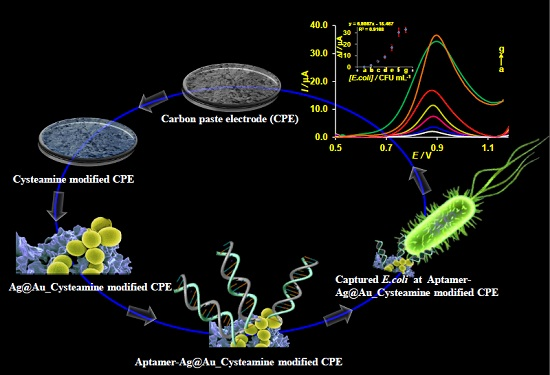
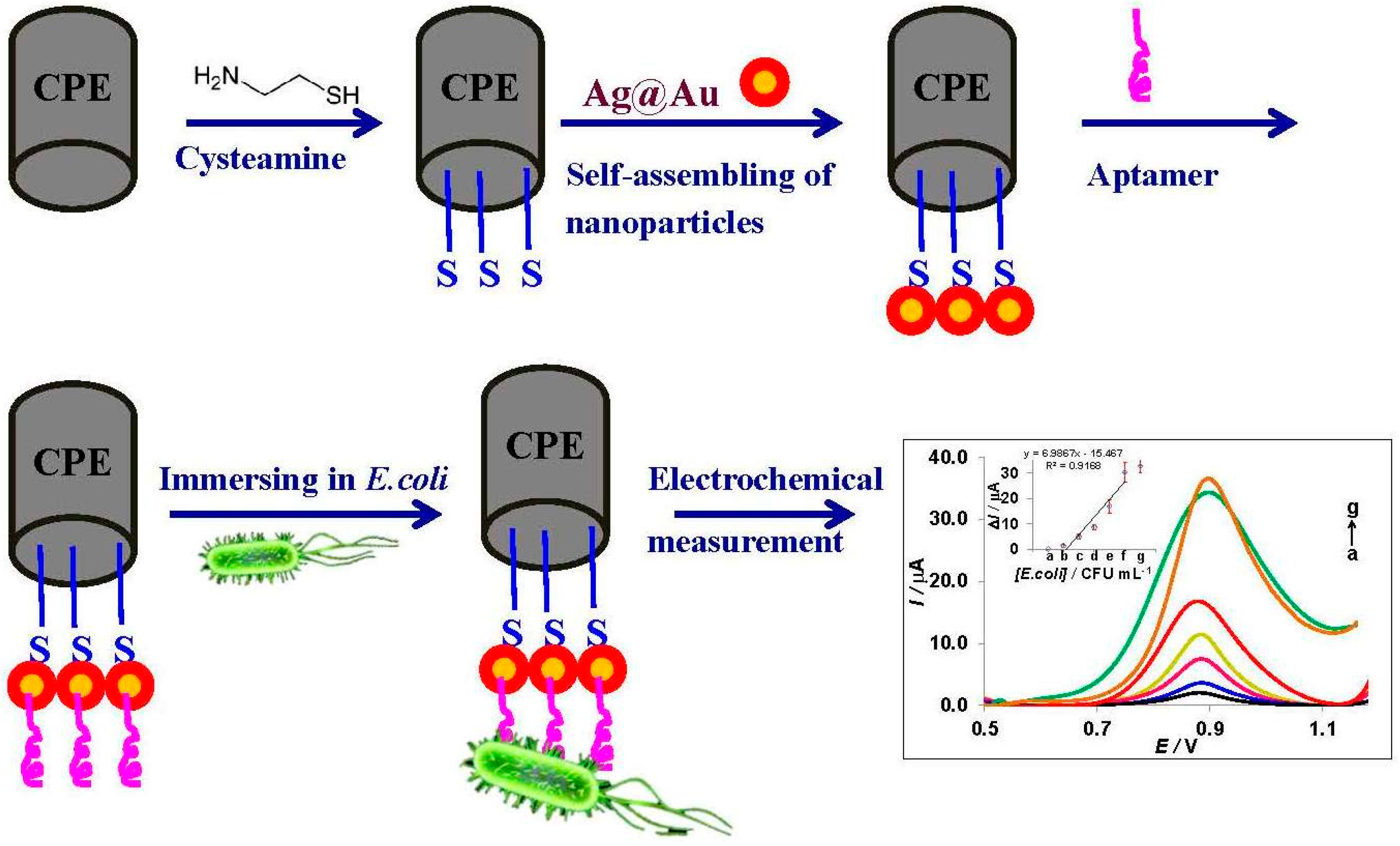
Unique Properties of Core Shell Ag@Au Nanoparticles for the Aptasensing of Bacterial Cells
Abstract
:1. Introduction
2. Experimental Section
3. Results and Discussion
3.1. Electrochemical Response of E. coli at Bare Electrode
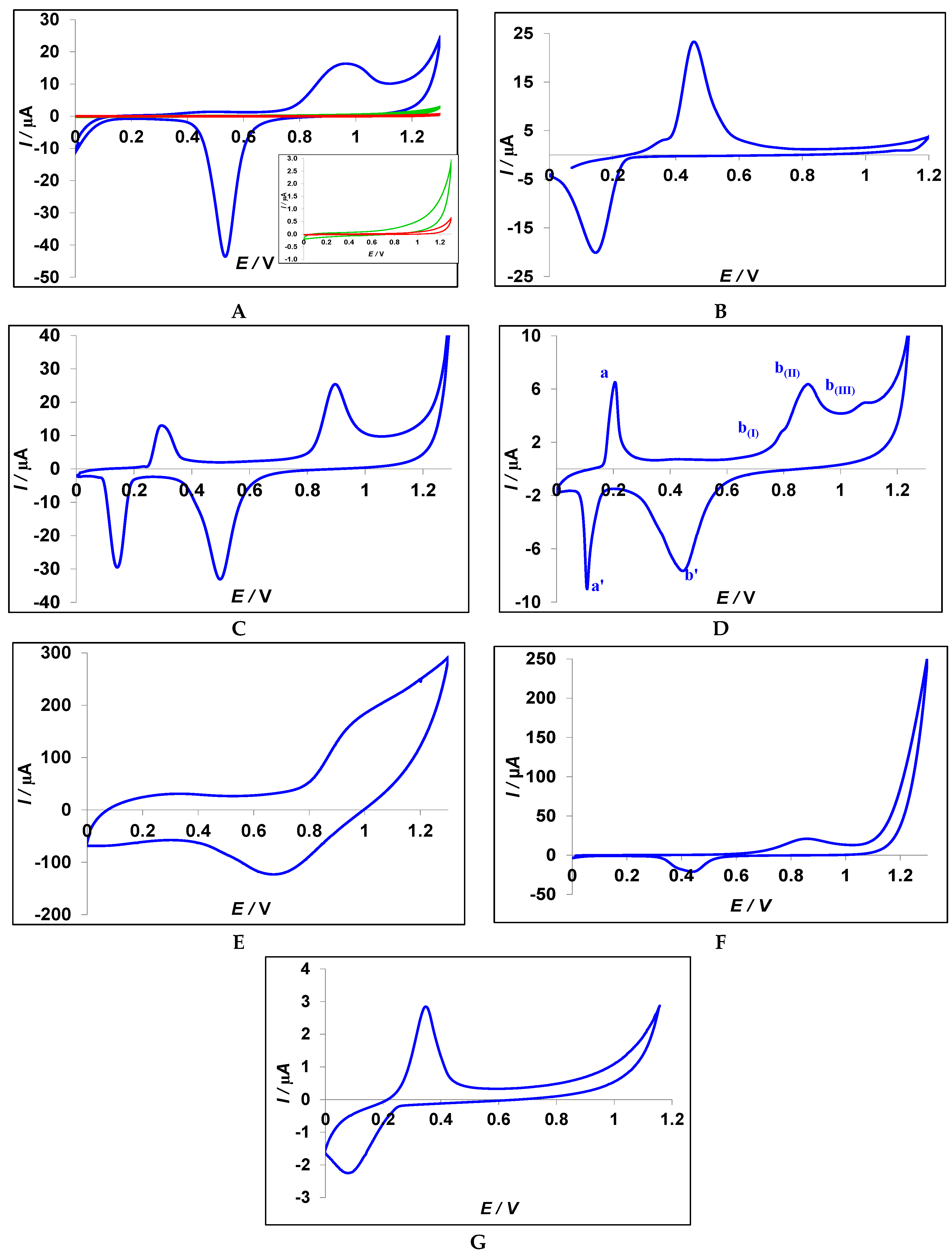
3.2. Characterization of the Nanoparticles
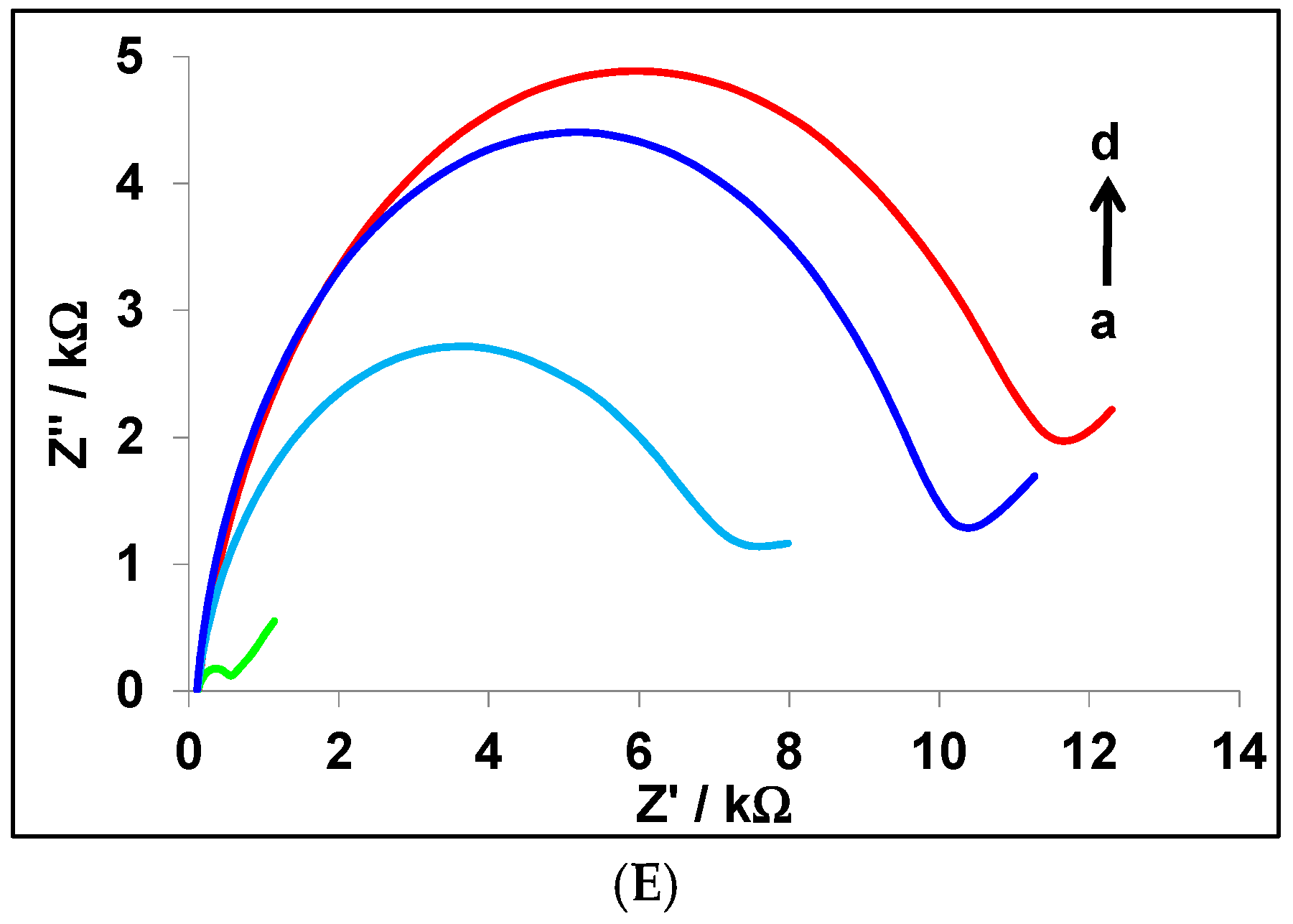
3.3. Characterization of the Modified CPE
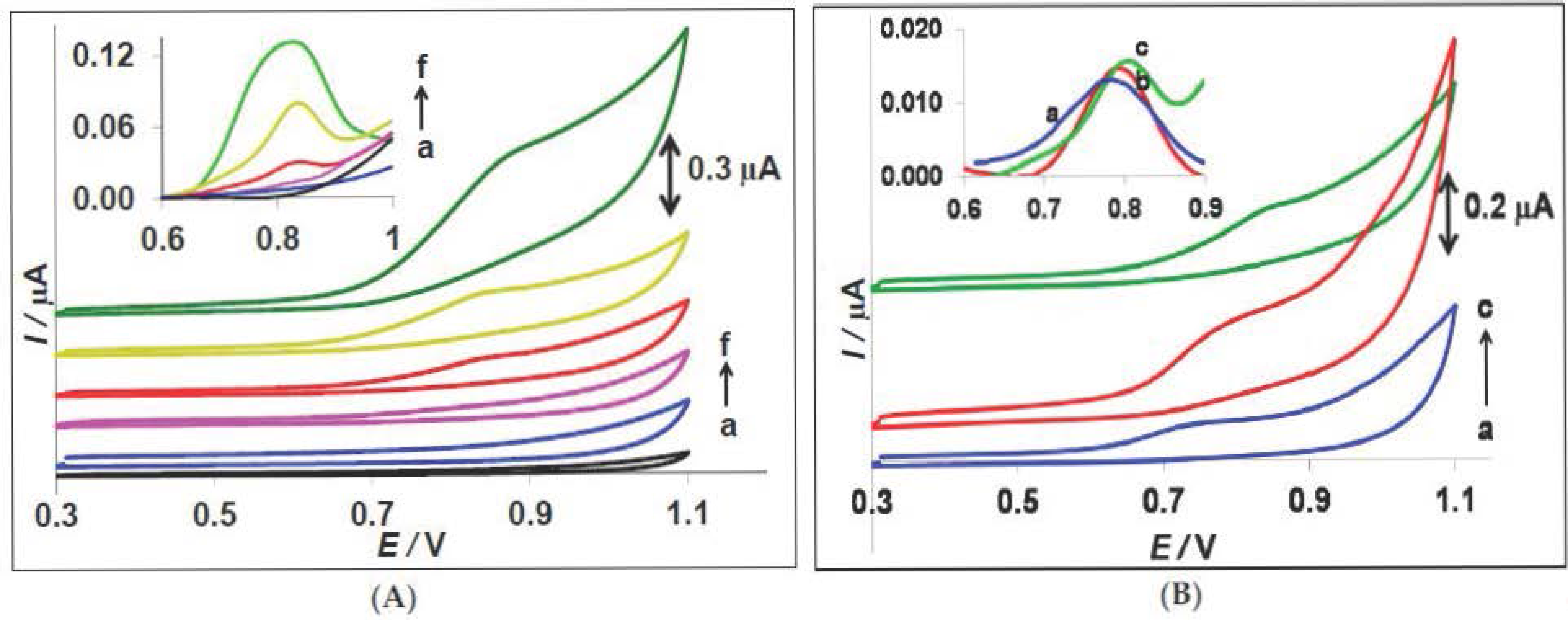
3.4. Electrochemical Detection of E. coli and the Role of Ag@Au Nanoparticles
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yin, Y.; Rioux, R.M.; Erdonmez, C.K.; Hughes, S.; Somorjai, G.A.; Alivisatos, A.P. Formation of hollow nanocrystals through the nanoscale kirkendall effect. Science 2004, 304, 711–714. [Google Scholar] [CrossRef] [PubMed]
- Campion, A.; Kambhampati, P. Surface-enhanced raman scattering. Chem. Soc. Rev. 1998, 27, 241–250. [Google Scholar] [CrossRef]
- Milliron, D.J.; Hughes, S.M.; Cui, Y.; Manna, L.; Li, J.; Wang, L.-W.; Paul Alivisatos, A. Colloidal nanocrystal heterostructures with linear and branched topology. Nature 2004, 430, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Toshima, N.; Harada, M.; Yonezawa, T.; Kushihashi, K.; Asakura, K. Structural analysis of polymer-protected palladium/platinum bimetallic clusters as dispersed catalysts by using extended X-ray absorption fine structure spectroscopy. J. Phys. Chem. 1991, 95, 7448–7453. [Google Scholar] [CrossRef]
- Ferrando, R.; Jellinek, J.; Johnston, R.L. Nanoalloys: From theory to applications of alloy clusters and nanoparticles. Chem. Rev. 2008, 108, 845–910. [Google Scholar] [CrossRef] [PubMed]
