Yeast Propagation Control: Low Frequency Electrochemical Impedance Spectroscopy as an Alternative for Cell Counting Chambers in Brewery Applications
Abstract
1. Introduction
2. Materials and Methods
2.1. Expression Host and Cultivation
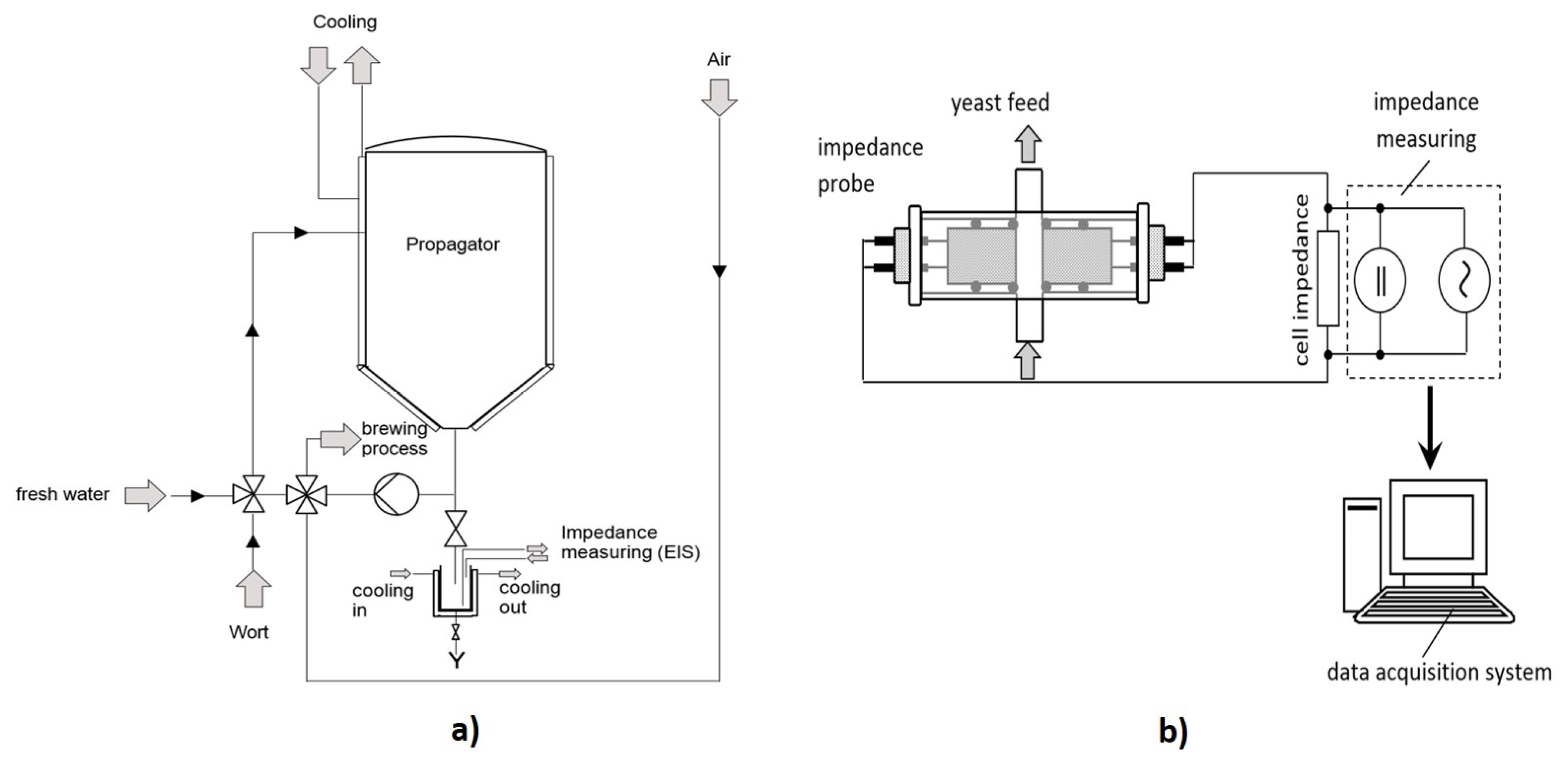
2.2. Process Analytics
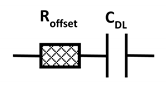
2.3. Impedance Measurements
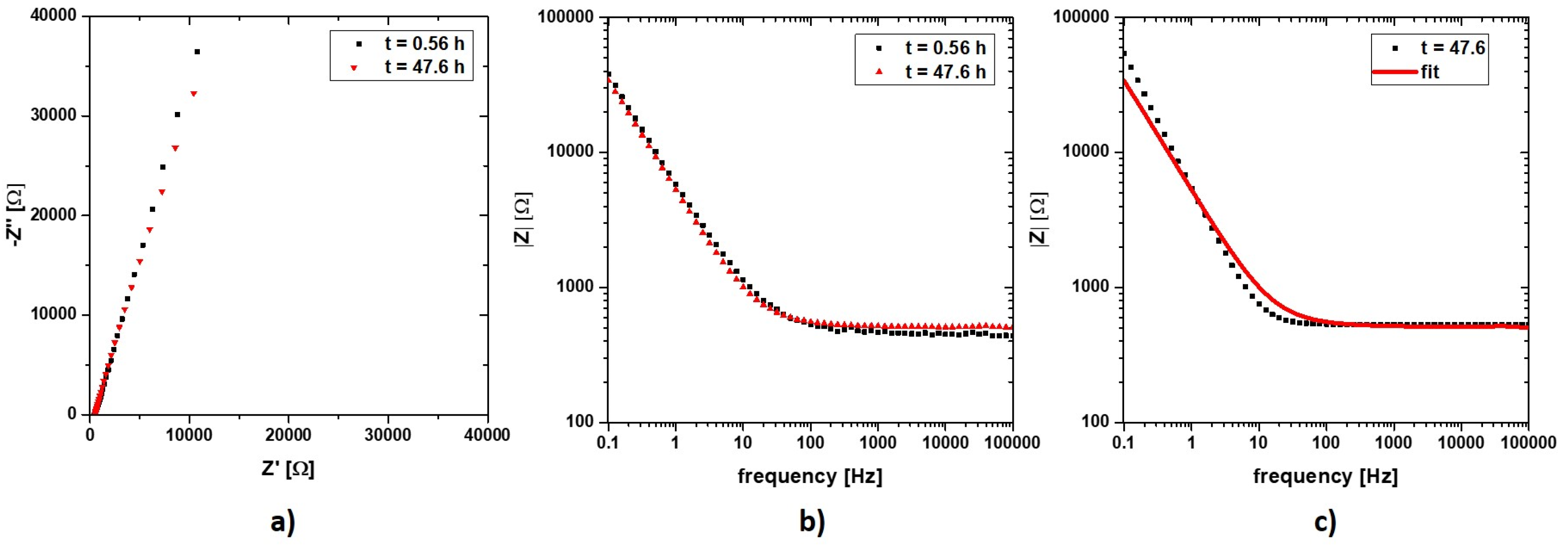
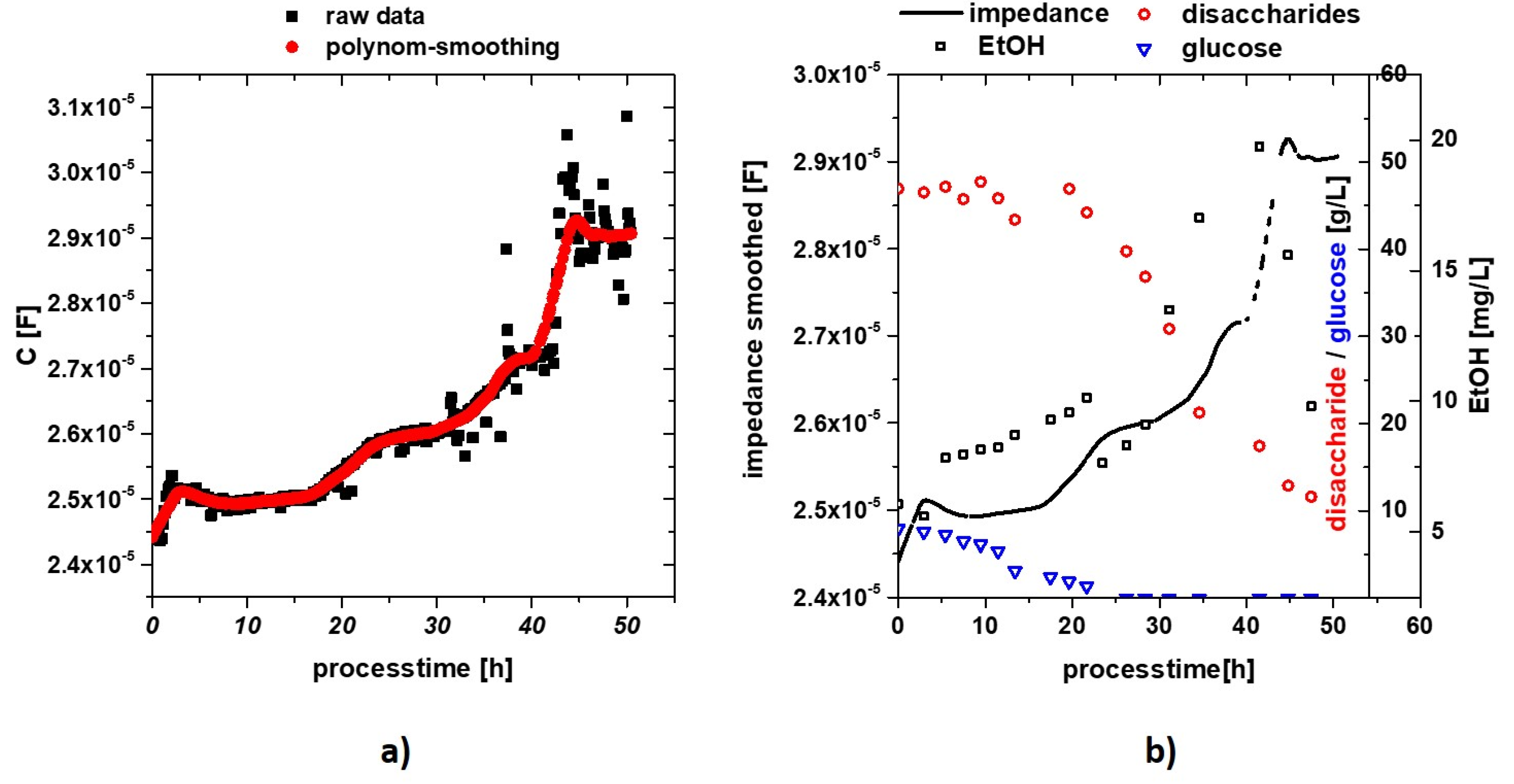
3. Results and Discussion
3.1. Labscale Propagation
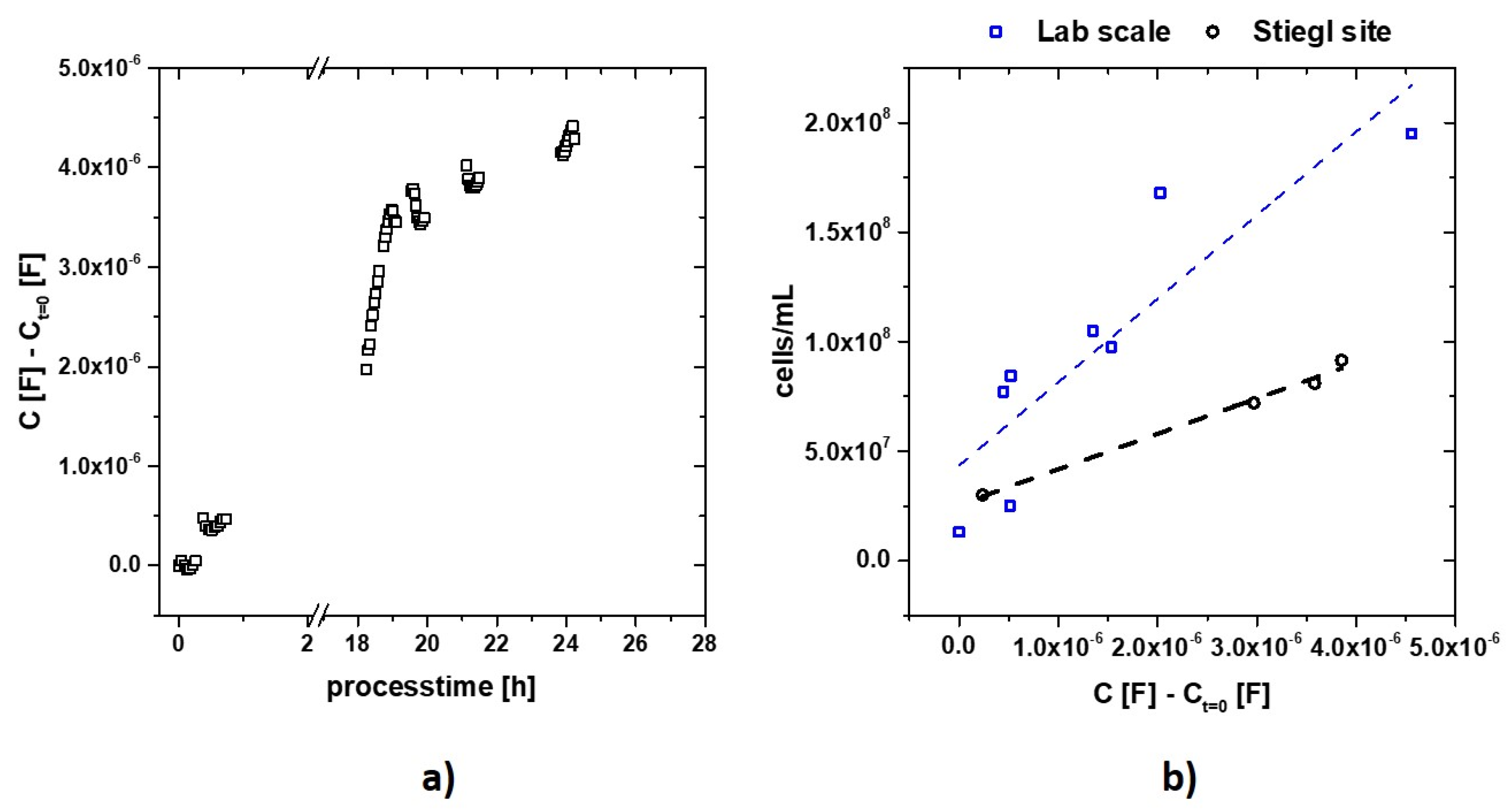
3.2. Impedance Measurement Atline at Stiegl Brewery
4. Conclusions and Outlook
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| VCC | viable cell concentration |
| FCM | flow cytometry |
| AC | alternating current |
| DC | direct current |
| OD | optical density |
| CDW | cell dry weight |
| C | capacitancelayer, |
| Roffset | offset resistance, |
| Z | impedance, |
| HPLC | high performance liquid chromatography. |
References
- Gavrilescu, M.; Chisti, Y. Biotechnology—A sustainable alternative for chemical industry. Biotechnol. Adv. 2005, 23, 471–499. [Google Scholar] [CrossRef] [PubMed]
- Salgado, A.; Folly, R.; Valdman, B. Biomass monitoring by use of a continuous on-line optical sensor. Sens. Actuators B Chem. 2001, 75, 24–28. [Google Scholar] [CrossRef]
- Clarke, D.; Blake-Coleman, B.; Carr, R.; Calder, M.; Atkinson, T. Monitoring reactor biomass. Trends Biotechnol. 1986, 4, 173–178. [Google Scholar] [CrossRef]
