Abstract
In recent years, reagentless aptamer biosensors, named aptasensors, have shown significant advancements. Particularly, electrochemical aptasensors could change the field of biosensors in this era, where digitalization seems to be a common goal of many fields. Biomedical devices are integrating electronic technologies for detecting pathogens, biomolecules, small molecules, and ions, and the physical-chemical properties of nucleic acid aptamers makes them very interesting for these devices. Aptamers can be easily synthesized and functionalized with functional groups for immobilization and with redox chemical groups that allow for the conversion of molecular interactions into electrical signals. Furthermore, non-labeled aptamers have also been utilized. This review presents the current challenges involved in aptasensor architectures based on gold electrodes as transducers.
1. Introduction
Biosensors are devices that facilitate the specific detection of analytes and produce detectable signals, correlating the presence of a target analyte such as proteins, DNA, glucose, cholesterol, toxins, hormones, bacteria, etc. Significant advances have been made in several areas of electrochemical biosensor design. Although a handful of biosensor solutions have successfully made a transition from laboratory benchtop to portable point-of-care devices, the sensitivity and selectivity of most electrochemical biosensors are not satisfactory. Contributions from electronics, biology, physics, chemistry, and materials science are enabling the advance of the biosensor field. In general, biosensors must quantify, identify, or characterize a biologically derived sample. The major challenges for the biosensor industry are the robustness, accuracy, and manufacture of cost-effective devices; yet, in recent years, this field has experienced continuous improvement.
Before developing a biosensor for commercial purposes, it is important to consider the analytical task and the specification requirements, such as sampling and sample preparation, type and concentration range of the analyte, composition of the sample matrix, background noise, regulations for the final application, and cost of the device. Normally, a biosensor is manufactured to be small and to be deployed in the field where samples cannot be processed as accurately as in a central laboratory. A good affinity-based biosensor must be developed to facilitate the binding of a probe to a specific target analyte and must produce a measurable signal that correlates with the presence of the analyte. Among the various signal readout options of biosensors, fluorescence and electrochemical-based devices are the most widely studied systems. Whereas optical detection methods have historically dominated new devices, electrochemical methods have several advantages, including the development of reagent-free detection applications. In this review, we focus on the latter due to their high impact on the digital world and the development of point-of-care (POC) devices and lab on-chip systems.
Potential probes for commercial electrochemical affinity-based biosensors include enzymes (e.g., oxidoreductases) and DNA molecules for genomic analysis and DNA or RNA aptamers for capturing target analytes. Many commercial devices employ enzymes, particularly glucose detectors in diabetic patients, representing approximately 85% of the biosensor market [1]. The glucose sensor was introduced in 1962 and has a long research record [2]. However, the discovery of new probes for electrochemical biosensors with excellent specificity and sensitivity to capture and detect target molecules other than glucose remains a challenge.
While enzymes have high selectivity for their substrates, they possess several disadvantages that affect the electron transfer efficiency—(1) they are large molecules, (2) the active site is usually deeply buried, affecting the electron transfer, and (3) they have a limited shelf life. To overcome these problems, efforts are dedicated to finding enzyme immobilization procedures and protein engineering in order to improve the electron transfer efficiency of enzymes. While solutions to these problems are solved case by case, the discovery of novel molecules for biosensors should be done in a systematic way compatible with high-throughput procedures. For example, several technologies have been developed in the field of drug discovery based on high parallelism processes for the interrogation of thousands to millions of molecules in a cost-effective timeframe. Hence, there is still a great opportunity in the biosensor field for better technologies that could detect a larger number of molecules with more specificity and bring novel assays to the clinic, and this demand can be readily approached with the aptamer technology. Aptamer-based biosensor technology is the fastest-growing aptamer field, and it promises to continuously produce novel clinical or companion drug diagnostic and personalized theranostic assays.
Aptamers are short single-stranded oligonucleotide molecules (ssDNA or ssRNA) with the ability to fold in a stable three-dimensional structure that allows them to interact sterically and through electrostatic interactions with a target molecule. In general, they are 15 to 80 nucleotides long. Aptamers are identified from synthetic ssDNA or ssRNA libraries through a process termed systematic evolution of ligands by exponential enrichment (SELEX) [3]. Aptamers were discovered 29 years ago, and since then, they have received extensive attention. So far, the accumulated data on aptamer functionality have demonstrated that protein molecules (e.g., enzymes and antibodies) are no longer the only entities that may be developed to bind to targets with high affinity and specificity. Aptamers have exceptional advantages—(1) they are chemically synthesized by solid-phase technology, which renders large-scale production cost-effective and confers reproducibility among different batches when compared to biological molecules, (2) they withstand long-term shelf storage at room temperature while preserving their activity, (3) it is easy to perform controlled modification, and (4) they have a high flexible structure, which renders ideal molecules for electrochemical biosensors [4].
To create an aptamer electrochemical biosensor, it is important to choose a conductive physicochemical transducer and, in some cases, to label the aptamer with good redox chemistry. Depending on the properties of the semiconductor, it will be necessary to follow a strategy to immobilize the nucleic acids onto the surface of the electrode. The most common electrode material for electrochemical nucleic acid sensors is gold, though other metals have been used as well. Gold electrodes have the advantage that thiol-modified aptamers and nucleic acids can be immobilized with a simple physisorption procedure.
Although therapeutic aptamer developments are facing skepticism [5], it is in the field of biosensors that aptamers could maximize their potential, and original research reports are showing how aptamers can be used to provide novel diagnostic solutions. However, in our opinion, there are still several limitations that aptamer electrochemical-based biosensors need to overcome, including the development of a systematic aptamer discovery process for biosensors, chemisorption of molecules on the conductive surface, quality of the semiconductor material, and improvement of the signal-to-noise ratio.
While many aspects of aptamers and their potential uses have been extensively reviewed elsewhere [4,6,7,8,9], this review is focused on the current limitations, which challenge the development of aptamer electrochemical-based biosensors, and the sensor architectures that have been developed so far.
2. Aptamer Selection for Electrochemical Biosensing Devices
One of the most promising features of aptamers is their potential for the development of reagentless electrochemical sensors. An interaction between an aptamer and its target molecule can be continuously monitored with the appropriate aptamer modification. However, one major problem is the lack of a reliable process to obtain aptamers to be specifically used in electrochemical sensors.
Combinatorial chemistry is an important technology for the biotechnological and pharmaceutical industries and for research laboratories to discover new compounds with useful activities. It is characterized by the synthesis of large searchable libraries of related compounds for rapid identification and isolation of functional molecules.
