Abstract
We present a generic quantitative chemical sensing methodology for assessing the concentration of a target material (TM) in an aqueous solution by using bioluminescent microbial bioreporters as the core sensing elements. Such bioreporters, genetically engineered to respond to the presence of a TM in their microenvironment by emitting bioluminescence, have previously been mostly designed to report the presence or absence of the TM in the sample. We extend this methodology to also assess the TM concentration, by exploiting the dose-dependency of the TM-induced luminescence. To overcome luminescence intensity variations due to bacterial batch differences and the ambient temperature, simultaneous measurements were carried out on sample solutions containing known concentrations of the TM. A “standard ratio” parameter, defined as the ratio between the two measurements, is shown to be independent of the bacterial batch and the temperature, and hence provides the conceptual basis for a generic quantitative chemical sensing methodology. Assessment of 2,4-dinitrotoluene (DNT) concentration in solutions is demonstrated with an accuracy of 2.5% over a wide dynamic range.
1. Introduction
We describe herein the underlying principles of a quantitative chemical sensing (QCS) methodology for assessing the concentration of a wide range of target materials (TMs) in aqueous solutions. The methodology is based on bioluminescent sensor bacteria (bioreporters) [1,2,3,4], genetically engineered to bioluminesce upon exposure to a TM. Bioreporters have been employed as the core sensing elements in sensors that detect the presence of a TM in a sample [5]. In a sensor apparatus that has previously implemented this sensing scheme, the bioreporters serve as minuscule self-contained biochemical laboratories integrated into an optoelectronic circuit [6,7]. By virtue of the fact that the sensor bacteria can be genetically engineered to respond to a wide range of TMs, this sensing methodology is inherently highly versatile. In this paper, we extend this methodology to quantify the concentration of the TM in the sample. To this end, we exploit the fact that the bioluminescent signal emitted by the bioreporters in response to their exposure to the TM is dose-dependent [8,9,10,11,12]. In addition, we describe and demonstrate schemes for avoiding inaccuracies due to signal fluctuations due to batch and temperature variations.
As classes of TMs of interest may be highly diverse, identification and quantification by state-of-the-art analytical chemistry methods need to be carried out in specially equipped central facilities, to which the tested samples are transported for analysis, often from remote locations [13]. In many scenarios, this modus operandi is ineffective and overly expensive. This brings forward the need for a generic general-purpose technology for a large-scale distribution of versatile and simple-to-operate chemical sensors, which can provide near-real-time information on TM distribution.
This challenge has been addressed by exploring sensor devices in which the core sensing elements are microbial bioreporters [6]. Bioreporters can be genetically engineered to respond to the presence of almost any TM in their microenvironment [8,14,15,16,17,18]. Example environments include health [19], agriculture [20], food industries [21] and environmental monitoring [22,23]. In most cases, the engineered bacteria harbor a plasmid-borne fusion between a molecular sensing element (a gene promoter) that detects the presence of the TM and a reporting element that emits a quantifiable signal such as bioluminescence or fluorescence [8]. As a result of this fusion, a dose-dependent response is generated upon exposure to the TM. Bioluminescent bioreporters provide a simple generic and versatile platform for constructing chemical sensors that operate in situ and recognize TMs in their microenvironment. However, sensors based on bacteria have hitherto been limited to detecting the presence of the TM, rather than yield its concentration in the inspected sample.
A modern bioreporter-based sensor is a two-unit module consisting of an analog sensing unit and a digital data processing and communication unit. The sensing unit contains the bioreporters, detects the light they emit, and produces an analog output signal. The digital unit is multifunctional: (i) it digitizes and processes the analog signals produced by the sensing unit and produces a digital record of the measurement, together with auxiliary relevant information (e.g., date and time); (ii) it manages the operation of the sensor; and (iii) it transmits the records produced by the sensor to remote receivers and receives operational instructions from a control center. Modern sensors are expected to operate either as standalone units or as nodes in sensor networks that are field deployable in large numbers, for both indoor and outdoor operation [7].
A quintessential example of a sensing unit that is based on bioreporters is the Bacterial Sensing Module (BSM) [7]. This module adopts the two-unit architecture described above and employs in its sensing unit bioreporters as the core sensing elements. The BSM is an autonomous self-contained unit designed for either standalone operation or as a node in a sensor network that monitors and maps the presence of TMs on the ground across wide areas in open environments. The module detects and processes the biological signal and transmits a digital record to a remote receiver. As a benchmark, the BSM has been configured to detect the presence of buried landmines underneath its footprint.
We present herein the underlying principles for extending the sensing methodology that employs bioluminescent bioreporters from the arena of presence detection to include quantitative assessment of the content of a selected TM in the inspected sample.
