Toward Non-Enzymatic Ultrasensitive Identification of Single Nucleotide Polymorphisms by Optical Methods
Abstract
:1. Introduction
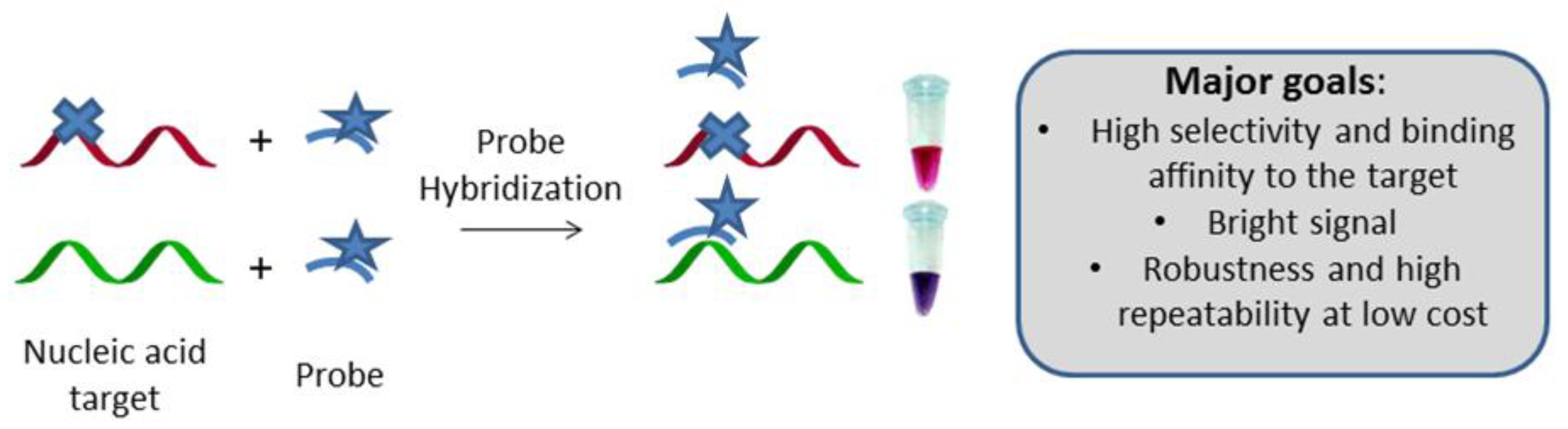
2. Enzyme-Based Detection of SNP
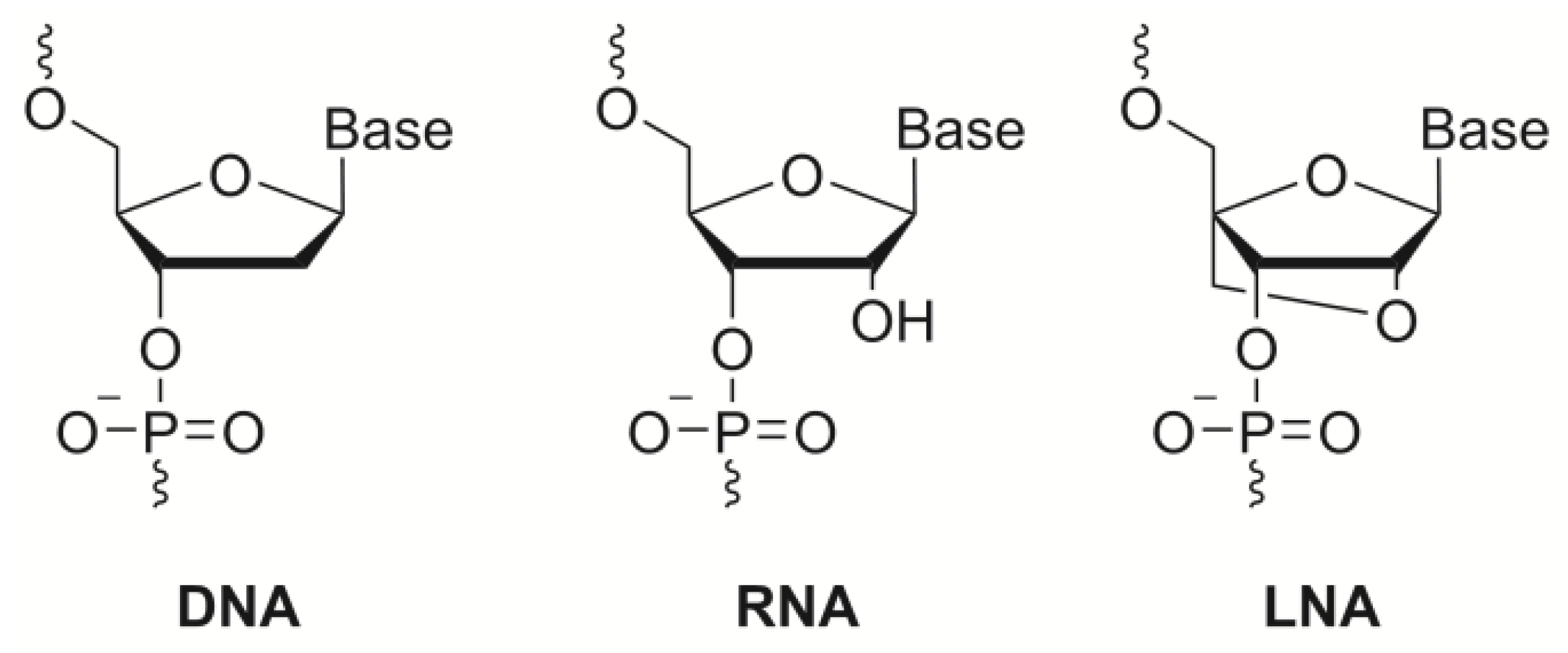
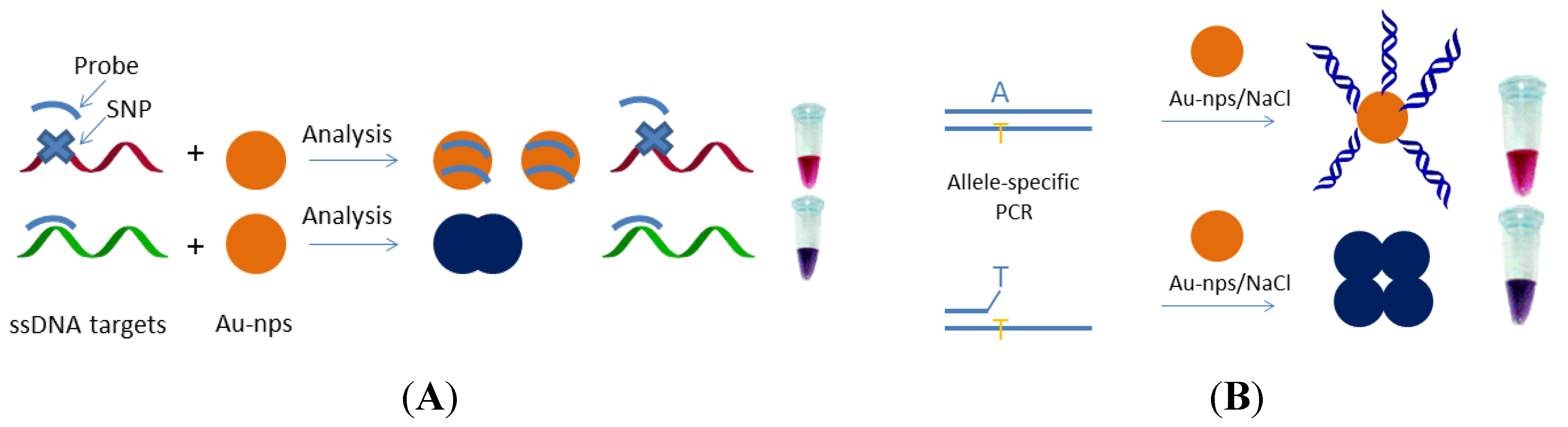
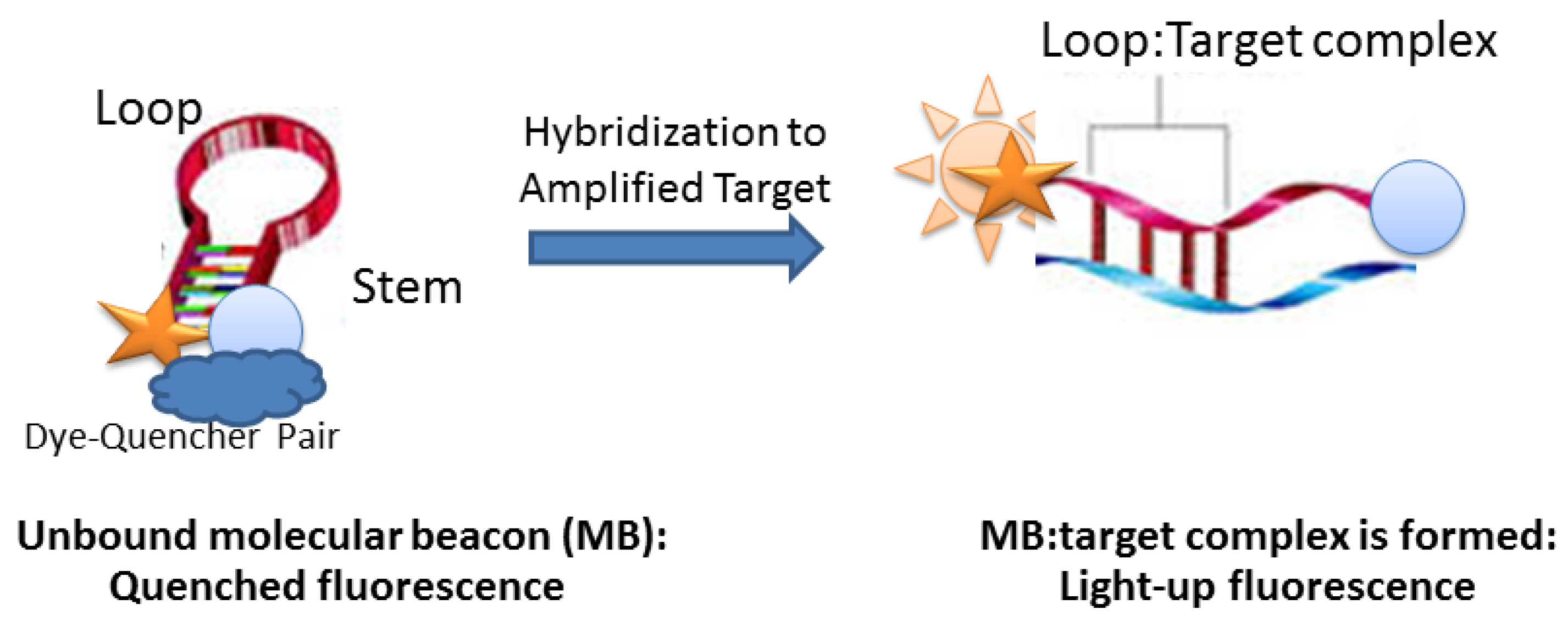
3. Toward Enzyme-Free Detection of SNP
3.1. Nano- and Magnetic-Particles in Amplification-Free SNP Diagnostics
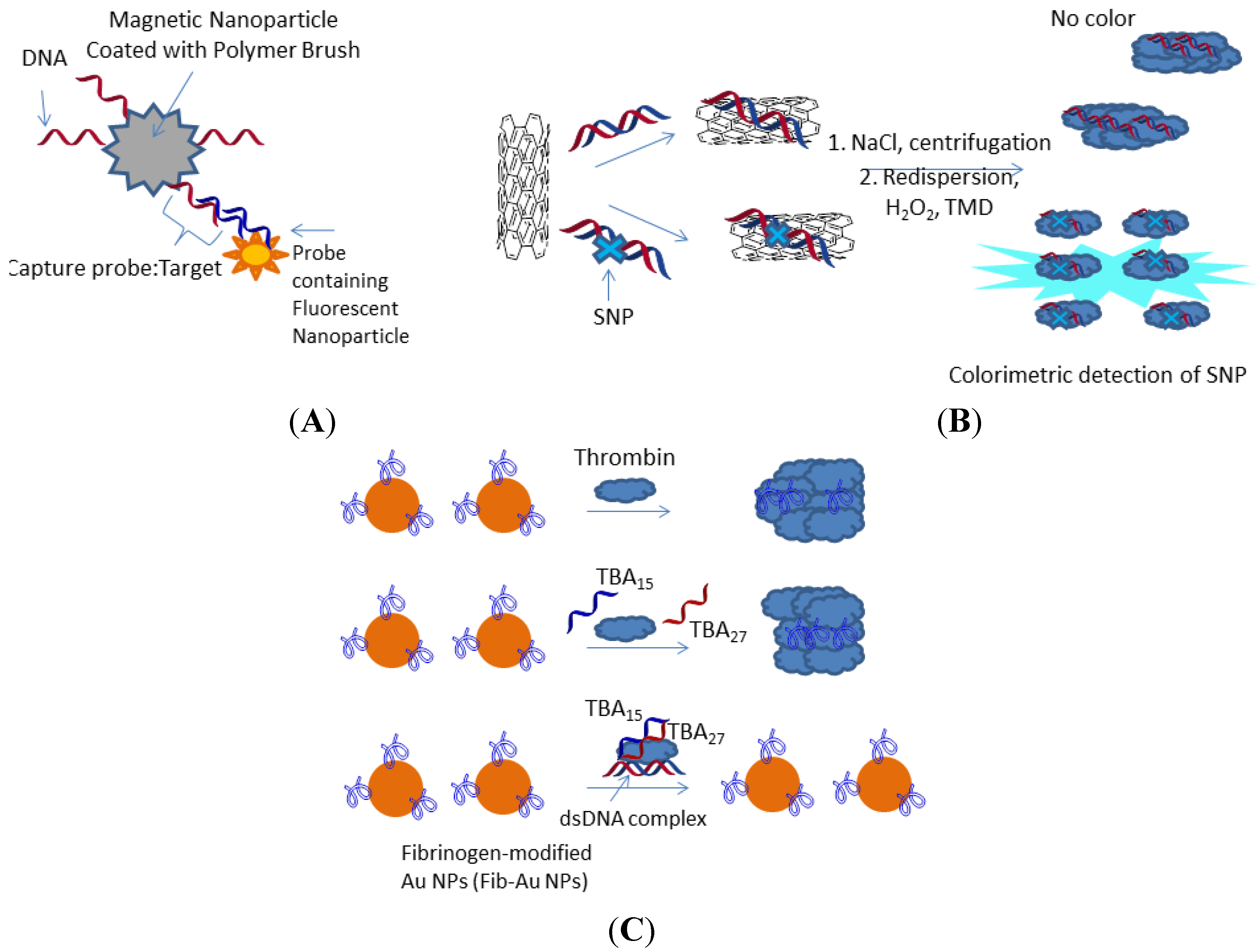
3.2. Nucleic Acid Aptamers and Proteins in SNP Diagnostics
3.3. Other Optical Methods for Sensing SNP
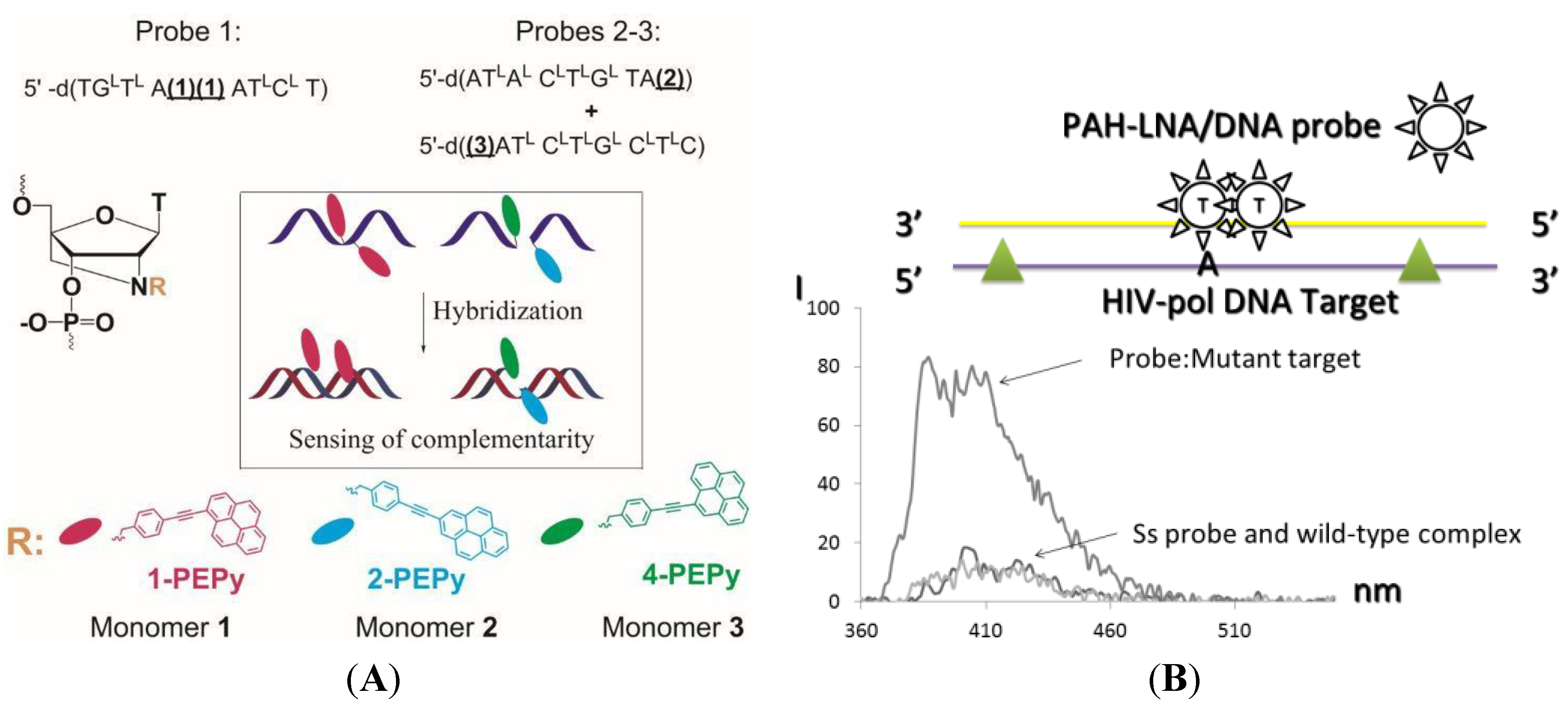
4. Conclusions
Acknowledgments
Conflicts of Interest
References and Notes
- Sachidanandam, R.; Weissman, D.; Schmidt, S.C.; Kakol, J.M.; Stein, L.D.; Marth, G.; Sherry, S.; Mullikin, J.C.; Mortimore, B.J.; Willey, D.L.; et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 2001, 409, 928–933. [Google Scholar] [CrossRef]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; et al. The sequence of the human genome. Science 2001, 291, 1304–1351. [Google Scholar] [CrossRef]
- Hacia, J.G. Resequencing and mutational analysis using oligonucleotide microarrays. Nat. Genet. 1999, 21, 42–47. [Google Scholar] [CrossRef]
- Syvanen, A.-C. Accessing genetic variation: Genotyping single nucleotide polymorphisms. Nat. Rev. Genet. 2001, 2, 930–942. [Google Scholar] [CrossRef]
- Kwok, P.Y. Methods for genotyping single nucleotide polymorphisms. Annu. Rev. Genomics Hum. Genet. 2001, 2, 235–258. [Google Scholar] [CrossRef]