Nucleic acids have become attractive compounds for combinatorial strategies because complex libraries of random sequence oligonucleotides comprising approximately 1015 different molecules can be produced by solid-phase chemical synthesis. SELEX and its variations [4] are the most widely used methodologies for interrogating this complex library. This methodology involves the progressive selection of aptamers by an iterative process where a synthetic library of aptamers is incubated with a target molecule of interest. The aptamer molecules binding to the target molecule are eluted from non-binding molecules by a user-defined partitioning methodology. The eluted species are then amplified by the polymerase chain reaction (PCR) technique, generating an enriched aptamer library with molecules that bind to some extent to the target of interest. This process is repeated as many times as necessary (usually 8–15 rounds) until aptamers with the desired properties are selected. While the described process seems to be easy to apply, this is far from simple [5] and it gets more complex when aptamers must be selected as a probe for an electrochemical sensor. In many electrochemical sensors, aptamers might need to be modified with at least an anchoring chemical group for surface immobilization and a redox-active dye to generate a measurable signal. Unfortunately, both modifications are not compatible with the SELEX process. At present, the PCR technique makes use of enzymes that are not able to recognize and incorporate modified nucleotides for surface anchoring or monomers containing redox chemical groups in the newly synthesized strand. Even if a recombinant enzyme capable of incorporating such modifications is obtained, it will be challenging to add these modifications in the correct position in the aptamer sequences. Hence, the aptamers, obtained through SELEX, that showed optimal activity during the selection process might lose their activity after the subsequent modifications for their use in an aptasensor device. Without the development of an aptamer selection process designed specifically to obtain molecules for electrochemical biosensors, the field of biosensors will continue to grow slowly and making improvements case by case. However, among all the possible electrosensitive probes, aptamers are unique because libraries of nucleic acids can be synthesized in-situ with solid-phase synthesizers in predefined locations on different solid supports and with extensive modifications [10,11,12], thus enabling non-SELEX procedures for aptamer identification. Re-engineering of aptamers obtained through SELEX is currently the most widely used methodology to adapt aptamers obtained in solution in a surface-attached signal transducer.
3. Immobilization of Aptamers onto Semiconductors
The most common electrode material for an electrochemical nucleic acid sensor is gold. However, other metals, carbon, and certain semiconductors have been used as well. Gold is of special interest for the biosensor field, since thiol-modified aptamers can be immobilized via simple chemisorption on this surface [13]. Whereas this immobilizing technique has become very popular, special care must be taken due to the adsorption of natural oligonucleotides on gold, which can dramatically affect the orientation and folding of the aptamers [13,14].
The DNA–gold interaction is governed by a number of non-covalent forces including electrostatic interactions, hydrophobic forces, and specific bonding between the chemical groups of purine and pyrimidine rings of the DNA bases and gold [14]. In addition, the absorption kinetics of DNA on gold is also dependent on other conditions such as pH or buffer composition [15]. These factors influence the immobilization process, and many efforts are being made to understand the nature of DNA-gold interactions.
Thiolated single-stranded DNA (HS-ssDNA) forms a compact layer over bare-gold, possibly due to the multiple contacts between DNA bases and the gold surface [16]. However, when the surface is treated with alkanethiols, such as 6-mercaptohexanol (MCH), HS-ssDNA shows an extended configuration rendering the probe accessible to its target molecule.
The salt concentration and pH of the solution can also alter the efficiency of ssDNA absorption. Self-assembled monolayers (SAMs) of HS-ssDNA are typically made at low pH and high concentration of salt [13,17]. Jiang et al. suggested that, at low pH, the adenine and cytosine bases are protonated, facilitating the binding to the gold surface due to a reduction in the repulsion forces between DNA and the gold surface. Dehydration of DNA bases at low pH may increase the short-range DNA-gold hydrophobic interactions [15]. On the other hand, Herne showed that a maximum HS-ssDNA coverage is achieved when using over 0.4 M KH2PO4, suggesting that ionic strength reduces the electrostatic repulsion between DNA strands in solution.
The DNA sequence [18] and length [19] also add complexity to the immobilization process. The first, and more obvious observation, is that sequence and length determine the structural flexibility of the aptamer. Therefore, aptamer conformation can expose either its bases or its negatively charged backbone.
Among the four natural nucleobases, thymine showed the lowest binding strength on gold surfaces in thermal desorption assays [20]. Demers showed that nucleobases desorb in the order T < C < A < G, which supports the empirical evidence that poly (T) absorbs more weakly on gold surface than poly (A) oligonucleotides [21]. Moreover, Kimura-Suda showed that the binding strength of poly (A) molecules to gold surfaces is strong enough to denature dsDNA molecules composed of poly (A) oligonucleotides and their complementary poly (T) strand. To increase the stability of HS-DNA on gold surfaces, oligonucleotides carrying three thiol groups have been developed [22].
Although chemisorption via metal-sulphur has proven to be a good method for attaching DNA molecules to gold, passivation with an alkanethiol is commonly used to reduce interactions between the gold surface and chemical groups of the oligonucleotide other than the DNA-thiol group. However, care must be taken to reduce the instability of this type of monolayers, which can show some sensitivity to buffers, enzymes, and changes in temperature and air [23,24,25,26]. The alkanethiol MCH is broadly used to displace non-specific binding of single-stranded DNAs from gold and to functionalize the surface with DNAs containing thiol end groups. MCH works by blocking nonspecific contacts between DNA backbone and the gold surface [16]. Schlenoff analyzed desorption of an octadecanethiol by increasing the temperature gradually and showed a 50% loss at 160 °C. They also showed that exposing the monolayer to a piranha solution for ten minutes was enough to remove more than 98% of the alkanethiol. Moreover, Schoenfisch provided further insight into the alkanethiol SAM structure upon air exposure and contributed with more data to evaluate the shelf-storage of alkanethiols SAM-functionalized gold surfaces. In addition, the formation of alkanethiols SAMs is also dependent of the cleanliness and structure of gold [27].