- Kiviharju, K.; Salonen, K.; Moilanen, U.; Eerikäinen, T. Biomass measurement online: The performance of in situ measurements and software sensors. J. Ind. Microbiol. Biotechnol. 2008, 35, 657–665. [Google Scholar] [CrossRef]
- Davey, H.M.; Kell, D.B. Flow cytometry and cell sorting of heterogeneous microbial populations: The importance of single-cell analyses. Microbiol. Rev. 1996, 60, 641–696. [Google Scholar] [CrossRef]
- Veal, D.A.; Deere, D.; Ferrari, B.; Piper, J.; Attfield, P.V. Fluorescence staining and flow cytometry for monitoring microbial cells. J. Immunol. Methods 2000, 243, 191–210. [Google Scholar] [CrossRef]
- Fiala, J.; Lloyd, D.; Rychtera, M.; Kent, C.; Al-Rubeai, M. Evaluation of cell numbers and viability of Saccharomyces cerevisiae by different counting methods. Biotechnol. Tech. 1999, 13, 787–795. [Google Scholar] [CrossRef]
- Lodolo, E.J.; Kock, J.L.; Axcell, B.C.; Brooks, M. The yeast Saccharomyces cerevisiae–the main character in beer brewing. FEMS Yeast Res. 2008, 8, 1018–1036. [Google Scholar] [CrossRef]
- Park, J.-Y.; Park, S.-M. DNA hybridization sensors based on electrochemical impedance spectroscopy as a detection tool. Sensors 2009, 9, 9513–9532. [Google Scholar] [CrossRef]
- Song, K.-M.; Lee, S.; Ban, C. Aptamers and their biological applications. Sensors 2012, 12, 612–631. [Google Scholar] [CrossRef]
- Lee, J.-O.; So, H.-M.; Jeon, E.-K.; Chang, H.; Won, K.; Kim, Y.H. Aptamers as molecular recognition elements for electrical nanobiosensors. Anal. Bioanal. Chem. 2008, 390, 1023–1032. [Google Scholar] [CrossRef] [PubMed]