2. Materials and Methods
2.1. Conceptual Basis
The methodology for using bioluminescent bioreporters based QCS is built upon the well-established fact that the response of the bioreporters to the presence of the TM is dose-dependent [5]. However, signal strength should not only be dose-dependent, but also be repeatable and reproducible. Thus, for a QCS methodology to be reliable, it is necessary that a series of measurements of samples containing the same concentration of a TM, carried out by multiple employment of the same sensor, or by a set of identical sensors, will yield the same result. It was found, however, that the bioluminescence signal produced by bacterial sensors exposed to identical solutions was variable, depending upon sensor batch, batch age, and ambient temperature [24,25].
This hurdle was overcome by employing a “comparative analysis” scheme: comparing the output of a sensor exposed to the inspected solution to the output of a sensor exposed to a standard solution, containing a known concentration of the TM. Both measurements were carried out simultaneously by identical sensors operating at the same temperature, using bioreporters of the same batch. As will be shown henceforth, this principle of operation enabled reliable quantitative biochemical sensing. As far as we are aware, this has not been previously reported for sensing applications employing bacterial bioreporters.
2.2. Experimental Methodology
The platform for developing the bioreporter-based quantitative assessment was BSM prototypes [7], adapted for quantitative sensing measurements. The BSM is a small autonomous unit designed to report the presence of a target chemical underneath its footprint. It is constructed of three units: (i) a sensing unit that harbors a bioluminescent bacterial sensor strain that enhances its light emission in the presence of its target compounds underneath the module, along with an optoelectronic (OE) circuit that detects and processes the bioluminescent signal; (ii) a digital unit that digitizes the analog signals received from the sensing unit and produces a digital record ready for transmission; and (iii) a communication unit that transmits the information produced by the module to the outside world and receives control instructions from the network’s manager. The electronic circuit is insulated from water vapors released by the humid environment of the bacteria, allowing it to convey and process very weak analog signals.
In quantitative assessment, each module consisted of four independent measurement channels operating in parallel. Each channel was equipped with a separate replaceable bacterial cassette for either the sample or the standard solution. A schematic illustration of the BSM adapted for quantitative sensing is presented in Figure 1. The four channels in the original design of the BSM can be used as two quantitative sensing systems, each consisting of a sample channel and a standard channel. Alternatively, the BSM can also be configured to three sample channels and one standard channel.
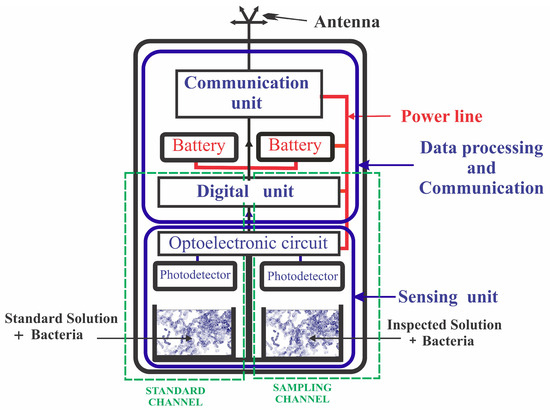
Figure 1.
Schematic illustration of the biosensing module (BSM) adapted for quantitative analysis.
The TM that served as the benchmark for testing the comparative analysis scheme was 2,4-dinitrotoluene (DNT) [8]. Traces of this compound emanate from 2,4,6-trinitrotoluene (TNT) in buried landmines and infiltrate the ground surface, serving as a chemical “signature” of the landmine [26,27]. To maintain identical bacterial preparations in the sample and standard solutions, a strict protocol for preparing the bacteria was carried out as follows: (i) a fresh bacterial culture of the species “tailored” to sense the selected TM was grown overnight in lysogeny broth (LB) at 37 °C, shaken at 200 rpm [28]; (ii) the culture was then diluted 100-fold in fresh LB and allowed to regrow under the same conditions to the mid-logarithmic growth phase (i.e., to an optical density at 600 nm (OD600) of 0.2) [29,30]; and (iii) 1 mL of the bacterial culture was loaded into the different cassettes and exposed to different concentrations of DNT. All four BSM channels were loaded with bacteria from the same batch.
As was pointed out above, the bioluminescence signal emitted by the bioreporters is proportional to the concentration of the TM in the inspected solution to which the bioreporters are exposed. This is demonstrated in the results section below.
However, as pointed out above, the strength of the bioluminescence signal emitted by the bioreporters in response to their exposure to the TM varies between measurements employing bacteria from different batches or under different environmental conditions. The raw bioluminescence signal, though dose-dependent, cannot therefore be used for a direct assessment of the TM concentration.