- Khlebtsov, N.G.; Dykman, L.A. Optical properties and biomedical applications of plasmonic nanoparticles. J. Quant. Spectrosc. Radiat. Transf. 2010, 111, 1–35. [Google Scholar] [CrossRef]
- Tang, D.; Yuan, R.; Chai, Y. Ligand-functionalized core/shell Ag@Au nanoparticles label-free amperometric immun-biosensor. Biotechnol. Bioeng. 2006, 94, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Y.; Zhong, Y.; Li, S. Enzyme-free hydrogen peroxide sensor based on Au@Ag@C core-double shell nanocomposites. Appl. Surf. Sci. 2015, 347, 428–434. [Google Scholar] [CrossRef]
- Eksi, H.; Güzel, R.; Güven, B.; Boyaci, I.H.; Solak, A.O. Fabrication of an electrochemical E. coli biosensor in biowells using bimetallic nanoparticle-labelled antibodies. Electroanalysis 2015, 27, 343–352. [Google Scholar] [CrossRef]
- Belluco, S.; Barco, L.; Roccato, A.; Ricci, A. Escherichia coli and enterobacteriaceae counts on poultry carcasses along the slaughterline: A systematic review and meta-analysis. Food Control 2016, 60, 269–280. [Google Scholar] [CrossRef]
- Søndergaard, R.V.; Christensen, N.M.; Henriksen, J.R.; Kumar, E.K.P.; Almdal, K.; Andresen, T.L. Facing the design challenges of particle-based nanosensors for metabolite quantification in living cells. Chem. Rev. 2015, 115, 8344–8378. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.A.; Shim, S.-H.; He, J.; Zhuang, X. Fast, three-dimensional super-resolution imaging of live cells. Nat. Methods 2011, 8, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Bergner, S.; Wegener, J.; Matysik, F.-M. Monitoring passive transport of redox mediators across a confluent cell monolayer with single-cell resolution by means of scanning electrochemical microscopy. Anal. Methods 2012, 4, 623–629. [Google Scholar] [CrossRef]