- Yang, L. Electrical impedance spectroscopy for detection of bacterial cells in suspensions using interdigitated microelectrodes. Talanta 2008, 74, 1621–1629. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Yu, G.; Kim, T.; Shin, K.; Yoon, J. Rapid bacterial detection with an interdigitated array electrode by electrochemical impedance spectroscopy. Electrochim. Acta 2012, 82, 126–131. [Google Scholar] [CrossRef]
- Schwan, H.P. Electrical and acoustic properties of biological materials and biomedical applications. Biomed. Eng. IEEE Trans. 1984, 12, 872–878. [Google Scholar] [CrossRef] [PubMed]
- Schwan, H.P.; Foster, K.R. RF-field interactions with biological systems: Electrical properties and biophysical mechanisms. Proc. IEEE 1980, 68, 104–113. [Google Scholar] [CrossRef]
- Yardley, J.E.; Kell, D.B.; Barrett, J.; Davey, C.L. On-line, real-time measurements of cellular biomass using dielectric spectroscopy. Biotechnol. Genet. Eng. Rev. 2000, 17, 3–36. [Google Scholar] [CrossRef]
- Dabros, M.; Dennewald, D.; Currie, D.J.; Lee, M.H.; Todd, R.W.; Marison, I.W.; von Stockar, U. Cole–Cole, linear and multivariate modeling of capacitance data for on-line monitoring of biomass. Bioprocess Biosyst. Eng. 2009, 32, 161–173. [Google Scholar] [CrossRef]
- Soley, A.; Lecina, M.; Gámez, X.; Cairo, J.; Riu, P.; Rosell, X.; Bragos, R.; Godia, F. On-line monitoring of yeast cell growth by impedance spectroscopy. J. Biotechnol. 2005, 118, 398–405. [Google Scholar] [CrossRef]
- Carvell, J.P.; Dowd, J.E. On-line measurements and control of viable cell density in cell culture manufacturing processes using radio-frequency impedance. Cytotechnology 2006, 50, 35–48. [Google Scholar] [CrossRef]