4. Nano- and Microelectrodes
Miniaturization and the need for high-throughput analytical assays are pushing the development of electrochemical sensing technologies using micro- and nanoelectrodes. Miniaturization of electrodes helps to reduce the undesired effects of the ohmic drop, decreases sample consumption, allows high mass transport density, reduces the double layer capacitance, enhances the signal-noise ratio, and might facilitate the measurement of samples in low-ionic strength solutions. However, while the background currents resulting from the charging and discharging of the electrode-electrolyte interface are reduced, the amount of signal is reduced as well. In addition, when using multi-electrode arrays, diffusion layers begin to overlap, generating a crosstalk effect among electrodes. Electrochemical laboratories normally use large electrodes with diameters in the millimeter scale, but micro- and nanoelectrodes are designed with a diameter below 100 μm. Micro- and nano electrodes become very useful when a high parallelism of analysis is needed and when a low sample volume is required. While aspects regarded to nanoelectrodes physical properties, fabrication and advancements in analytical instrumentation have been reviewed elsewhere [28,29,30], many efforts are oriented to enhance electrode signal using novel redox molecules or improving experimental procedures.
In most cases, reagentless aptamer electrochemical biosensors use probes modified or labeled with a redox molecule such as Methylene blue (MB) and Ferrocene [31,32]. These approaches rely on the conformational change that an aptamer suffers upon target binding. Changes in the folding of the aptamer modify the electron transfer efficiency between the redox-tag and the electrode surface. However, many laboratories exploit the intrinsic property of MB to bind to dsDNA via intercalation [33].
To tackle the problem of low signals in micro/nanoelectrodes, it is necessary to build more sensitive electronics [28] or develop better signal sources. Following the latter, Bonnet and coworkers [34], developed a probe bearing three MB molecules that can be coupled into a DNA strand during standard solid-phase synthesis. This novel poly-MB probe was used to detect bacteria in a sandwich assay using gold nanoparticles and microorganisms captured on magnetic beads. Although no aptasensor was used in this work, we consider the poly-MB an original solution to overcome the low signal problem in nano- or microelectrodes devices.
Another simple experimental procedure to enhance the signal from microelectrodes came from the work of Juan Liu et al., which suggested that dissolved oxygen interfered with electrochemical analysis over microelectrodes even when a highly sensitive potentiostat is used and showed that a better signal was obtained when the buffers were deoxygenated with nitrogen [35]. It is worth to mention that among electrochemists is very common to degass the buffers with an inert gas, but the effect of oxygen on microelectrode aptasensors was not previously assayed.
5. Aptasensor Electrochemistry Architectures
The ability of aptamers to detect an analyte has been tested in a variety of sensing technologies, including optical, enzymatic, and electrochemical approaches. For the purpose of this review, we will limit our discussion to the advancement in the field of electrochemical sensors using planar gold electrodes. Table 1 summarizes the pioneering works reviewed below that have led to the current sensor architectures in continuous improvement.

Table 1.
Aptasensor architectures described in this review. MB: Methylene blue, Fc: Ferrocene, HRP: horse radish peroxidase.
The standard architecture (Figure 1) includes the anti-thrombin aptamer, one of the most widely studied aptamers, discovered in 1992 [55], to modulate blood coagulation. Based on the sequence of the thrombin aptamer, Xiao et al. built an aptasensor by attaching an MB moiety to the DNA aptamer [36]. This aptamer adopted a G-quartet conformation, which was necessary for thrombin binding. Xiao also showed that the aptasensor was able to detect 64 nM of thrombin in a simple saline buffer and calf serum using a 1.6 mm diameter gold electrode. Upon thrombin binding, the current was significantly reduced where the maximum and minimum current peaks were observed in the nanoampere range. The aptasensor showed a linear dynamic range (LDR) from 6.4 nM to 768 nM, whereas normal thrombin concentration in serum is around 1.3 micromolar. Rade et al. reported a similar aptasensor using an aptamer labeled with a ferrocene redox molecule and with a limit of detection (LOD) of 0.5 nM. However, in this work, the result of thrombin binding to its aptamer was an on-signal, possibly due to the shorter sequence used to build the sensor [37]. A more sensitive thrombin aptasensor was developed by Liu et al. using a sandwich assay with two thrombin aptamers [56]. In this report, an ethynylferrocene aptamer was responsible for generating an on-signal reaching a LOD of 84 μM using 2 mm diameter gold electrodes and measuring currents in the microampere order.
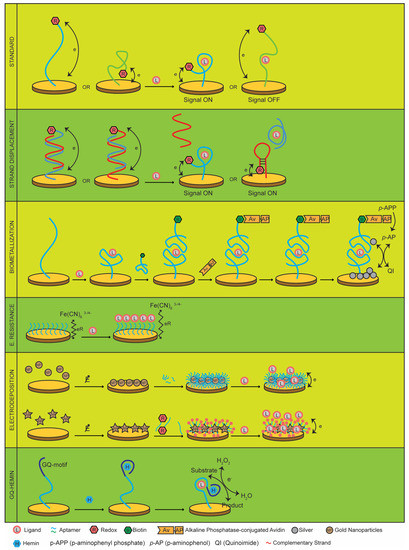
Figure 1.
Biosensor architectures presented in this review.
In order to build a rapid cocaine detection process, Baker et al. made an electrochemical aptasensor based on an aptamer published in 2001 [40,57]. They were able to detect up to 90 μM of cocaine using 1 mm-diameter gold electrodes and measured currents in the nanoampere range. In addition, they also detected cocaine in adulterated samples often used to mask or cut cocaine. Although ligand-receptor physical-chemical properties are important, other important aspects of biosensors, including packing density and alternate current (AC) signal, affect their performance and need to be studied. White et al. observed a six-fold signal enhancement when the molecular crowding over the electrode was reduced [58]. By contrast, the effect of AC frequency did not show a significant impact on the sensor performance. In addition, they analyzed the sensor performance using alkanethiols of different lengths, which generate monolayers of different thicknesses, and they observed a better performance with the thickest monolayer. This observation supports a previous report aimed mainly at understanding the effects of monolayer thickness on electrochemical signals [59]. These standard aptasensor architectures were also used to detect the cytokine tumor necrosis factor α (TNF-α) [46] with a LOD of 10 ng/mL in whole blood, c-reactive protein [50] with a LDR from 1 to 100 pM, L-tryptophan [53] with a LDR from 0.1 to 10 mM, and luteinizing hormone with a LDR between 5 to 500 nM [54].