- Nemes, P.; Knolhoff, A.M.; Rubakhin, S.S.; Sweedler, J.V. Metabolic differentiation of neuronal phenotypes by single-cell capillary electrophoresis–electrospray ionization-mass spectrometry. Anal. Chem. 2011, 83, 6810–6817. [Google Scholar] [CrossRef] [PubMed]
- Drescher, D.; Giesen, C.; Traub, H.; Panne, U.; Kneipp, J.; Jakubowski, N. Quantitative imaging of gold and silver nanoparticles in single eukaryotic cells by laser ablation icp-ms. Anal. Chem. 2012, 84, 9684–9688. [Google Scholar] [CrossRef] [PubMed]
- Collins, J.M.; Lam, R.T.S.; Yang, Z.; Semsarieh, B.; Smetana, A.B.; Nettikadan, S. Targeted delivery to single cells in precisely controlled microenvironments. Lab Chip 2012, 12, 2643–2648. [Google Scholar] [CrossRef] [PubMed]
- Trouillon, R.; Passarelli, M.K.; Wang, J.; Kurczy, M.E.; Ewing, A.G. Chemical analysis of single cells. Anal. Chem. 2012, 85, 522–542. [Google Scholar] [CrossRef] [PubMed]
- Hernández, R.; Vallés, C.; Benito, A.M.; Maser, W.K.; Xavier Rius, F.; Riu, J. Graphene-based potentiometric biosensor for the immediate detection of living bacteria. Biosens. Bioelectron. 2014, 54, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Zelada-Guillén, G.A.; Riu, J.; Düzgün, A.; Rius, F.X. Immediate detection of living bacteria at ultralow concentrations using a carbon nanotube based potentiometric aptasensor. Angew. Chem. Int. Ed. 2009, 48, 7334–7337. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Kwa, T.; Revzin, A. Simultaneous detection of cell-secreted tnf-α and ifn-γ using micropatterned aptamer-modified electrodes. Biomaterials 2012, 33, 7347–7355. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Morris, M.D.; Macazo, F.C.; Schoukroun-Barnes, L.R.; White, R.J. The current and future role of aptamers in electroanalysis. J. Electrochem. Soc. 2014, 161, H301–H313. [Google Scholar] [CrossRef]
