative detection of bacillus bacterial spores using surface-enhanced raman scattering. Anal. Chem. 2013, 85, 3297–3302. [Google Scholar] [CrossRef]
- He, L.; Deen, B.D.; Pagel, A.H.; Diez-Gonzalez, F.; Labuza, T.P. Concentration, detection and discrimination of bacillus anthracis spores in orange juice using aptamer based surface enhanced raman spectroscopy. Analyst 2013, 138, 1657–1659. [Google Scholar] [CrossRef]
- Schirhagl, R.; Latif, U.; Podlipna, D.; Blumenstock, H.; Dickert, F.L. Natural and biomimetic materials for the detection of insulin. Anal. Chem. 2012, 84, 3908–3913. [Google Scholar] [CrossRef]
- Schirhagl, R.; Lieberzeit, P.A.; Dickert, F.L. Chemosensors for viruses based on artificial immunoglobulin copies. Adv. Mater. 2010, 22, 2078–2081. [Google Scholar] [CrossRef] [PubMed]
- Schirhagl, R.; Podlipna, D.; Lieberzeit, P.A.; Dickert, F.L. Comparing biomimetic and biological receptors for insulin sensing. Chem. Commun. 2010, 46, 3128–3130. [Google Scholar] [CrossRef] [PubMed]
- Schirhagl, R.; Seifner, A.; Husain, F.; Cichna-Markl, M.; Lieberzeit, P.; Dickert, F. Antibodies and their replicae in microfluidic sensor systems—Labelfree quality assessment in food chemistry and medicine. Sens. Lett. 2010, 8, 399–404. [Google Scholar] [CrossRef]
- Yaqub, S.; Latif, U.; Dickert, F.L. Plastic antibodies as chemical sensor material for atrazine detection. Sens. Actuators B Chem. 2011, 160, 227–233. [Google Scholar] [CrossRef]
- Jia, M.; Zhang, Z.; Li, J.; Ma, X.; Chen, L.; Yang, X. Molecular imprinting technology for microorganism analysis. TrAC Trends Anal. Chem. 2018, 106, 190–201. [Google Scholar] [CrossRef]
- Mujahid, A.; Mustafa, G.; Dickert, F.L. Label-free bioanalyte detection from nanometer to micrometer dimensions—Molecular imprinting and qcms †. Biosensors 2018, 8, 52. [Google Scholar] [CrossRef]
- Alexander, C.; Vulfson, E.N. Spatially functionalized polymer surfaces produced via cell-mediated lithography. Adv. Mater. 1997, 9, 751–755. [Google Scholar] [CrossRef]
- Findeisen, A.; Wackerlig, J.; Samardzic, R.; Pitkänen, J.; Anttalainen, O.; Dickert, F.L.; Lieberzeit, P.A. Artificial receptor layers for detecting chemical and biological agent mimics. Sens. Actuators B Chem. 2012, 170, 196–200. [Google Scholar] [CrossRef]
- Latif, U.; Can, S.; Hayden, O.; Grillberger, P.; Dickert, F.L. Sauerbrey and anti-sauerbrey behavioral studies in qcm sensors—Detection of bioanalytes. Sens. Actuators B Chem. 2013, 176, 825–830. [Google Scholar] [CrossRef]
- Martínez-Salas, E.; Martín, J.A.; Vicente, M. Relationship of escherichia coli density to growth rate and cell age. J. Bacteriol. 1981, 147, 97–100. [Google Scholar] [CrossRef]
- Howard, G.T. Microbial biodegradation of polyurethane. Recent Dev. Polym. Recycl. 2011, 215, 238. [Google Scholar]
- Xu, Z.; Yuan, Y.J. Quantification of staphylococcus aureus using surface acoustic wave sensors. RSC Adv. 2019, 9, 8411–8414. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Latif, U.; Can, S.; Sussitz, H.F.; Dickert, F.L. Molecular Imprinted Based Quartz Crystal Microbalance Sensors for Bacteria and Spores. Chemosensors 2020, 8, 64. https://doi.org/10.3390/chemosensors8030064
Latif U, Can S, Sussitz HF, Dickert FL. Molecular Imprinted Based Quartz Crystal Microbalance Sensors for Bacteria and Spores. Chemosensors. 2020; 8(3):64. https://doi.org/10.3390/chemosensors8030064
Chicago/Turabian StyleLatif, Usman, Serpil Can, Hermann F. Sussitz, and Franz L. Dickert. 2020. "Molecular Imprinted Based Quartz Crystal Microbalance Sensors for Bacteria and Spores" Chemosensors 8, no. 3: 64. https://doi.org/10.3390/chemosensors8030064
APA StyleLatif, U., Can, S., Sussitz, H. F., & Dickert, F. L. (2020). Molecular Imprinted Based Quartz Crystal Microbalance Sensors for Bacteria and Spores. Chemosensors, 8(3), 64. https://doi.org/10.3390/chemosensors8030064