An ATP-specific aptamer discovered in solution [60] was also adapted to an electrochemical aptasensor [41]. This aptamer was modified with a 3’thiol group and 5´ ferrocene (Fc) group for immobilization and detection, respectively, using a 2 mm diameter gold electrode. Interestingly, Zuo et al. employed an ATP-specific aptamer hybridized with its complementary DNA sequence, which placed the Fc moiety far from the gold electrode. Upon ATP binding, the denatured duplex displaced the complementary strand of the aptamer, followed by a conformational change of the aptamer, which placed the ferrocene closer to the electrode surface, generating an on-signal current. Figure 1 shows a schematic of the strand displacement architecture. A similar approach was used by Lu et al., though it differed in that the Fc moiety was attached to the complementary strand of the aptamer [61]. They obtained an LDR from 10 to 80 nM in 1.6 mm diameter gold electrodes. While the complementary DNA was attached to the surface through a thiol group, the aptamer was not. Hence, upon ATP binding, the aptamer strand was displaced from its complementary sequence and washed out. This denaturation process induced a conformational change of the complementary DNA, which was tailor-made by the user to place the Fc molecule close to the electrode surface through a hairpin structure for electron transfer maximization. This concept was also exploited to detect ATP in rat brains using long concatemers based on one MB-labeled complementary DNA hybridized with two aptamers [62]. This work showed an ATP aptasensor with an LDR from 0.1 nM to 1 mM, using a 2 mm diameter gold electrode. Another aptasensor based on a similar approach was developed by Yu et al. to improve the signal gain and lower the limit of detection of an ampicillin-specific aptamer [49]. This aptasensor showed a LDR between 0.2 and 15000 μM. Recently, Munzar and coworkers described a method for screening optimal aptamer-complementary strands based on the DNA microarray technology in order to improve strand displacement assays [63]. Interestingly, this work shows that complementary strands can enhance the binding activity of single stranded aptamers that were previously discovered in solution.
Many discovered aptamers contain guanine (G)-rich stretches that are able to self-assemble into a secondary structure called G-quadruplex (GQ) [64]. Monovalent cations, such as sodium and potassium, play an important role in stabilizing GQ structures. When DNA containing G-rich stretches is deprived of these cations, GQ secondary structures tend to disassemble. Mono- and divalent ions play an important role in single cells and multicellular organisms. Sensing devices for specific ions could unveil new molecular mechanisms of cells. Wu et al. exploited the cation-dependence of QGs to build a potassium sensor with a thiolated-DNA containing a G-rich stretch and a ferrocene group for electrochemical detection of the GQ dissembling upon potassium deprivation [42].
So far, the described aptasensors are based on the modulation of electron transfer efficiency between a transducer and an electroactive moiety linked to an oligonucleotide. However, other sensors have been developed to measure changes in the charge transfer resistance induced upon a target binding to a non-labeled aptamer (Figure 1). Accordingly, an aptasensor has been constructed for cancer diagnosis by exploiting specific biomarkers expressed on cancer-cell surfaces [43]. In this work, an aptamer selected against a cell line of human leukemic lymphoblasts (CCRF-CEM) in solution [65] was modified with a thiol group and assembled onto a gold electrode. Interestingly, the aptamer was not labeled with a redox molecule, since they took advantage of the insulating properties of cells and their effects on electron transfer of free Fe(CN)6 3-/4- in solution. With this approach, they obtained an LDR from 1 × 104 to 1 × 107 cells/mL in a 2 mm diameter gold electrode. The electron transfer resistance strategy was also applied to develop an aptasensor against the mycotoxin aflatoxin M1 [44]. In this work, a 4 mm diameter gold electrode was coated with streptavidin molecules and an aflatoxin-specific aptamer synthesized with a 3´terminal biotin moiety for immobilization. Upon aflatoxin M1 binding, the aptasensor increased the electron transfer resistance between the dissolved K3[Fe(CN)6] in solution and the electrode surface. Pandey et al. showed an LDR between 1 to 105 parts per trillion. A similar strategy was used to detect B. anthracis spores [51].
Biometallization (Figure 1) is another approach that could increase the sensitivity and enhance the signal-to-noise ratio of aptasensors at the expense of greater labor for obtaining results [66]. Briefly, this process involves the use of a biotinylated aptamer and a streptavidin conjugated to an alkaline phosphatase enzyme (AP). A signal is obtained when an AP substrate is added to the enzyme and the product reduces metal ions in solution such as silver. Reduced silver is then deposited on the electrode surface, and a signal is obtained applying stripping voltammetry. Degefa et al. utilized biometallization using two thrombin-specific aptamer probes, one thiol-modified aptamer to form a monolayer over a 150 nm diameter gold electrode and a second biotin-labeled aptamer as a probe to bind a neutravidin-conjugated AP for electrochemical detection [39]. The two aptamers resulted in an aptamer–protein–aptamer sandwich, and the enzyme-labeled neutravidin was added for enzymatic deposition of silver ions and linear sweep voltammetric read-out signal. The AP enzyme allowed the conversion of p-aminophenyl phosphate to p-aminophenol, a compound that reduces silver ions and forms silver metal on the electrode surface and quinoimide as the result of p-aminophenol oxidation. Interestingly, with this small electrode, Degefa showed an LDR from 1 μM to 1 nM.
Another aptasensor architecture developed by Zhu et al. consisted of the electrodeposition of gold nanoparticles (Figure 1) over a gold screen-printed electrode to enhance the electrochemical detection of kanamycin [45]. Basically, self-assembled 2,5-di-(2-thienyl)-1H-pyrrole-1-(p-benzoic acid) (DPB)-gold nanoparticles (DPB-AuNP) were electrodeposited on a gold electrode, and an NH2-modified aptamer selected to bind kanamycin was then covalently conjugated to the DPB-AuNP. This aptasensor showed an LDR up to 9 μM of kanamycin, the kanamycin also being the redox molecule. Similarly, Liu et al. electrodeposited dendritic gold nanostructures on 25 μm-diameter gold microelectrodes in order to improve the sensitivity of these small transducers [35]. Small-scale electrodes offer many advantages in detriment of the surface available for electron transfer. Therefore, gold nanostructures were used to increase the transducer surface. In this work, Liu et al. challenged their aptasensor architecture with two aptamer targets, ATP, and the antibiotic tobramycin. Interestingly, they noted the interference of dissolved oxygen and suggested a deoxygenation procedure to improve the signal-to-noise ratio. Without electrodepositing gold nanoparticles, Chen et al. developed an aptasensor using a dual gold nanoparticle conjugates in order to overcome the problem of sensitivity in conventional aptasensors [67]. Basically, they linked a first nanoparticle coated with a thiolated-oligonucleotide sequence hybridized to its complementary DNA sequence, which belonged to an ochratoxin A-specific aptamer. The addition of ochratoxin A denatured the DNA duplex, displacing the aptamer strand. Then, a second nanoparticle coated with a thio-Fc-modified DNA sequence complementary to the DNA strand on the first nanoparticle was added. With this architecture, Chen obtained an LDR from 0.001 to 500 ppb for ochratoxin A detection.