2.3. Details of the Quantitative Chemical Sensing Methodology
For performing QCS based on genetically engineered bioluminescent bioreporters, it is imperative that the results are reproducible, irrespective of either the bacterial batch used or the ambient environmental conditions. Thus, it is required to define a measurable quantity that can be used for performing comparison analysis of the target material in the sample, independent of the bacteria batch and the operating temperature. To this end, the measurement of the bioluminescence signal produced by exposing the sensor to the inspected sample was augmented by a simultaneous measurement of the bioluminescence signal produced by exposing bioreporters from the same batch to a solution containing a known concentration of the TM (standard solution). Both measurements were carried out in parallel under the same conditions, using adjacent sensor channels in the same sensor module or using sensor channels in different but identical sensor modules. The measurable quantity employed to normalize the measured luminescence signal was the “standard ratio” (SR), defined as the ratio between the maximal output signals produced by the sample channel and the standard channel. The block diagram for implementing the scheme for obtaining the SR is presented schematically in Figure 2.
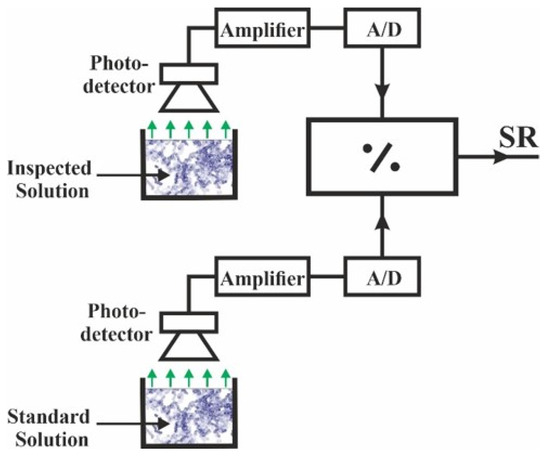
Figure 2.
Block diagram for implementing the scheme for obtaining the standard ratio.
2.4. Reagents
The following chemicals and reagents were used: 2,4-dinitrotoluene (DNT) (Sigma Aldrich, St. Louis, MO, USA); nalidixic acid sodium salt (Sigma Aldrich, St. Louis, MO, USA). The bacterial strains employed in the course of this study and their sources are listed in Section 2.5 below.
2.5. Bacterial Strains and Plasmids
Three Escherichia coli bioreporter strains were used in the course of this study (Table 1), in all of which the reporter entity was the luxCDABE gene cassette of Photorhabdus luminescens (Pl):
- DNT bioreporter strain azoR, which contains two plasmids: The first is pBR-P2G2-Pl, in which the sensing element is an enhanced version of the E. coli azoR gene promoter, fused to the Pl reporter [31,32]. The second plasmid, pACYC-yhaJ-G2, contains an enhanced copy of the yhaJ transcriptional activator [33].
- DNT bioreporter strain yqjF, also harboring two plasmids: pBR-C55-Pl, in which the sensing element is an enhanced version of the yqjF gene promoter (C55), fused to the Pl reporter [33], and plasmid pACYC-yhaJ-G2, as above.
- Genotoxicity bioreporter strain recA [34], harboring plasmid pBR-recA-Pl, with the DNA damage-inducible recA gene promoter as the sensing element.

Table 1.
Bacterial bioreporter strains and plasmids used in this study.
Table 1.
Bacterial bioreporter strains and plasmids used in this study.
| Bioreporter Designation | Target Material(s) | Plasmid 1 | Plasmid 2 | Host Bacterium |
|---|---|---|---|---|
| azoR | DNT | pBR-P2G2-Pl [33] | pACYC-yhaJ(G2) [33] | E. coli BW25113 ΔpykF [35] |
| yqjF | DNT | pBR-C5-Pl [33] | pACYC-yhaJ(G2) [33] | E. coli BW25113 ΔpykF [35] |
| recA | Genotoxic agents | pBR-recA-Pl [34] | - | E. coli RFM443 [34] |
3. Experimental Results
A set of DNT measurements employing the protocol described above is presented in Figure 3. The protocol was employed in identical sensor channels loaded with DNT bioreporters of the same batch. The sensors were exposed to 1 mL samples containing different concentrations of DNT mixed with the bioreporters. The results presented in Figure 3 demonstrate the fact that the bioluminescence produced by the bioreporters is dose-dependent, substantiating the underlying hypothesis at the basis of employing bioluminescent bioreporters for performing quantitative chemical sensing.