- Kirk, B.W.; Feinsod, M.; Favis, R.; Kliman, R.M.; Barany, F. Single nucleotide polymorphism seeking long term association with complex disease. Nucleic Acids Res. 2002, 30, 3295–3311. [Google Scholar]
- Koshkin, A.; Rajwanshi, V.K.; Wengel, J. Novel Convenient Syntheses of LNA [2.2.1] Bicyclo Nucleosides. Tetrahedron Lett. 1998, 39, 4381–4384. [Google Scholar] [CrossRef]
- Hari, Y.; Obika, S.; Sasaki, M. Effective synthesis of C-nucleosides with 2'-4'-BNA modification. Tetrahedron 2002, 58, 3051–3063. [Google Scholar] [CrossRef]
- Latorra, D.; Arar, K.; Hurley, J.M. Design considerations and effects of LNA in PCR primers. Mol. Cell. Probes 2003, 17, 253–259. [Google Scholar] [CrossRef]
- Di Giusto, D.A.; King, G.C. Strong positional preference in the interaction of LNA oligonucleotides with DNA polymerase and proofreadingexonuclease activities: Implications for genotying assays. Nucleic Acids Res. 2004, 32, e32. [Google Scholar] [CrossRef]
- Levin, J.D.; Fiala, D.; Samala, M.F.; Kahn, J.D.; Peterson, R.J. Position-dependent effects of locked nucleic acid (LNA) on DNA sequencing and PCR primers. Nucleic Acids Res. 2006, 34, e142. [Google Scholar] [CrossRef]
- Rupp, J.; Solbach, W.; Gieffers, J. Single-nucleotide-polymorphism-specific PCR for quantifcation and discriminationof Chlamydia pneumoniae genotypes by use of a “locked” nucleic acid. Appl. Environ. Microbiol. 2006, 72, 3785–3787. [Google Scholar] [CrossRef]