A continuous real-time aptasensor was proposed by Ferguson et al. to detect the chemotherapeutic drug doxorubicin (DOX) in biological fluids [47]. The aptamer was conjugated to a gold surface through a thiol chemical group and the binding of DOX was detected through a MB moiety located at the terminal end of the aptamer. Microfluidic was adapted to this aptasensor in order to generate a continuous flow diffusion filter, which prevented blood cells and high molecular weight molecules to interfere with the aptasensor and allowed the molecules with high diffusivity, such as DOX, to reach the electrode. In a simple buffer, the aptasensor showed an LDR of 0.1 to 10 μM, and the authors were able to detect 0.3 μM of DOX in human whole blood, which is within the drug´s therapeutic range, using a technique that self-corrects signal drift. Sensor base-drift is an important source of inaccuracy when complex samples such as blood are analyzed, and efforts are being oriented to overcome base-drift problems for these types of aptasensors [68]. Using a dual redox reporter, Li et al. developed an aptasensor to correct the signal drift and detect cocaine, kanamycin, and DOX in whole blood. This approach relied on the use of aptamer probes modified with a MB reporter on the distal end of the oligonucleotide and an anthraquinone (AQ) reference reporter placed proximal to the electrode surface. In these sensors, the MB electron transfer efficiency was modulated by the aptamer conformation; by contrast, AQ electron transfer was independent of the aptamer´s conformation and was only affected by the molecular composition of its environment. In addition, MB and AQ potentials did not overlap, allowing both to be simultaneously monitored.
Another class of aptasensors was introduced by Shen el al., who took advantage of DNAzymes with peroxidase-like activity, which involves GQ-based structures bound to a hemin molecule [38]. This GQ-Hemin complex (Figure 1) was used for electrocatalysis reduction of hydrogen peroxidase (H2O2). In this work, a thrombin-specific aptamer was modified with a thiol group for anchoring onto a 2 mm diameter gold electrode, and a GQ motif was added for hemin complexing. Shen et al. observed a correlation between thrombin concentration and current upon H2O2 addition and obtained a LDR between 0.1 nM and 1 μM. In a similar architecture, Sun et al. used the TLS11a aptamer that targets human liver hepatocellular carcinoma cells (HepG2) to create a sandwich-type aptasensor to detect cancer cells [48]. Their strategy involved the attachment of a thiolated-TLS11a aptamer onto a 3 mm-diameter gold electrode, and gold nanoparticles were constructed for detection. These nanoparticles were covered with horse radish peroxidase (HRP) and the thiolated-TLS11a carrying a GQ for hemin binding. Both, the HRP and the DNAzyme, were used to amplify the electrochemical signal by reducing hydroquinone in the presence of H2O2. This aptasensor showed a LDR from 1 × 102 to 1 × 107 cells/mL. An HRP-mimicked DNAzyme strategy was also used to detect hemagglutinin (HA) protein, one of the factors for transmitting the avian influenza virus from birds to humans [52]. Lee et al. combined gold nanoparticles with a three-way junction oligonucleotide (3WJ) to easily attach and detach specific properties. Thus, a thiol group, an HA-specific aptamer, and a GQ motif for hemin binding were added to the 3WJ. This aptasensor showed a LOD of 1 pM of HA.
Two important parameters characterizing an electrochemical aptasensor are the LDR and the LOD. These parameters are usually difficult to modulate and are associated primarily to the nature of an aptamer, making it difficult to optimize the aptasensor performance. To control these parameters, Schoukroun-Barnes et al. proposed a heterogenous aptasensor using several aptamers at different ratios but targeting equal targets with different affinities [69] to tune the dynamic range and the sensitivity of aptasensors.
6. Conclusions
In the last few years, we have witnessed novel developments in electrochemical biosensors and their translation from bench to commercial devices; however, a key is the lack of systematic screening technology to discover novel molecules for biosensing applications. Aptamers are an interesting class of oligonucleotides, larger than small molecules but smaller than antibodies, with specific binding affinities toward a variety of targets [64,70,71]. Most aptamers show large conformation changes upon ligand binding, which makes them ideal molecules for electrochemical biosensing, and many original developments have been published by academic laboratories. In general, it is possible to classify the aptasensor architectures according to their different designs—(1) Standard, (2) Strand Displacement, (3) Electrotransfer resistance, (4) Electrodeposition, (5) GQ-Hemin, (6) Continuous, (7) Real-time, and (8) Biometallization. However, commercialization of electrochemical biosensors has lagged behind academic output, probably due to technical barriers and cost-effective manufacturing methods. Some technical barriers include sensitivity, signal-to-noise ratio, reproducibility, and shelf life. Furthermore, cost-effective manufacturing methods include access to raw materials, variability among material batches and their effect on the device performance, and the compatibility of the sensing elements with standard electronic manufacturing procedures. Although many challenges must still be overcome, aptamers have great potential as probes for commercial electrochemical affinity-based biosensors.
Funding
This work was financially supported by the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, Argentina), Aplife Biotech (Argentina), and Universidad Argentina de la Empresa (Argentina).
Acknowledgments
The authors apologize for omissions in citations and coverage. We also apologize to the many investigators whose relevant work in this area could not be covered because of space limitations.
Conflicts of Interest
The authors have no commercial associations that might pose a conflict of interest in connection with the submitted manuscript. All funding sources supporting the work and all institutional or corporate affiliations are acknowledged and all authors have approved the manuscript and agreed with the submission to Chemosensors.