- Gerngross, T.U. Advances in the production of human therapeutic proteins in yeasts and filamentous fungi. Nat. Biotech. 2004, 22, 1409–1414. [Google Scholar] [CrossRef]
- Buckholz, R.G.; Gleeson, M.A.G. Yeast Systems for the Commercial Production of Heterologous Proteins. Nat. Biotech. 1991, 9, 1067–1072. [Google Scholar] [CrossRef] [PubMed]
- Cereghino, G.P.L.; Cregg, J.M. Applications of yeast in biotechnology: Protein production and genetic analysis. Curr. Opin. Biotechnol. 1999, 10, 422–427. [Google Scholar] [CrossRef]
- Konakovsky, V.; Yagtu, A.C.; Clemens, C.; Müller, M.M.; Berger, M.; Schlatter, S.; Herwig, C. Universal Capacitance Model for Real-Time Biomass in Cell Culture. Sensors 2015, 15, 22128–22150. [Google Scholar] [CrossRef] [PubMed]
- Ehgartner, D.; Sagmeister, P.; Herwig, C.; Wechselberger, P. A novel real-time method to estimate volumetric mass biodensity based on the combination of dielectric spectroscopy and soft-sensors. J. Chem. Technol. Biotechnol. 2015, 90, 262–272. [Google Scholar] [CrossRef]
- Ferreira, A.P.; Vieira, L.M.; Cardoso, J.P.; Menezes, J.C. Evaluation of a new annular capacitance probe for biomass monitoring in industrial pilot-scale fermentations. J. Biotechnol. 2005, 116, 403–409. [Google Scholar] [CrossRef]
- Mishima, K.; Mimura, A.; Takahara, Y.; Asami, K.; Hanai, T. On-line monitoring of cell concentrations by dielectric measurements. J. Ferment. Bioeng. 1991, 72, 291–295. [Google Scholar] [CrossRef]
- Radke, S.M.; Alocilja, E.C. Design and fabrication of a microimpedance biosensor for bacterial detection. Sens. J. IEEE 2004, 4, 434–440. [Google Scholar] [CrossRef]