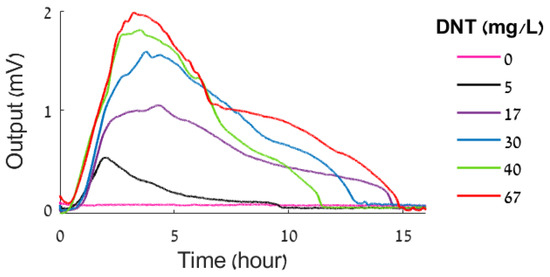
Figure 3.
Luminescence as a function of DNT concentration.
As pointed out above, the bioluminescence signal emitted by the bioreporters in response to their exposure to the TM varies between measurements employing bacteria from different batches or under different environmental conditions. This is demonstrated in Figure 4 (blue symbols), in which measurements of the maximal response of the azoR bioreporter, exposed to DNT (30 mg/L), are presented. Marked variations were observed in sensor outputs produced from identical samples by bioreporters from either five different batches (Figure 4a) or a single batch at different temperatures (Figure 4b). The SR values for these measurements were obtained by augmenting these measurements with measurements of standard solutions containing 17 mg/L DNT. Each of these measurements was carried out in parallel to the original (30 mg/L) measurement, using bacteria from the same batch at the same operating temperature. The results are presented in Figure 4 (red symbols). As can be seen, the SR is indeed independent of both the bioreporter batch (Figure 4a) and the operating temperature (Figure 4b).
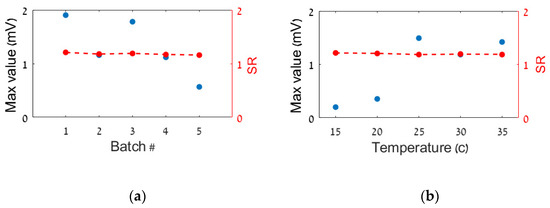
Figure 4.
The standard ratio (SR) computed for two series of measurements: (a) different batches of bioreporters; (b) different temperatures. In blue are the maximal outputs of each BSM channel exposed to 30 mg/L of DNT and in red are the computed values of the standard ratio.
Employing the SR concept for performing quantitative assessment of a given TM in the inspected sample is achieved by composing an empirical calibration curve which depicts the SR vs. the TM concentration. In all measurements, the sensor is loaded with an equal quantity of bioreporters from the same batch, and all the measurements are carried out at the same temperature. One measurement for which the concentration of the TM is approximately at the center of the concentration range is designated as the standard measurement. Following the procedure carried out in the setup presented in Figure 4, the SR vs. the concentration is evaluated for each measurement, and the SR values are then depicted on the calibration curve.
Assessment of the TM concentration is performed by the following steps: (i) load the inspected sample and the standard sample into their containers in the sensor together with the bioreporter solution; (ii) monitor the evolution of the emitted bioluminescence with time; (iii) extract the maximum point of the signal; (iv) calculate the SR by computing the ratio between the measured maxima from step iii and the maximum of the standard solution; (v) pin the SR to the respective ordinate of the calibration curve; and (vi) retrieve the concentration of the TM from the calibration curve.
3.1. Employing the Calibration Curve
An example of two DNT calibration curves for the yqjF and azoR bioreporters are presented in Figure 5,. The calibration curves were obtained from a set of measurements performed with a sensor of the type presented in Figure 1, following the procedure described in Section 2.3. For each measurement, a mixture of 1 mL LB containing DNT at the required concentration plus the bioreporters was loaded into the sensor container. For each DNT concentration, at least four different measurements were conducted. The variance between the SR of similar measurements did not exceed 2.5%. The DNT concentration in the standard solution was chosen to be in the approximate middle of the dynamic range for which the bacterial strain was found to be effective.
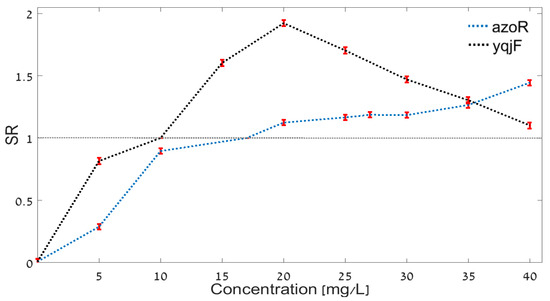
Figure 5.
Calibration curves for two strains of bioreporters: yqjF (black); azoR (blue). The mean values of the SR and their standard deviation (denoted by the error bars) are presented (STD = 0.025). The gray dashed line denotes the standard solution (for which SR = 1).