- Le, T.T.; Adamiak, B.; Benton, D.J.; Johnson, C.J.; Sharma, S.; Fenton, R.; McCauley, J.W.; Iqbal, M.; Cass, A.E.G. Aptamer-based biosensors for the rapid visual detection of flu viruses. Chem. Commun. 2014, 50, 15533–15536. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Hu, Y.; Duan, J.; Yuan, W.; Wang, C.; Xu, H.; Yang, X.-D. Novel aptamer-nanoparticle bioconjugates enhances delivery of anticancer drug to muc1-positive cancer cells in vitro. PLoS ONE 2011, 6, e24077. [Google Scholar] [CrossRef] [PubMed]
- Farokhzad, O.C.; Jon, S.; Khademhosseini, A.; Tran, T.-N.T.; LaVan, D.A.; Langer, R. Nanoparticle-aptamer bioconjugates: A new approach for targeting prostate cancer cells. Cancer Res. 2004, 64, 7668–7672. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.Z.; Bagalkot, V.; Vasilliou, C.C.; Gu, F.; Alexis, F.; Zhang, L.; Shaikh, M.; Yuet, K.; Cima, M.J.; Langer, R.; et al. Superparamagnetic iron oxide nanoparticle–aptamer bioconjugates for combined prostate cancer imaging and therapy. ChemMedChem 2008, 3, 1311–1315. [Google Scholar] [CrossRef] [PubMed]
- Medley, C.D.; Bamrungsap, S.; Tan, W.; Smith, J.E. Aptamer-conjugated nanoparticles for cancer cell detection. Anal. Chem. 2011, 83, 727–734. [Google Scholar] [CrossRef] [PubMed]
- Kashefi-Kheyrabadi, L.; Mehrgardi, M.A. Aptamer-conjugated silver nanoparticles for electrochemical detection of adenosine triphosphate. Biosens. Bioelectron. 