- Johnson, M.P.; Haupt, L.M.; Griffiths, L.R. Locked nucleic acid (LNA) single nucleotide polymorphism (SNP) genotype analysis and validation using real-time PCR. Nucleic Acids Res. 2004, 32, e55. [Google Scholar] [CrossRef]
- You, Y.; Moreira, B.; Behlke, M.A.; Owczarzy, R. Design of LNA probes that improve mismatch discrimination. Nucleic Acids Res. 2006, 34, e60. [Google Scholar] [CrossRef]
- Mouritzen, P.; Nielsen, A.T.; Pfundheller, H.M.; Choleva, Y.; Kongsbak, L.; Møller, S. Single nucleotide polymorphism genotyping using locked nucleic acid (LNATM). Expert Rev. Mol. Diagn. 2003, 3, 27–38. [Google Scholar] [CrossRef]
- Crowley, E.; Di Nicolantonio, F.; Loupaki, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef]
- Didelot, A.; Kotsopoulos, S.K.; Lupo, A.; Pekin, D.; Li, X.; Atochin, I.; Srinivasan, P.; Zhong, Q.; Olson, J.; Link, D.R.; et al. Multiplex Picoliter-Droplet Digital PCR for Quantitative Assessment of DNA Integrity in Clinical Samples. Clin. Chem. 2013, 59, 815–823. [Google Scholar] [CrossRef]
- stergaard, M.E.; Hrdlicka, P.J. Pyrene-functionalized oligonucleotides and locked nucleic acids (LNAs): Tools for fundamental research, diagnostics, and nanotechnology. Chem. Soc. Rev. 2011, 40, 5771–5788. [Google Scholar] [CrossRef]
- Xiao, Y.; Lou, X.; Uzawa, T.; Plakos, K.J.I.; Plaxco, K.W.; Soh, H.T. An Electrochemical Sensor for Single Nucleotide Polymorphism Detection in Serum Based on a Triple-Stem DNA Probe. J. Am. Chem. Soc. 2009, 131, 15311–15316. [Google Scholar] [CrossRef]