It can be seen in Figure 5 that employing the calibration curve for assessing the concentration of the DNT in the inspected solution is not necessarily straightforward. Consider first the calibration curve for the yqjF strain (in black). Due to the fact that the calibration curve increases monotonically for DNT concentrations below 20 mg/L, and decreases for higher concentrations, there is no one-to-one correspondence between the measured SR and the concentration. For example, consider a sample for which it was found that SR = 1.25. The calibration curve indicates that the concentration can be either 12 mg/L or 36.6 mg/L. Consider also the calibration curve of the azoR strain (in blue). Note that here, in the concentration range between 20 mg/L and 30 mg/L, the calibration curve becomes almost horizontal, and hence the concentration of the TM in the inspected solution retrieved by pinning the SR on the calibration curve is bound to be inaccurate. For example, if the SR is found to be between 1.1 and 1.125, the azoR calibration curve indicates that the concentration is between 20 mg/L and 30 mg/L. Both problems can be overcome by considering the two calibration curves as complementary to each other. To this end, both curves are depicted together in Figure 5. A preliminary result can be obtained from the azoR curve, whereas fine tuning of this result can be obtained from the yqjF curve. For concentrations between 25 and 30 mg/L, the response of the yqjF bacteria varies significantly more than the azoR bioreporter’s response. Thus, for this concentration range the calibration curve of yqjF bacteria was calculated, so that for concentrations of 25–30 mg/L a separation could be obtained based on the SR value.
Note that in principle it is possible to genitically engineer several different bacteria strains for every target material. Moreover, different bacteria strains that are genetically engineered for the same target material can be optimal for different segments of the concentration dynamic range. This can be exploited as a general principle for improving the accuracy of the bacterial quantitative assessment methodology
3.2. Quantitative Assessment in a Mixture of Different Target Materials
In real environemntal samples, such as in drinking water [36], it is highly likely the sample will contain more than one TM. The question then arises of whether the methodology presented here can be used for evaluating the concentration of a TM in a sample containing a mixture of different materials. This question was investigated by evaluating the SR in a solution containing a mixture of two TMs, DNT (30 mg/L) and nalidixic acid (NA, 10 mg/L) [37]. The SR values for both compounds were evaluated using the azoR and the recA bioreporters, respectively. The results, presented in Figure 6, show that each of the bioreporters responded only to its deignated TM, either singly or in a mixture, but not to the other chemical.
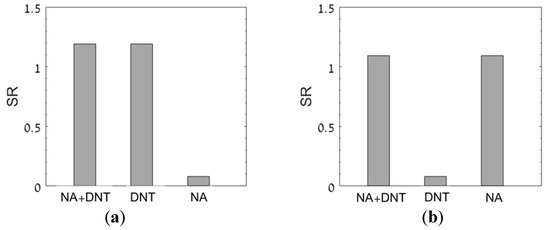
Figure 6.
SR values measured by bioreprter strains azoR (a) and recA (b) for solutions containing either DNT (30 mg/L), NA (10 mg/L), or both. The standard solutions contained either 17 mg/L DNT or 5 mg/L NA.
The exact DNT concentration was retrieved from the azoR calibration curve (Figure 5) and the exact NA concentration was retrieved from a similar calibration curve constructed for the recA strain prior to the measurement.
4. Summary and Conclusions
We present in this paper a generic quantitative chemical sensing methodology for assessment of TM concentrations by employing bacterial chemical sensing. The methodology is based on bioluminescent bioreporters that serve as the core sensing elements of the sensor apparatus. Hitherto, bacterial chemical sensing has been employed to detect the presence of the TM in the inspected sample. Herein, we present and provide proof of concept of a methodology for performing a quantitative assessment of the TM concentration. The methodology exploits the fact that the strength of the bioluminescence signal emitted by the bioreporters is dose-dependent. However, this signal varies between different bioreporter batches and is also temperature-dependent. We overcame this problem by introducing the SR parameter: the ratio between the maximal bioluminescence signals emitted by the sample and a standard solution, respectively. The SR was found to be independent of both the bioreporter batch and the operating temperature, and thus enabled quantitative assessment of the TM.
As pointed out above, in bacterial chemical sensing systems the fundamental sensing operation is performed by bioreporters which are bacteria that are installed in an optoelectronic (OE) circuit and function as self-contained biochemical laboratories that are genetically engineered to respond to their exposure to a selected TM by emitting light. The complex sensing operation is performed inside the bioreporter cell and, in principle, almost any chemical substance can be sensed by a bacterial chemical sensing system provided it is loaded with bioreporters that were genetically engineered to respond to this substance.