2012, 37, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Chung, J.; Song, M.Y.; Jurng, J.; Kim, B.C. Aptamer cocktails: Enhancement of sensing signals compared to single use of aptamers for detection of bacteria. Biosens. Bioelectron. 2014, 54, 195–198. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Song, M.Y.; Jurng, J.; Kim, B.C. Isolation and characterization of DNA aptamers against Escherichia coli using a bacterial cell-systematic evolution of ligands by exponential enrichment approach. Anal. Biochem. 2013, 436, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-J.; Gu, M.-M.; Zheng, T.-T.; Zhu, J.-J. Synthesis of gelatin-stabilized gold nanoparticles and assembly of carboxylic single-walled carbon nanotubes/Au composites for cytosensing and drug uptake. Anal. Chem. 2009, 81, 6641–6648. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Ci, Y.-X.; Lou, J.-L.; Zhang, X.-Q. Voltammetric behavior of mammalian tumor cells and bioanalytical applications in cell metabolism. Bioelectrochem. Bioenerg. 1999, 48, 217–222. [Google Scholar] [CrossRef]
- Nonner, W.; Eisenberg, B. Electrodiffusion in ionic channels of biological membranes. J. Mol. Liq. 2000, 87, 149–162. [Google Scholar] [CrossRef]
- Schaadt, D.M.; Feng, B.; Yu, E.T. Enhanced semiconductor optical absorption via surface plasmon excitation in metal nanoparticles. Appl. Phys. Lett. 2005, 86, 063106. [Google Scholar] [CrossRef]
- Liu, M.; Guyot-Sionnest, P. Synthesis and optical characterization of Au/Ag core/shell nanorods. J. Phys. Chem. B 2004, 108, 5882–5888. [Google Scholar] [CrossRef]
- Shen, X.S.; Wang, G.Z.; Hong, X.; Zhu, W. Nanospheres of silver nanoparticles: Agglomeration, surface morphology control and application as sers substrates. Phys. Chem. Chem. Phys. 2009, 11, 7450–7454. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lin, Y.; Zhao, B. Spontaneous agglomeration of silver nanoparticles deposited on carbon film surface. J. Nanopart. Res. 2002, 4, 345–349. [Google Scholar] [CrossRef]