References
- Newman, J.D.; Turner, A.P. Home Blood Glucose Biosensors: A Commercial Perspective. Biosens. Bioelectron. 2005, 20, 2435–2453. [Google Scholar] [CrossRef] [PubMed]
- Clark, L.C., Jr.; Lyons, C. Electrode Systems for Continuous Monitoring in Cardiovascular Surgery. Ann. N. Y. Acad. Sci. 1962, 102, 29–45. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic Evolution of Ligands by Exponential Enrichment: Rna Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. Selex—A (R)Evolutionary Method to Generate High-Affinity Nucleic Acid Ligands. Biomol. Eng. 2007, 24, 381–403. [Google Scholar] [CrossRef] [PubMed]
- Rozenblum, G.T.; Lopez, V.G.; Vitullo, A.D.; Radrizzani, M. Aptamers: Current Challenges and Future Prospects. Expert Opin. Drug Discov. 2016, 11, 127–135. [Google Scholar] [CrossRef]
- Nimjee, S.M.; Rusconi, C.P.; Sullenger, B.A. Aptamers: An Emerging Class of Therapeutics. Annu. Rev. Med. 2005, 56, 555–583. [Google Scholar] [CrossRef]
- Schoukroun-Barnes, L.R.; Macazo, F.C.; Gutierrez, B.; Lottermoser, J.; Liu, J.; White, R.J. Reagentless, Structure-Switching, Electrochemical Aptamer-Based Sensors. Annu. Rev. Anal. Chem. (Palo Alto Calif.) 2016, 9, 163–181. [Google Scholar] [CrossRef]
- Kim, Y.S.; Raston, N.H.; Gu, M.B. Aptamer-Based Nanobiosensors. Biosens. Bioelectron. 2016, 76, 2–19. [Google Scholar]
- Hianik, T.; Wang, J. Electrochemical Aptasensors–Recent Achievements and Perspectives. Electroanalysis 2009, 21, 1223–1235. [Google Scholar] [CrossRef]
- Dufva, M. Fabrication of High Quality Microarrays. Biomol. Eng. 2005, 22, 173–184. [Google Scholar] [CrossRef]
- Hughes, T.R.; Mao, M.; Jones, A.R.; Burchard, J.; Marton, M.J.; Shannon, K.W.; Lefkowitz, S.M.; Ziman, M.; Schelter, J.M.; Meyer, M.R.; et al. Expression Profiling Using Microarrays Fabricated by an Ink-Jet Oligonucleotide Synthesizer. Nat. Biotechnol. 2001, 19, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Maurer, K.; Cooper, J.; Caraballo, M.; Crye, J.; Suciu, D.; Ghindilis, A.; Leonetti, J.A.; Wang, W.; Rossi, F.M.; Stover, A.G.; et al. Electrochemically Generated Acid and Its Containment to 100 Micron Reaction Areas for the Production of DNA Microarrays. PLoS ONE 2006, 1, E34. [Google Scholar] [CrossRef] [PubMed]
- Herne, T.M.; Tarlov, M.J. Characterization of DNA Probes Immobilized on Gold Surfaces. J. Am. Chem. Soc. 1997, 119, 8916–8920. [Google Scholar] [CrossRef]
- Koo, K.M.; Sina, A.A.I.; Carrascosa, L.G.; Shiddiky, M.J.A.; Trau, M. DNA–Bare Gold Affinity Interactions: Mechanism and Applications in Biosensing. Anal. Methods 2015, 7, 7042–7054. [Google Scholar] [CrossRef]
- Nelson, E.M.; Rothberg, L.J. Kinetics and Mechanism of Single-Stranded DNA Adsorption onto Citrate-Stabilized Gold Nanoparticles in Colloidal Solution. Langmuir 2011, 27, 1770–1777. [Google Scholar] [CrossRef] [PubMed]
- Levicky, R.; Herne, T.M.; Tarlov, M.J.; Satija, S.K. Using Self-Assembly to Control the Structure of DNA Monolayers on Gold: A Neutron Reflectivity Study. J. Am. Chem. Soc. 1998, 120, 9787–9792. [Google Scholar] [CrossRef]
- Jiang, H.; Materon, E.M.; Sotomayor Mdel, P.; Liu, J. Fast Assembly of Non-Thiolated DNA on Gold Surface at Lower PH. J. Colloid. Interface Sci. 2013, 411, 92–97. [Google Scholar] [CrossRef]
- Tao, N.J.; Derose, J.A.; Lindsay, S.M. Self-Assembly of Molecular Superstructures Studied by In Situ Scanning Tunneling Microscopy: DNA Bases on Gold (111). J. Phys. Chem. 1993, 97, 910–919. [Google Scholar] [CrossRef]
- Steel, A.B.; Levicky, R.L.; Herne, T.M.; Tarlov, M.J. Immobilization of Nucleic Acids at Solid Surfaces: Effect of Oligonucleotide Length on Layer Assembly. Biophys. J. 2002, 79, 975–981. [Google Scholar] [CrossRef]
- Demers, L.M.; Ostblom, M.; Zhang, H.; Jang, N.H.; Liedberg, B.; Mirkin, C.A. Thermal Desorption Behavior and Binding Properties of DNA Bases and Nucleosides on Gold. J. Am. Chem. Soc. 2002, 124, 11248–11249. [Google Scholar] [CrossRef]
- Kimura-Suda, H.; Petrovykh, D.Y.; Tarlov, M.J.; Whitman, L.J. Base-Dependent Competitive Adsorption of Single-Stranded DNA on Gold. J. Am. Chem. Soc. 2003, 125, 9014–9015. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jin, R.; Mirkin, C.A.; Letsinger, R.L. Multiple Thiol-Anchor Capped DNA-Gold Nanoparticle Conjugates. Nucleic. Acids. Res. 2002, 30, 1558–1562. [Google Scholar] [CrossRef] [PubMed]
- Nuzzo, R.G.; Fusco, F.A.; Allara, D.L. Spontaneously Organized Molecular Assemblies. 3. Preparation and Properties of Solution Adsorbed Monolayers of Organic Disulfides on Gold Surfaces. J. Am. Chem. Soc. 1987, 109, 2358–2368. [Google Scholar] [CrossRef]
- Schlenoff, J.B.; Li, M.; Ly, H. Stability and Self-Exchange in Alkanethiol Monolayers. J. Am. Chem. Soc. 1995, 117, 12528–12536. [Google Scholar] [CrossRef]
- Schoenfisch, M.H.; Pemberton, J.E. Air Stability of Alkanethiol Self-Assembled Monolayers on Silver and Gold Surfaces. J Am. Chem. Soc. 1998, 120, 4502–4513. [Google Scholar] [CrossRef]
- Petrovykh, D.Y.; Kimura-Suda, H.; Whitman, L.J.; Tarlov, M.J. Quantitative Analysis and Characterization of DNA Immobilized on Gold. J. Am. Chem. Soc. 2003, 125, 5219–5226. [Google Scholar] [CrossRef]