- Yang, L.; Li, Y.; Griffis, C.L.; Johnson, M.G. Interdigitated microelectrode (IME) impedance sensor for the detection of viable Salmonella typhimurium. Biosens. Bioelectron. 2004, 19, 1139–1147. [Google Scholar] [CrossRef]
- Liu, J.; Li, H.; Zhang, F.; Li, X.; Wang, L.; Chen, Y. Online impedance monitoring of yeast cell culture behaviors. Microelectron. Eng. 2011, 88, 1711–1713. [Google Scholar] [CrossRef]
- Lei, K.F. Review on impedance detection of cellular responses in micro/nano environment. Micromachines 2014, 5, 1–12. [Google Scholar] [CrossRef]
- Yang, L.; Ruan, C.; Li, Y. Detection of viable Salmonella typhimurium by impedance measurement of electrode capacitance and medium resistance. Biosens. Bioelectron. 2003, 19, 495–502. [Google Scholar] [CrossRef]
- Gonzalez, J.; Santana, A.F.; Mirza-Rosca, J. Effect of bacterial biofilm on 316 SS corrosion in natural seawater by EIS. Corros. Sci. 1998, 40, 2141–2154. [Google Scholar] [CrossRef]
- Bayoudh, S.; Othmane, A.; Ponsonnet, L.; Ouada, H.B. Electrical detection and characterization of bacterial adhesion using electrochemical impedance spectroscopy-based flow chamber. Colloids Surf. A Physicochem. Eng. Asp. 2008, 318, 291–300. [Google Scholar] [CrossRef]
- Wu, J.; Ben, Y.; Chang, H.-C. Particle detection by electrical impedance spectroscopy with asymmetric-polarization AC electroosmotic trapping. Microfluid. Nanofluidics 2005, 1, 161–167. [Google Scholar] [CrossRef]