- Danscher, G.; Hansen, H.J.; Møller-Madsen, B. Energy dispersive x-ray analysis of tissue gold after silver amplification by physical development. Histochemistry 1984, 81, 283–285. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.-Z.; Zhang, D.-P.; Wu, W.-L.; Yang, Z.-S. Selective determination of dopamine in the presence of high concentration of ascorbic acid using nano-au self-assembly glassy carbon electrode. Colloids Surf. B 2008, 62, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Love, J.C.; Estroff, L.A.; Kriebel, J.K.; Nuzzo, R.G.; Whitesides, G.M. Self-assembled monolayers of thiolates on metals as a form of nanotechnology. Chem. Rev. 2005, 105, 1103–1170. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jiang, X.; Wang, E.; Dong, S. Attachment of gold nanoparticles to glassy carbon electrode and its application for the direct electrochemistry and electrocatalytic behavior of hemoglobin. Biosens. Bioelectron. 2005, 21, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.-J.; Luo, T.-J.M. Electrochemical properties of silver nanoparticle doped aminosilica nanocomposite. Int. J. Electrochem. 2011, 2011. [Google Scholar] [CrossRef]
- Tang, M.H.; Hahn, C.; Klobuchar, A.J.; Ng, J.W.D.; Wellendorff, J.; Bligaard, T.; Jaramillo, T.F. Nickel-silver alloy electrocatalysts for hydrogen evolution and oxidation in an alkaline electrolyte. Phys. Chem. Chem. Phys. 2014, 16, 19250–19257. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Tu, C.Q.; Chen, Y.F.; Li, W.Y.; Houlachi, G. Cyclic voltammetric studies of the behavior of lead-silver anodes in zinc electrolytes. J. Mater. Eng. Perform. 2013, 22, 1672–1679. [Google Scholar] [CrossRef]