From a system perspective, the sensing unit of the bacterial chemical sensing system is an optoelectronic circuit in which the bioreporters function as the light sources. The light signal produced by the bioreporters upon their exposure to a sample that contains the selected TM is detected by the photodetectors in the OE circuit, which converts it to an analog electrical output signal. As such, the sensing unit of the bacterial chemical sensing system is a simple OE circuit that can sense the presence of any TM, provided it was loaded with bioreporters engineered to respond to that TM. Hence, performing a quantitative assessment of the TM concentration in the inspected solution necessitatesthe inclusion of an additional sensing channel in the system that measures the signal emitted by a solution that contains the “standard solution”, and then compute the SR, and extract the concentration of the TM in the inspected solution from the calibration curve.
Thus, the underlying architecture of a bacterial sensing system that assesses the concentration of a selected TM is independent of the specific TM it was designed to inspect. It is adapted to its application by loading it with bioreporters that were genetically engineered to respond to the relevant TM (or a group of relevant TMs). Such systems can be constructed in different configurations so that they fit a variety of modes of operation that due to their simplicity can be deployed in large numbers in different environments, outdoors or indoors. As stationary systems, they can be deployed and operate in large numbers at the location where the inspected samples are retrieved, and transmit their findings across the communication network to a distant monitoring center that also controls their operation. The methodology can also be implemented in mobile systems (e.g., the BSMs that were configured to perform quantitative assessment of the selected TM in the inspected solution that were described above in Section 2.2 and Figure 3). An ensemble of such modules can be employed for mapping on location in real time the spatial distribution of the concentration of the TMs across the inspected area by processing a set of sample solutions that were retrieved on location.
In summary, due to its simplicity and versatility, the bacterial quantitative sensing methodology described in this paper has the potential to form the basis for quantitative chemical analysis technology that can be employed on location. As such, it can be the basis for a wide range of applications in which it is required to assess the concentration of TMs in solutions without the need to export the inspected solution to a remote central analysis laboratory. Such applications span across a wide spectrum of fields, including in medical care (in particular for remote medical clinics in the Global South), in precision agriculture for monitoring the fertilizers and pesticides in the soil, in chemical industry plants where it is required to perform chemical analysis in different locations of the production line, and in environmental monitoring of hazardous materials and more. This research focused on assessing the concentrations of TMs in solutions, but the versatility and strong performance of the bioluminescent bioreporters encourages the employment of bacterial quantitative assessment in a variety of additional settings. This includes scenarios such as assessing the concentration of the target material in the soil underneath the footprint of the bacterial sensor (e.g., the BSM).
Author Contributions
Conceptualization, A.J.A. and S.B.; methodology, A.J.A., Y.U. and Y.K.; software, Y.U.; validation, Y.U. and Y.K.; formal analysis, Y.U. and Y.K.; investigation, Y.U.; resources, B.S. and E.S.; data curation, Y.U.; writing—original draft preparation, A.J.A., Y.U. and Y.K.; writing—review and editing, S.B., B.S., E.S. and Y.K; supervision, A.J.A. and S.B.; project administration, Y.K.; funding acquisition, A.J.A. and S.B. All authors have read and agreed to the published version of the manuscript.
Funding
Internal Hebrew University Funding. Work in the Belkin laboratory was also partially supported by the Minerva Center for Bio-hybrid complex systems.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Acknowledgments