- Cagnin, S.; Caraballo, M.; Guiducci, C.; Martini, P.; Ross, M.; Santaana, M.; Danley, D.; West, T.; Lanfranchi, G.; et al. Overview of Electrochemical DNA Biosensors: New Approaches to Detect the Expression of Life. Sensors 2009, 9, 3122–3148. [Google Scholar] [CrossRef]
- Huang, T.J.; Liu, M.; Knight, L.D.; Grody, W.W.; Miller, J.F.; Ho, C.-M. An electrochemical detection scheme for identification of single nucleotide polymorphisms using hairpin-forming probes. Nucleic Acids Res. 2002, 30, e55. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, X.Y.; Yang, W.; Chen, J.; Fu, F.F. Ultra-sensitive electrochemical detection of single nucleotide polymorphisms based on an electrically controllable magnetic gold electrode. Chem. Commun. 2013, 49, 996–998. [Google Scholar]
- Guo, Y.; Chen, J.; Chen, G. A label-free electrochemical biosensor for detection of HIV related gene based on interaction between DNA and protein. Sens.Actuators B. Chem. 2013, 184, 113–117. [Google Scholar] [CrossRef]
- Wei, F.; Lillehoj, P.B.; Ho, C.-M. DNA Diagnostics: Nanotechnology-Enhanced Electrochemical Detection of Nucleic Acids. Pediatr. Res. 2010, 67, 458–468. [Google Scholar]
- Astakhova, I.K.; Kumar, T.S.; Campbell, M.A.; Ustinov, A.V.; Korshun, V.A.; Wengel, J. Branched DNA nanostructures efficiently stabilised and monitored by novel pyrene-perylene 2'-α-L-amino-LNA FRET pairs. Chem. Commun. 2013, 49, 511–513. [Google Scholar] [CrossRef]
- Li, H.; Rothberg, L.J. Label-Free Colorimetric Detection of Specific Sequences in Genomic DNA Amplified by the Polymerase Chain Reaction. J. Am. Chem. Soc. 2004, 126, 10958–10961. [Google Scholar] [CrossRef]