- Rouya, E.; Cattarin, S.; Reed, M.L.; Kelly, R.G.; Zangari, G. Electrochemical characterization of the surface area of nanoporous gold films. J. Electrochem. Soc. 2012, 159, K97–K102. [Google Scholar] [CrossRef]
- Tremiliosi-Filho, G.; Dall’Antonia, L.H.; Jerkiewicz, G. Growth of surface oxides on gold electrodes under well-defined potential, time and temperature conditions. J. Electroanal. Chem. 2005, 578, 1–8. [Google Scholar] [CrossRef]
- Gasparotto, L.H.S.; Gomes, J.F.; Tremiliosi-Filho, G. Cyclic-voltammetry characteristics of poly(vinyl pyrrolidone) (pvp) on single-crystal pt surfaces in aqueous h2so4. J. Electroanal. Chem. 2011, 663, 48–51. [Google Scholar] [CrossRef]
- Ebato, H.; Gentry, C.A.; Herron, J.N.; Mueller, W.; Okahata, Y.; Ringsdorf, H.; Suci, P.A. Investigation of specific binding of antifluorescyl antibody and fab to fluorescein lipids in langmuir-blodgett deposited films using quartz crystal microbalance methodology. Anal. Chem. 1994, 66, 1683–1689. [Google Scholar] [CrossRef] [PubMed]
- Okahata, Y.; Kawase, M.; Niikura, K.; Ohtake, F.; Furusawa, H.; Ebara, Y. Kinetic measurements of DNA hybridization on an oligonucleotide-immobilized 27-mhz quartz crystal microbalance. Anal. Chem. 1998, 70, 1288–1296. [Google Scholar] [CrossRef] [PubMed]
- Lavrich, D.J.; Wetterer, S.M.; Bernasek, S.L.; Scoles, G. Physisorption and chemisorption of alkanethiols and alkyl sulfides on Au(111). J. Phys. Chem. B 1998, 102, 3456–3465. [Google Scholar] [CrossRef]
- Fischer, D.; Curioni, A.; Andreoni, W. Decanethiols on gold: The structure of self-assembled monolayers unraveled with computer simulations. Langmuir 2003, 19, 3567–3571. [Google Scholar] [CrossRef]