- Yang, Z.; Gonzalez-Cortes, A.; Jourquin, G.; Viré, J.-C.; Kauffmann, J.-M.; Delplancke, J.-L. Analytical Application of Self Assembled Monolayers on Gold Electrodes: Critical Importance of Surface Pretreatment. Biosens. Bioelectron. 1995, 10, 789–795. [Google Scholar] [CrossRef]
- Arrigan, D.W.M. Nanoelectrodes, Nanoelectrode Arrays and Their Applications. Analyst 2004, 129, 1157–1165. [Google Scholar] [CrossRef]
- Dawson, K.; O’riordan, A. Electroanalysis at the Nanoscale. Annu. Rev. Anal. Chem. (Palo Alto Calif.) 2014, 7, 163–181. [Google Scholar] [CrossRef]
- Galvin, P.; Padmanathan, N.; Razeeb, K.M.; Rohan, J.F.; Nagle, L.C.; Wahl, A.; Moore, E.; Messina, W.; Twomey, K.; Ogurtsov, V. Nanoenabling Electrochemical Sensors for Life Sciences Applications. J. Mater. Res. 2017, 32, 2883–2904. [Google Scholar] [CrossRef]
- Fan, C.; Plaxco, K.W.; Heeger, A.J. Electrochemical Interrogation of Conformational Changes as a Reagentless Method for the Sequence-Specific Detection of DNA. Proc. Natl. Acad. Sci. USA 2003, 100, 9134–9137. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, S.; Uto, Y.; Kondo, H.; Ihara, T.; Takagi, M. Electrochemically Active DNA Probes: Detection of Target DNA Sequences at Femtomole Level by High-Performance Liquid Chromatography with Electrochemical Detection. Anal. Biochem. 1994, 218, 436–443. [Google Scholar] [CrossRef] [PubMed]
- Erdem, A.; Kerman, K.; Meric, B.; Ozsoz, M. Methylene Blue as a Novel Electrochemical Hybridization Indicator. Electroanalysis 2001, 13, 219–223. [Google Scholar] [CrossRef]
- Bonnet, R.; Farre, C.; Valera, L.; Vossier, L.; Leon, F.; Dagland, T.; Pouzet, A.; Jaffrezic-Renault, N.; Fareh, J.; Fournier-Wirth, C.; et al. Highly Labeled Methylene Blue-Ds DNA Silica Nanoparticles for Signal Enhancement of Immunoassays: Application to the Sensitive Detection of Bacteria in Human Platelet Concentrates. Analyst 2018, 143, 2293–2303. [Google Scholar] [CrossRef]
- Liu, J.; Wagan, S.; Davila Morris, M.; Taylor, J.; White, R.J. Achieving Reproducible Performance of Electrochemical, Folding Aptamer-Based Sensors on Microelectrodes: Challenges and Prospects. Anal. Chem. 2014, 86, 11417–11424. [Google Scholar] [CrossRef]
- Xiao, Y.; Lubin, A.A.; Heeger, A.J.; Plaxco, K.W. Label-Free Electronic Detection of Thrombin in Blood Serum by Using an Aptamer-Based Sensor. Angew. Chem. Int. Ed. Engl. 2005, 44, 5456–5459. [Google Scholar] [CrossRef]
- Radi, A.E.; Acero Sanchez, J.L.; Baldrich, E.; O’sullivan, C.K. Reagentless, Reusable, Ultrasensitive Electrochemical Molecular Beacon Aptasensor. J. Am. Chem. Soc. 2006, 128, 117–124. [Google Scholar] [CrossRef]
- Shen, B.; Wang, Q.; Zhu, D.; Luo, J.; Cheng, G.; He, P.; Fang, Y. G-Quadruplex-Based Dnazymes Aptasensor for the Amplified Electrochemical Detection of Thrombin. Electroanalysis 2010, 22, 2985–2990. [Google Scholar] [CrossRef]
- Degefa, T.H.; Hwang, S.; Kwon, D.; Park, J.H.; Kwak, J. Aptamer-Based Electrochemical Detection of Protein Using Enzymatic Silver Deposition. Electrochim. Acta 2009, 54, 6788–6791. [Google Scholar] [CrossRef]
- Baker, B.R.; Lai, R.Y.; Wood, M.S.; Doctor, E.H.; Heeger, A.J.; Plaxco, K.W. An Electronic, Aptamer-Based Small-Molecule Sensor for the Rapid, Label-Free Detection of Cocaine In Adulterated Samples and Biological Fluids. J. Am. Chem. Soc. 2006, 128, 3138–3139. [Google Scholar] [CrossRef]
- Zuo, X.; Song, S.; Zhang, J.; Pan, D.; Wang, L.; Fan, C. A Target-Responsive Electrochemical Aptamer Switch (TREAS) for Reagentless Detection of Nanomolar Atp. J. Am. Chem. Soc. 2007, 129, 1042–1043. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.S.; Chen, C.R.; Shen, G.L.; Yu, R.Q. Reversible Electronic Nanoswitch Based on DNA G-Quadruplex Conformation: A Platform for Single-Step, Reagentless Potassium Detection. Biomaterials 2008, 29, 2689–2696. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Guo, M.; Nie, Z.; Xiao, X.; Yao, S. Aptamer-Based Electrochemical Sensor for Label-Free Recognition and Detection of Cancer Cells. Electroanalysis 2009, 21, 1321–1326. [Google Scholar] [CrossRef]
- Pandey, A.K.; Rajput, Y.S.; Sharma, R.; Singh, D. Immobilized Aptamer on Gold Electrode Senses Trace Amount of Aflatoxin M1. Appl. Nanosci. 2017, 7, 893–903. [Google Scholar] [CrossRef]
- Zhu, Y.; Chandra, P.; Song, K.M.; Ban, C.; Shim, Y.B. Label-Free Detection Of Kanamycin Based on the Aptamer-Functionalized Conducting Polymer/Gold Nanocomposite. Biosens. Bioelectron. 2012, 36, 29–34. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, Q.; Revzin, A. An Aptasensor for Electrochemical Detection of Tumor Necrosis Factor in Human Blood. Analyst 2013, 138, 4321–4326. [Google Scholar] [CrossRef]
- Ferguson, B.S.; Hoggarth, D.A.; Maliniak, D.; Ploense, K.; White, R.J.; Woodward, N.; Hsieh, K.; Bonham, A.J.; Eisenstein, M.; Kippin, T.E.; et al. Real-Time, Aptamer-Based Tracking of Circulating Therapeutic Agents in Living Animals. Sci. Transl. Med. 2013, 5, 213ra165. [Google Scholar] [CrossRef]
- Sun, D.; Lu, J.; Chen, Z.; Yu, Y.; Mo, M. A Repeatable Assembling and Disassembling Electrochemical Aptamer Cytosensor for Ultrasensitive and Highly Selective Detection of Human Liver Cancer Cells. Anal. Chim. Acta 2015, 885, 166–173. [Google Scholar] [CrossRef]