- K’Owino, I.O.; Sadik, O.A. Impedance spectroscopy: A powerful tool for rapid biomolecular screening and cell culture monitoring. Electroanalysis 2005, 17, 2101–2113. [Google Scholar] [CrossRef]
- Kim, Y.-H.; Park, J.-S.; Jung, H.-I. An impedimetric biosensor for real-time monitoring of bacterial growth in a microbial fermentor. Sens. Actuators B Chem. 2009, 138, 270–277. [Google Scholar] [CrossRef]
- Slouka, C.; Brunauer, G.; Kopp, J.; Strahammer, M.; Fricke, J.; Fleig, J.; Herwig, C. Low-Frequency Electrochemical Impedance Spectroscopy as a Monitoring Tool for Yeast Growth in Industrial Brewing Processes. Chemosensors 2017, 5, 24. [Google Scholar] [CrossRef]
- Slouka, C.; Wurm, D.J.; Brunauer, G.; Welzl-Wachter, A.; Spadiut, O.; Fleig, J.; Herwig, C. A Novel Application for Low Frequency Electrochemical Impedance Spectroscopy as an Online Process Monitoring Tool for Viable Cell Concentrations. Sensors 2016, 16, 1900. [Google Scholar] [CrossRef]
- Veiter, L.; Herwig, C. The filamentous fungus Penicillium chrysogenum analysed via flow cytometry—A fast and statistically sound insight into morphology and viability. Appl. Microbiol. Biotechnol. 2019, 103, 6725–6735. [Google Scholar] [CrossRef]
- De Deken, R. The Crabtree effect: A regulatory system in yeast. Microbiology 1966, 44, 149–156. [Google Scholar] [CrossRef]
- Seward, R.; Willetts, J.C.; Dinsdale, M.G.; Lloyd, D. The effects of ethanol, hexan-1-ol, and 2-phenylethanol on cider yeast growth, viability, and energy status; synergistic inhibition. J. Inst. Brew. 1996, 102, 439–443. [Google Scholar] [CrossRef]