This research was supported by the Attract program of the European Union within the framework of the SENSEI project.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hastings, J.W. [13] Bacterial Bioluminescence: An Overview. In Methods in Enzymology; Bioluminescence and Chemiluminescence; Academic Press: New York, NY, USA, 1978; Volume 57, pp. 125–135. [Google Scholar]
- Meighen, E.A. Molecular Biology of Bacterial Bioluminescence. Microbiol. Rev. 1991, 55, 123–142. [Google Scholar] [CrossRef] [PubMed]
- Nwankwo, C.E.; Chigor, V.N.; Akani, N.P. Bacterial Bioluminescent Biosensors: Principle and Applications. AIBM 2019, 15, 156–159. [Google Scholar] [CrossRef]
- Su, L.; Jia, W.; Hou, C.; Lei, Y. Microbial Biosensors: A Review. Biosens. Bioelectron. 2011, 26, 1788–1799. [Google Scholar] [CrossRef]
- van der Meer, J.R.; Belkin, S. Where Microbiology Meets Microengineering: Design and Applications of Reporter Bacteria. Nat. Rev. Microbiol. 2010, 8, 511–522. [Google Scholar] [CrossRef]
- Kabessa, Y.; Korouma, V.; Ilan, H.; Yagur-Kroll, S.; Belkin, S.; Agranat, A.J. Simultaneous Quantification of the Fluorescent Responses of an Ensemble of Bacterial Sensors. Biosens. Bioelectron. 2013, 49, 394–398. [Google Scholar] [CrossRef]
- Agranat, A.J.; Kabessa, Y.; Shemer, B.; Shpigel, E.; Schwartsglass, O.; Atamneh, L.; Uziel, Y.; Ejzenberg, M.; Mizrachi, Y.; Garcia, Y.; et al. An Autonomous Bioluminescent Bacterial Biosensor Module for Outdoor Sensor Networks, and Its Application for the Detection of Buried Explosives. Biosens. Bioelectron. 2021, 185, 113253. [Google Scholar] [CrossRef]
- Yagur-Kroll, S.; Lalush, C.; Rosen, R.; Bachar, N.; Moskovitz, Y.; Belkin, S. Escherichia coli Bioreporters for the Detection of 2,4-Dinitrotoluene and 2,4,6-Trinitrotoluene. Appl. Microbiol. Biotechnol. 2014, 98, 885–895. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhang, X.; Tefsen, B.; Wells, M. From Speciation to Toxicity: Using a “Two-in-One” Whole-Cell Bioreporter Approach to Assess Harmful Effects of Cd and Pb. Water Res. 2022, 217, 118384. [Google Scholar] [CrossRef]
- Zhang, X.; Qin, B.; Deng, J.; Wells, M. Whole-Cell Bioreporters and Risk Assessment of Environmental Pollution: A Proof-of-Concept Study Using Lead. Environ. Pollut. 2017, 229, 902–910. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, Y.; Wells, M. A Case Study Comparing Lead-Response in Different Bioreporters: What Constitutes a Good Biosensor? Case Stud. Chem. Environ. Eng. 2022, 5, 100192. [Google Scholar] [CrossRef]
- Mabekou, S.S.; Lee, S.C.; Dinh, T.H.; Won, K.; Mitchell, R.J. Enhanced Sensitivity and Responses to Viologens from a Whole-cell Bacterial Bioreporter Treated with Branched Polyethyleneimines. J. Appl. Microbiol. 2017, 123, 1478–1487. [Google Scholar] [CrossRef] [PubMed]
- Cooks, R.G.; Busch, K.L.; Glish, G.L. Mass Spectrometry: Analytical Capabilities and Potentials. Science 1983, 222, 273–291. [Google Scholar] [CrossRef] [PubMed]
- Applegate, B.M.; Kehrmeyer, S.R.; Sayler, G.S. A Chromosomally Based Tod-luxCDABE Whole-Cell Reporter for Benzene, Toluene, Ethybenzene, and Xylene (BTEX) Sensing. Appl. Env. Microbiol. 1998, 64, 2730–2735. [Google Scholar] [CrossRef] [PubMed]
- Trang, P.T.K.; Berg, M.; Viet, P.H.; Van Mui, N.; Van Der Meer, J.R. Bacterial Bioassay for Rapid and Accurate Analysis of Arsenic in Highly Variable Groundwater Samples. Env. Sci. Technol. 2005, 39, 7625–7630. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Youn, C.H.; Kim, B.C.; Gu, M.B. An Oxidative Stress-Specific Bacterial Cell Array Chip for Toxicity Analysis. Biosens. Bioelectron. 2007, 22, 2223–2229. [Google Scholar] [CrossRef]
- Melamed, S.; Lalush, C.; Elad, T.; Yagur-Kroll, S.; Belkin, S.; Pedahzur, R. A Bacterial Reporter Panel for the Detection and Classification of Antibiotic Substances. Microb. Biotechnol. 2012, 5, 536–548. [Google Scholar] [CrossRef]
- Checa, S.K.; Zurbriggen, M.D.; Soncini, F.C. Bacterial Signaling Systems as Platforms for Rational Design of New Generations of Biosensors. Curr. Opin. Biotechnol. 2012, 23, 766–772. [Google Scholar] [CrossRef]
- Thacharodi, A.; Chinnadurai, J.; Thacharodi, D. Biomonitoring of Heavy Metal Pollution by Bioluminescent Bacterial Biosensors. Indian. J. Sci. Technol. 2019, 12, 9. [Google Scholar] [CrossRef]