- Yu, Z.-G.; Sutlief, A.L.; Lai, R.Y. Towards The Development Of A Sensitive and Selective Electrochemical Aptamer-Based Ampicillin Sensor. Sens. Actuators B Chem. 2018, 258, 722–729. [Google Scholar] [CrossRef]
- Jarczewska, M.; Rebis, J.; Gorski, L.; Malinowska, E. Development of DNA Aptamer-Based Sensor for Electrochemical Detection of C-Reactive Protein. Talanta 2018, 189, 45–54. [Google Scholar] [CrossRef]
- Mazzaracchio, V.; Neagu, D.; Porchetta, A.; Marcoccio, E.; Pomponi, A.; Faggioni, G.; D’amore, N.; Notargiacomo, A.; Pea, M.; Moscone, D.; et al. A Label-Free Impedimetric Aptasensor for the Detection of Bacillus Anthracis Spore Simulant. Biosens. Bioelectron. 2019, 126, 640–646. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Park, S.Y.; Jang, H.; Kim, G.H.; Lee, Y.; Park, C.; Mohammadniaei, M.; Lee, M.H.; Min, J. Fabrication of Electrochemical Biosensor Consisted of Multi-Functional DNA Structure/Porous Au Nanoparticle for Avian Influenza Virus (H5N1) in Chicken Serum. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 99, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Idili, A.; Gerson, J.; Parolo, C.; Kippin, T.; Plaxco, K.W. An Electrochemical Aptamer-Based Sensor for the Rapid and Convenient Measurement of L-Tryptophan. Anal. Bioanal. Chem. 2019, 411, 4629–4635. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Kinghorn, A.B.; Voliotis, M.; Prague, J.K.; Veldhuis, J.D.; Tsaneva-Atanasova, K.; Mcardle, C.A.; Li, R.H.W.; Cass, A.E.G.; Dhillo, W.S.; et al. Measuring Luteinising Hormone Pulsatility with A Robotic Aptamer-Enabled Electrochemical Reader. Nat. Commun. 2019, 10, 852. [Google Scholar] [CrossRef]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection Of Single-Stranded DNA Molecules that Bind and Inhibit Human Thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef]
- Liu, Q.; Hu, Q.; Li, L.; Kong, J.; Zhang, X. Click Chemistry-Based Aptasensor for Highly Sensitive Electrochemical Detection of Thrombin. Anal. Methods 2017, 9, 3825–3830. [Google Scholar] [CrossRef]
- Stojanovic, M.N.; De Prada, P.; Landry, D.W. Aptamer-Based Folding Fluorescent Sensor for Cocaine. J. Am. Chem. Soc. 2001, 123, 4928–4931. [Google Scholar] [CrossRef]
- White, R.J.; Phares, N.; Lubin, A.A.; Xiao, Y.; Plaxco, K.W. Optimization of Electrochemical Aptamer-Based Sensors via Optimization of Probe Packing Density and Surface Chemistry. Langmuir 2008, 24, 10513–10518. [Google Scholar] [CrossRef]
- Lai, R.Y.; Seferos, D.S.; Heeger, A.J.; Bazan, G.C.; Plaxco, K.W. Comparison of the Signaling and Stability of Electrochemical DNA Sensors Fabricated from 6- Or 11-Carbon Self-Assembled Monolayers. Langmuir 2006, 22, 10796–10800. [Google Scholar] [CrossRef]
- Huizenga, D.E.; Szostak, J.W. A DNA Aptamer that Binds Adenosine and Atp. Biochemistry 1995, 34, 656–665. [Google Scholar] [CrossRef]
- Lu, Y.; Li, X.; Zhang, L.; Yu, P.; Su, L.; Mao, L. Aptamer-Based Electrochemical Sensors with Aptamer-Complementary DNA Oligonucleotides as Probe. Anal. Chem. 2008, 80, 1883–1890. [Google Scholar] [CrossRef]
- Jiang, Y.; Ma, W.; Ji, W.; Wei, H.; Mao, L. Aptamer Superstructure-Based Electrochemical Biosensor for Sensitive Detection of Atp in Rat Brain With In Vivo Microdialysis. Analyst 2019, 144, 1711–1717. [Google Scholar] [CrossRef] [PubMed]
- Munzar, J.D.; Ng, A.; Juncker, D. Comprehensive Profiling of the Ligand Binding Landscapes of Duplexed Aptamer Families Reveals Widespread Induced Fit. Nat. Commun. 2018, 9, 343. [Google Scholar] [CrossRef] [PubMed]
- Chiorcea-Paquim, A.-M.; Oliveira-Brett, M.A. Guanine Quadruplex Electrochemical Aptasensors. Chemosensors 2016, 4, 13. [Google Scholar] [CrossRef]
- Shangguan, D.; Tang, Z.; Mallikaratchy, P.; Xiao, Z.; Tan, W. Optimization and Modifications of Aptamers Selected from Live Cancer Cell Lines. Chembiochem 2007, 8, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Kim, E.; Kwak, J. Electrochemical Detection of DNA Hybridization Using Biometallization. Anal. Chem. 2005, 77, 579–584. [Google Scholar] [CrossRef]
- Chen, W.; Yan, C.; Cheng, L.; Yao, L.; Xue, F.; Xu, J. An Ultrasensitive Signal-On Electrochemical Aptasensor for Ochratoxin a Determination Based on DNA Controlled Layer-By-Layer Assembly of Dual Gold Nanoparticle Conjugates. Biosens. Bioelectron. 2018, 117, 845–851. [Google Scholar] [CrossRef]
- Li, H.; Arroyo-Curras, N.; Kang, D.; Ricci, F.; Plaxco, K.W. Dual-Reporter Drift Correction to Enhance the Performance of Electrochemical Aptamer-Based Sensors in Whole Blood. J. Am. Chem. Soc. 2016, 138, 15809–15812. [Google Scholar] [CrossRef]
- Schoukroun-Barnes, L.R.; Glaser, E.P.; White, R.J. Heterogeneous Electrochemical Aptamer-Based Sensor Surfaces for Controlled Sensor Response. Langmuir 2015, 31, 6563–6569. [Google Scholar] [CrossRef]
- Evtugyn, G.; Hianik, T. Electrochemical Immuno- and Aptasensors for Mycotoxin Determination. Chemosensors 2019, 7, 10. [Google Scholar] [CrossRef]
- Mehlhorn, A.; Rahimi, P.; Joseph, Y. Aptamer-Based Biosensors for Antibiotic Detection: A Review. Biosensors 2018, 8, 54. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).