- Jung, Y.-L.; Jung, C.; Parab, H.; Cho, D.Y.; Park, H.G. Colorimetric SNP genotyping based on allele-specific PCR by using a thiol-labeled primer. ChemBioChem 2011, 12, 1387–1390. [Google Scholar] [CrossRef]
- Veigas, B.; Machado, D.; Perdigão, J.; Portugal, I.; Couto, I.; Viveiros, M.; Baptista, P.V. Au-nanoprobes for detection of SNPs associated with antibiotic resistance in Mycobacterium tuberculosis. Nanotechnology 2010, 21, 415101–415108. [Google Scholar]
- Valentini, P.; Fiammengo, R.; Sabella, S.; Gariboldi, M.; Maiorano, G.; Cingolani, R.; Pompa, P.P. Gold-nanoparticle-based colorimetric discrimination of cancer-related point mutations with picomolar sensitivity. ACS Nano 2013, 7, 5530–5538. [Google Scholar] [CrossRef]
- Vicent, M.; Xu, Y.; Kong, H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004, 5, 795–800. [Google Scholar] [CrossRef]
- Marras, S.A.; Kramer, F.R.; Tyagi, S. Genotyping SNPs with Molecular Beacons. Methods Mol. Biol. 2002, 212, 111–128. [Google Scholar]
- Barreiro, L.B.; Henriques, R.; Mhlanga, M.M. High-throughput SNP genotyping: Combining tag SNPs and molecular beacons. Methods Mol. Biol. 2009, 578, 255–276. [Google Scholar] [CrossRef]
- Neo, J.L.; Uttamchandani, M. Visual SNP genotyping using asymmetric PCR and split DNA enzymes. Analyst 2011, 136, 1569–1572. [Google Scholar] [CrossRef]