- Mulvaney, P. Surface plasmon spectroscopy of nanosized metal particles. Langmuir 1996, 12, 788–800. [Google Scholar] [CrossRef]
- Link, S.; Wang, Z.L.; El-Sayed, M.A. Alloy formation of gold−silver nanoparticles and the dependence of the plasmon absorption on their composition. J. Phys. Chem. B 1999, 103, 3529–3533. [Google Scholar] [CrossRef]
- Queirós, R.B.; de-los-Santos-Álvarez, N.; Noronha, J.P.; Sales, M.G.F. A label-free DNA aptamer-based impedance biosensor for the detection of E. coli outer membrane proteins. Sens. Actuators B Chem. 2013, 181, 766–772. [Google Scholar] [CrossRef]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamidi-Asl, E.; Dardenne, F.; Pilehvar, S.; Blust, R.; De Wael, K. Unique Properties of Core Shell Ag@Au Nanoparticles for the Aptasensing of Bacterial Cells. Chemosensors 2016, 4, 16. https://doi.org/10.3390/chemosensors4030016
Hamidi-Asl E, Dardenne F, Pilehvar S, Blust R, De Wael K. Unique Properties of Core Shell Ag@Au Nanoparticles for the Aptasensing of Bacterial Cells. Chemosensors. 2016; 4(3):16. https://doi.org/10.3390/chemosensors4030016
Chicago/Turabian StyleHamidi-Asl, Ezat, Freddy Dardenne, Sanaz Pilehvar, Ronny Blust, and Karolien De Wael. 2016. "Unique Properties of Core Shell Ag@Au Nanoparticles for the Aptasensing of Bacterial Cells" Chemosensors 4, no. 3: 16. https://doi.org/10.3390/chemosensors4030016
APA StyleHamidi-Asl, E., Dardenne, F., Pilehvar, S., Blust, R., & De Wael, K. (2016). Unique Properties of Core Shell Ag@Au Nanoparticles for the Aptasensing of Bacterial Cells. Chemosensors, 4(3), 16. https://doi.org/10.3390/chemosensors4030016