- Fleig, J. The grain boundary impedance of random microstructures: Numerical simulations and implications for the analysis of experimental data. Solid State Ion. 2002, 150, 181–193. [Google Scholar] [CrossRef]
- Christensen, P.; Stryhn, H.; Hansen, C. Discrepancies in the determination of sperm concentration using Bürker-Türk, Thoma and Makler counting chambers. Theriogenology 2005, 63, 992–1003. [Google Scholar] [CrossRef] [PubMed]
- Wasserberg, D.; Zhang, X.; Breukers, C.; Connell, B.J.; Baeten, E.; van den Blink, D.; Èlia, S.; Bloem, A.C.; Nijhuis, M.; Wensing, A.M. All-printed cell counting chambers with on-chip sample preparation for point-of-care CD4 counting. Biosens. Bioelectron. 2018, 117, 659–668. [Google Scholar] [CrossRef] [PubMed]
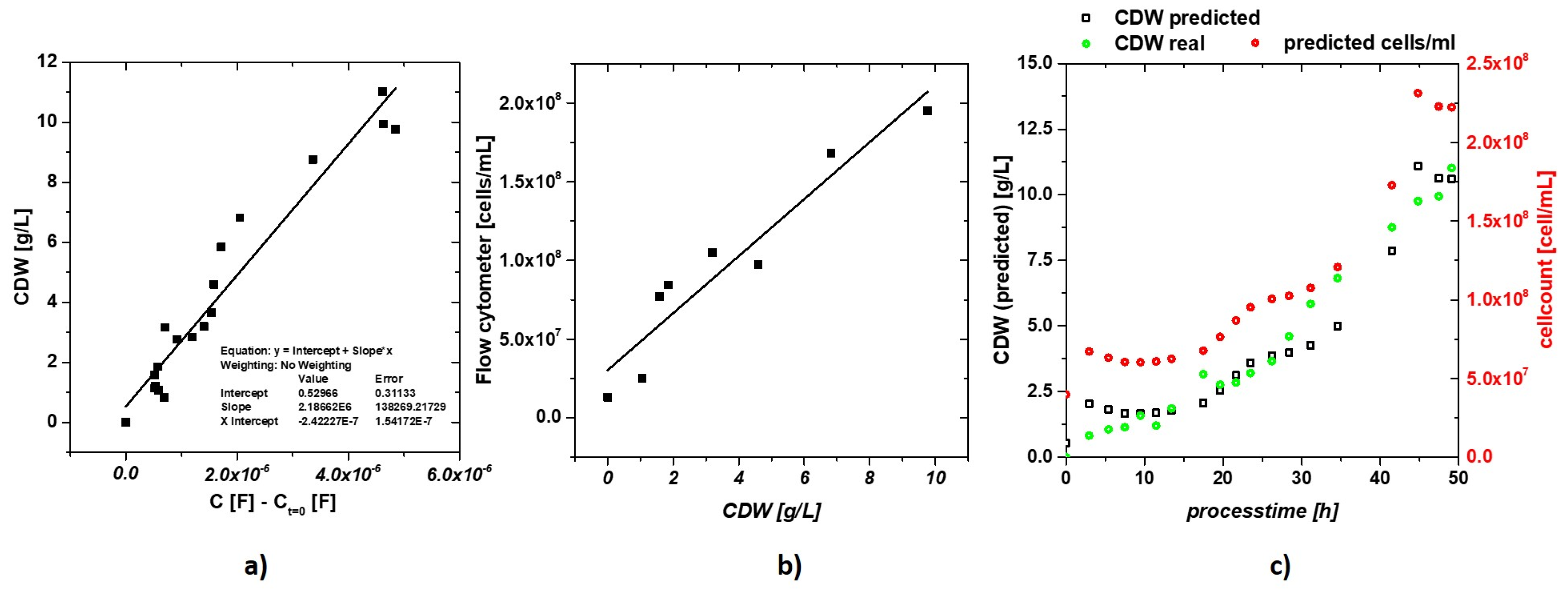
| Sample | Batch Time [h] | Glucose [mmol/L] | EtOH [mg/L] | OD600 [AU] | CDW [g/L] |
|---|---|---|---|---|---|
| wort (4 L) | 59.4 | 507 | 0.6 | ||
| (1 L pure yeast) | 0.2 | 28,317 | 6.8 | 4.74 | |
| Batch start | 0 | 47.6 | 6069 | 1.4 | 0 |
| S1 | 2.93 | 57.1 | 5614 | 2.3 | 0.82 |
| S2 | 5.4 | 55.6 | 7828 | 2.6 | 1.06 |
| S3 | 7.48 | 49.4 | 7968 | 2.9 | 1.14 |
| S4 | 9.47 | 40.6 | 8167 | 3.0 | 1.58 |
| S5 | 11.47 | 36.3 | 8243 | 3.4 | 1.2 |
| S6 | 13.42 | 31.2 | 8715 | 3.6 | 1.86 |
| S7 | 17.47 | 18.7 | 9293 | 4.3 | 3.16 |
| S8 | 19.62 | 11.9 | 9568 | 4.9 | 2.76 |
| S9 | 21.64 | 5.8 | 10,135 | 5.3 | 2.84 |
| S10 | 23.48 | 2.2 | 7638 | 5.9 | 3.2 |
| S11 | 26.21 | 0.4 | 8323 | 6.6 | 3.66 |
| S12 | 28.38 | 0.1 | 9116 | 6.9 | 4.6 |
| S13 | 31.13 | 0.0 | 13,495 | 7.7 | 5.84 |
| S14 | 34.56 | 0.0 | 17,018 | 8.6 | 6.82 |
| S15 | 41.49 | 0.0 | 19,751 | 11.7 | 8.76 |