- Wabuyele, M.B.; Farquar, H.; Stryjewski, W.; Hammer, R.P.; Soper, S.; Cheng, Y.W.; Barany, F. Approaching real-time molecular diagnostics: Single-pair fluorescence resonance energy transfer (spFRET) detection for the analysis of low abundant point mutations in K-ras oncogenes. J. Am. Chem. Soc. 2003, 125, 6937–6945. [Google Scholar]
- Hall, J.G.; Eis, P.S.; Law, S.M.; Reynaldo, L.P.; Prudent, J.R.; Marshall, D.J.; Allawi, H.T.; Mast, A.L.; Dahlberg, J.E.; Kwiatkowski, R.W.; et al. Sensitive detection of dna polymorphisms by the serial invasive signal amplification reaction. Proc. Natl. Acad. Sci. USA 2000, 97, 8272–8277. [Google Scholar] [CrossRef]
- Spittle, C.; Ward, M.R.; Nathanson, K.L.; Gimotty, P.A.; Rappaport, E.; Brose, M.S.; Medina, A.; Letrero, R.; Herlyn, M.; Edwards, R.H.; et al. Application of a BRAF Pyrosequencing Assay for Mutation Detection and Copy Number Analysis in Malignant Melanoma. J. Mol. Diagn. 2007, 9, 464–471. [Google Scholar] [CrossRef]
- Gessi, M.; Pietsch, T. The Diagnostic Role and Clinical Relevance of Determination of BRAF Status in Brain Tumors. Pers. Med. 2013, 10, 405–412. [Google Scholar] [CrossRef]
- Wang, Y.; Cottman, M.; Schiffman, J.D. Molecular inversion probes: A novel microarray technology and its application in cancer research. Cancer Genet. 2012, 205, 341–355, and references cited therein. [Google Scholar] [CrossRef]
- Mirkin, C.A.; Letsinger, R.L.; Mucic, R.C.; Storhoff, J.J. A DNA-Based Method for Rationally Assembling Nanoparticles into Macroscopic Materials. Nature 1996, 382, 607–609. [Google Scholar] [CrossRef]