- Pini, F.; East, A.K.; Appia-Ayme, C.; Tomek, J.; Karunakaran, R.; Mendoza-Suárez, M.; Edwards, A.; Terpolilli, J.J.; Roworth, J.; Downie, J.A.; et al. Bacterial Biosensors for in Vivo Spatiotemporal Mapping of Root Secretion. Plant Physiol. 2017, 174, 1289–1306. [Google Scholar] [CrossRef]
- Luo, J.; Liu, X.; Tian, Q.; Yue, W.; Zeng, J.; Chen, G.; Cai, X. Disposable Bioluminescence-Based Biosensor for Detection of Bacterial Count in Food. Anal. Biochem. 2009, 394, 1–6. [Google Scholar] [CrossRef]
- Tecon, R.; Van der Meer, J.R. Bacterial Biosensors for Measuring Availability of Environmental Pollutants. Sensors 2008, 8, 4062–4080. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, A.; Malachowsky, K.; Thonnard, J.E.; Bienkowski, P.R.; White, D.C.; Sayler, G.S. Optical Biosensor for Environmental On-Line Monitoring of Naphthalene and Salicylate Bioavailability with an Immobilized Bioluminescent Catabolic Reporter Bacterium. Appl. Env. Microbiol. 1994, 60, 1487–1494. [Google Scholar] [CrossRef] [PubMed]
- Dewhurst, R.E.; Wheeler, J.R.; Chummun, K.S.; Mather, J.D.; Callaghan, A.; Crane, M. The Comparison of Rapid Bioassays for the Assessment of Urban Groundwater Quality. Chemosphere 2002, 47, 547–554. [Google Scholar] [CrossRef]
- Halmi, M.I.E.; Jirangon, H.; Johari, W.L.W.; Abdul Rachman, A.R.; Shukor, M.Y.; Syed, M.A. Comparison of Microtox and Xenoassay Light as a Near Real Time River Monitoring Assay for Heavy Metals. Sci. World J. 2014, 2014, e834202. [Google Scholar] [CrossRef]
- Dubey, A.C.; Barnard, R.L.; Lowe, C.J.; McFee, J.E. Detection and Remediation Technologies for Mines and Minelike Targets. Detect. Remediat. Technol. Mines Minelike Targets 1996, 2765, 385–396. [Google Scholar]
- Sylvia, J.M.; Janni, J.A.; Klein, J.D.; Spencer, K.M. Surface-Enhanced Raman Detection of 2,4-Dinitrotoluene Impurity Vapor as a Marker To Locate Landmines. Anal. Chem. 2000, 72, 5834–5840. [Google Scholar] [CrossRef]
- Shahzadi, I.; Al-Ghamdi, M.A.; Nadeem, M.S.; Sajjad, M.; Ali, A.; Khan, J.A.; Kazmi, I. Scale-up Fermentation of Escherichia coli for the Production of Recombinant Endoglucanase from Clostridium thermocellum. Sci. Rep. 2021, 11, 7145. [Google Scholar] [CrossRef]
- Stevenson, K.; McVey, A.F.; Clark, I.B.N.; Swain, P.S.; Pilizota, T. General Calibration of Microbial Growth in Microplate Readers. Sci. Rep. 2016, 6, 38828. [Google Scholar] [CrossRef] [PubMed]
- Sezonov, G.; Joseleau-Petit, D.; D’Ari, R. Escherichia coli Physiology in Luria-Bertani Broth. J. Bacteriol. 2007, 189, 8746–8749. [Google Scholar] [CrossRef]
- Henshke, Y.; Shemer, B.; Belkin, S. The Escherichia coli azoR Gene Promoter: A New Sensing Element for Microbial Biodetection of Trace Explosives. Curr. Res. Biotechnol. 2021, 3, 21–28. [Google Scholar] [CrossRef]
- Shpigel, E.; Shemer, B.; Elad, T.; Glozman, A.; Belkin, S. Bacterial Bioreporters for the Detection of Trace Explosives: Performance Enhancement by DNA Shuffling and Random Mutagenesis. Appl. Microbiol. Biotechnol. 2021, 105, 4329–4337. [Google Scholar] [CrossRef] [PubMed]
- Shemer, B.; Shpigel, E.; Glozman, A.; Yagur-Kroll, S.; Kabessa, Y.; Agranat, A.J.; Belkin, S. Genome-Wide Gene-Deletion Screening Identifies Mutations That Significantly Enhance Explosives Vapor Detection by a Microbial Sensor. New Biotechnol. 2020, 59, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Yagur-Kroll, S.; Bilic, B.; Belkin, S. Strategies for Enhancing Bioluminescent Bacterial Sensor Performance by Promoter Region Manipulation. Microb. Biotechnol. 2010, 3, 300–310. [Google Scholar] [CrossRef]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-Frame, Single-Gene Knockout Mutants: The Keio Collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef]
- US EPA, O. Online Water Quality Monitoring Resources. Available online: https://www.epa.gov/waterqualitysurveillance/online-water-quality-monitoring-resources (accessed on 2 January 2022).
- Biran, A.; Ben Yoav, H.; Yagur-Kroll, S.; Pedahzur, R.; Buchinger, S.; Shacham-Diamand, Y.; Reifferscheid, G.; Belkin, S. Microbial Genotoxicity Bioreporters Based on sulA Activation. Anal. Bioanal. Chem. 2011, 400, 3013–3024. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).