| S16 | 44.80 | 0.0 | 15,599 | 12.1 | 9.76 |
| S17 | 47.44 | 0.0 | 9806 | 14.3 | 9.94 |
| S18 | 49.09 | 0.0 | 16.1 | 11.02 |
| Sample No. | Time [h] | C [F] – Ct=0 [F] | [cell/mL] |
|---|---|---|---|
| 1 | 0.4 | 2.36E-07 | 2.94E+07 |
| 2 | 18.7 | 2.97E-06 | 7.36E+07 |
| 3 | 19.7 | 3.58E-06 | 8.35E+07 |
| 4 | 21.3 | 3.86E-06 | 8.80E+07 |
| 5 | 24.0 | 4.26E-06 | 9.45E+07 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brunauer, G.C.; Spadiut, O.; Gruber, A.; Slouka, C. Yeast Propagation Control: Low Frequency Electrochemical Impedance Spectroscopy as an Alternative for Cell Counting Chambers in Brewery Applications. Chemosensors 2020, 8, 27. https://doi.org/10.3390/chemosensors8020027
Brunauer GC, Spadiut O, Gruber A, Slouka C. Yeast Propagation Control: Low Frequency Electrochemical Impedance Spectroscopy as an Alternative for Cell Counting Chambers in Brewery Applications. Chemosensors. 2020; 8(2):27. https://doi.org/10.3390/chemosensors8020027
Chicago/Turabian StyleBrunauer, Georg Christoph, Oliver Spadiut, Alfred Gruber, and Christoph Slouka. 2020. "Yeast Propagation Control: Low Frequency Electrochemical Impedance Spectroscopy as an Alternative for Cell Counting Chambers in Brewery Applications" Chemosensors 8, no. 2: 27. https://doi.org/10.3390/chemosensors8020027
APA StyleBrunauer, G. C., Spadiut, O., Gruber, A., & Slouka, C. (2020). Yeast Propagation Control: Low Frequency Electrochemical Impedance Spectroscopy as an Alternative for Cell Counting Chambers in Brewery Applications. Chemosensors, 8(2), 27. https://doi.org/10.3390/chemosensors8020027