- Kang, J.-H.; Asami, Y.; Murata, M.; Kitazaki, H.; Sadanaga, N.; Tokunaga, E.; Shiotani, S.; Okada, S.; Maehara, Y.; Niidome, T.; et al. Gold nanoparticle-based colorimetric assay for cancer diagnosis. Biosens. Bioelectron. 2010, 25, 1869–1874. [Google Scholar] [CrossRef]
- Murphy, D.; O’Brien, P.; Redmond, G. Sub-picomole colorimetric single nucleotide polymorphism discrimination using oligonucleotide–nanoparticle conjugates. Analyst 2004, 129, 970–974. [Google Scholar] [CrossRef]
- Bao, Y.P.; Huber, M.; Wei, T.F.; Marla, S.S.; Storhoff, J.J.; Müller, U.R. SNP identification in unamplified human genomic DNA with gold nanoparticle probes. Nucleic Acids Res. 2005, 33, e15. [Google Scholar] [CrossRef]
- Ihara, T.; Tanaka, S.; Chikaura, Y.; Jyo, A. Preparation of DNA-modified nanoparticles and preliminary study for colorimetric SNP analysis using their selective aggregations. Nucleic Acids Res. 2004, 32, e105. [Google Scholar] [CrossRef]
- Thomson, D.A.C.; Tee, E.H.-L.; Tran, N.T.D.; Monteiro, M.J.; Cooper, M.A. Oligonucleotide and polymer functionalized nanoparticles for amplification-free detection of DNA. Biomacromolecules 2012, 13, 1981–1989. [Google Scholar] [CrossRef]
- Song, Y.; Wang, X.; Zhao, C.; Qu, K.; Ren, J.; Qu, X. Label-Free Colorimetric Detection of Single Nucleotide Polymorphism by Using Single-Walled Carbon Nanotube Intrinsic Peroxidase-Like Activity. Chem. Eur. J. 2010, 16, 3617–3621. [Google Scholar] [CrossRef]
- ELISA-inspired technology developed in so-called Simoa instrument by Quanterix, founded 2007 by D. Walt and N. Naclerio.
- Gold, L.; Polisky, B.; Uhlenbeck, O.; Yarus, M. Diversity of Oligonucleotide Functions. Annu. Rev. Biochem. 1995, 64, 763–797. [Google Scholar] [CrossRef]
- Huang, C.-C.; Huang, Y.F.; Cao, Z.; Tan, W.; Chang, H.T. Aptamer-modified gold nanoparticles for colorimetric determination of platelet-derived growth factors and their receptors. Anal. Chem. 2005, 77, 5735–5741. [Google Scholar] [CrossRef]
- Zhao, W.; Chiuman, W.; Brook, M.A.; Li, Y. Simple and rapid colorimetric biosensors based on DNA aptamer and noncrosslinking gold nanoparticle aggregation. ChemBioChem 2007, 8, 727–731. [Google Scholar] [CrossRef]
- Chen, C.-K.; Shiang, Y.-C.; Huang, C.-C.; Chang, H.-T. Using self-assembled aptamers and fibrinogen-conjugated gold nanoparticles to detect DNA based on controlled thrombin activity. Biosens. Bioelectron. 2011, 26, 3464–3468. [Google Scholar] [CrossRef]
- Jian, J.-W.; Huang, C.-C. Colorimetric Detection of DNA by Modulation of Thrombin Activity on Gold Nanoparticles. Chem. Eur. J. 2011, 17, 2374–2380. [Google Scholar] [CrossRef]
- Capper, D.; Berghoff, A.-S.; von Deimling, A.; Preusser, M. Clinical neuropathology practice news 2-2012: BRAF V600E testing. Clin. Neuropathol. 2012, 31, 64–66. [Google Scholar] [CrossRef]
- Umemoto, T.; Hrdlicka, P.; Babu, B.R.; Wengel, J. Sensitive SNP Dual-Probe Assays Based on Pyrene-Functionalized 2′-Amino-LNA: Lessons to Be Learned. ChemBioChem 2007, 8, 2240–2248. [Google Scholar] [CrossRef]
- Astakhova, I.K.; Samokhina, E.; Babu, B.R.; Wengel, J. Novel (Phenylethynyl)pyrene–LNA Constructs for Fluorescence SNP Sensing in Polymorphic Nucleic Acid Targets. ChemBioChem 2012, 13, 1509–1519. [Google Scholar]
- Kumar, T.S.; Myznikova, A.; Samokhina, E.; Astakhova, I.K. Rapid genotyping using pyrene-perylene locked nucleic acid complexes. Artif. XNA DNA PNA 2013, 4, 58–68. [Google Scholar] [CrossRef]
- Breaker, R.R. DNA enzymes. Nat. Biotechnol. 1997, 15, 427–431. [Google Scholar] [CrossRef]
- Tang, S.; Tong, P.; Li, H.; Gu, F.; Zhang, L. The three-way junction DNAzyme based probe for label-free colorimetric detection of DNA. Biosens. Bioelectron. 2013, 41, 397–402. [Google Scholar] [CrossRef]
- Kolpashnikov, D.M. Split DNA Enzyme for Visual Single Nucleotide Polymorphism Typing. J. Am. Chem. Soc. 2008, 130, 2934–2935. [Google Scholar] [CrossRef]
- Blatt, L.; Chowrira, B.; Haeberli, P.; Jadhav, V.; Kossen, K.; Mcswiggen, J.; Seiwert, S.; Usman, N.; Vaish, N.; Zinnen, S. Allosteric nucleic acid sensor molecules. WO2003089650 A2, 2003. [Google Scholar]
- Kitamura, Y.; Ihara, T.; Tsujimura, Y.; Osawa, Y.; Jyo, A. Colorimetric allele analysis based on the DNA-directed cooperative formation of luminous lanthanide complexes. Nucleic Acids Symp. Ser. 2006, 50, 105–106. [Google Scholar] [CrossRef]
- Carlson, J.C.T.; Meimetis, L.G.; Hilderbrand, S.A.; Weissleder, R. BODIPY–Tetrazine Derivatives as Superbright Bioorthogonal Turn-on Probes. Angew. Chem. Int. Ed. 2013, 52, 6917–6920. [Google Scholar] [CrossRef]
- Mao, F.; Leung, W.-Y.; Xin, X. Characterization of EvaGreen and the implication of its physicochemical properties for qPCR applications. BMC Biotechnol. 2007, 7, 76. [Google Scholar] [CrossRef]
- Astakhova, I.K.; Wengel, J. Scaffolding along Nucleic Acid Duplexes Using 2′-Amino-Locked Nucleic Acids. Acc. Chem. Res. 2014, 47, 1768–1777. [Google Scholar] [CrossRef]
- Wirpsza, L.; Pillai, S.; Batish, M.; Marras, S.; Krasnoperov, L.; Mustaev, A. Highly Bright Avidin-based Affinity Probes Carrying Multiple Lanthanide Chelates. J. Photochem. Photobiol. B. 2012, 116, 22–29. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Astakhova, K. Toward Non-Enzymatic Ultrasensitive Identification of Single Nucleotide Polymorphisms by Optical Methods. Chemosensors 2014, 2, 193-206. https://doi.org/10.3390/chemosensors2030193
Astakhova K. Toward Non-Enzymatic Ultrasensitive Identification of Single Nucleotide Polymorphisms by Optical Methods. Chemosensors. 2014; 2(3):193-206. https://doi.org/10.3390/chemosensors2030193
Chicago/Turabian StyleAstakhova, Kira. 2014. "Toward Non-Enzymatic Ultrasensitive Identification of Single Nucleotide Polymorphisms by Optical Methods" Chemosensors 2, no. 3: 193-206. https://doi.org/10.3390/chemosensors2030193
APA StyleAstakhova, K. (2014). Toward Non-Enzymatic Ultrasensitive Identification of Single Nucleotide Polymorphisms by Optical Methods. Chemosensors, 2(3), 193-206. https://doi.org/10.3390/chemosensors2030193



