Recent Advances in Functionalization Strategies for Biosensor Interfaces, Especially the Emerging Electro-Click: A Review
Abstract
:1. Introduction
2. Functionalization Technologies of Biosensor Interfaces
2.1. Nanostructured Biosensor Interface
2.1.1. Carbon Nanomaterials
Graphene Nanomaterials
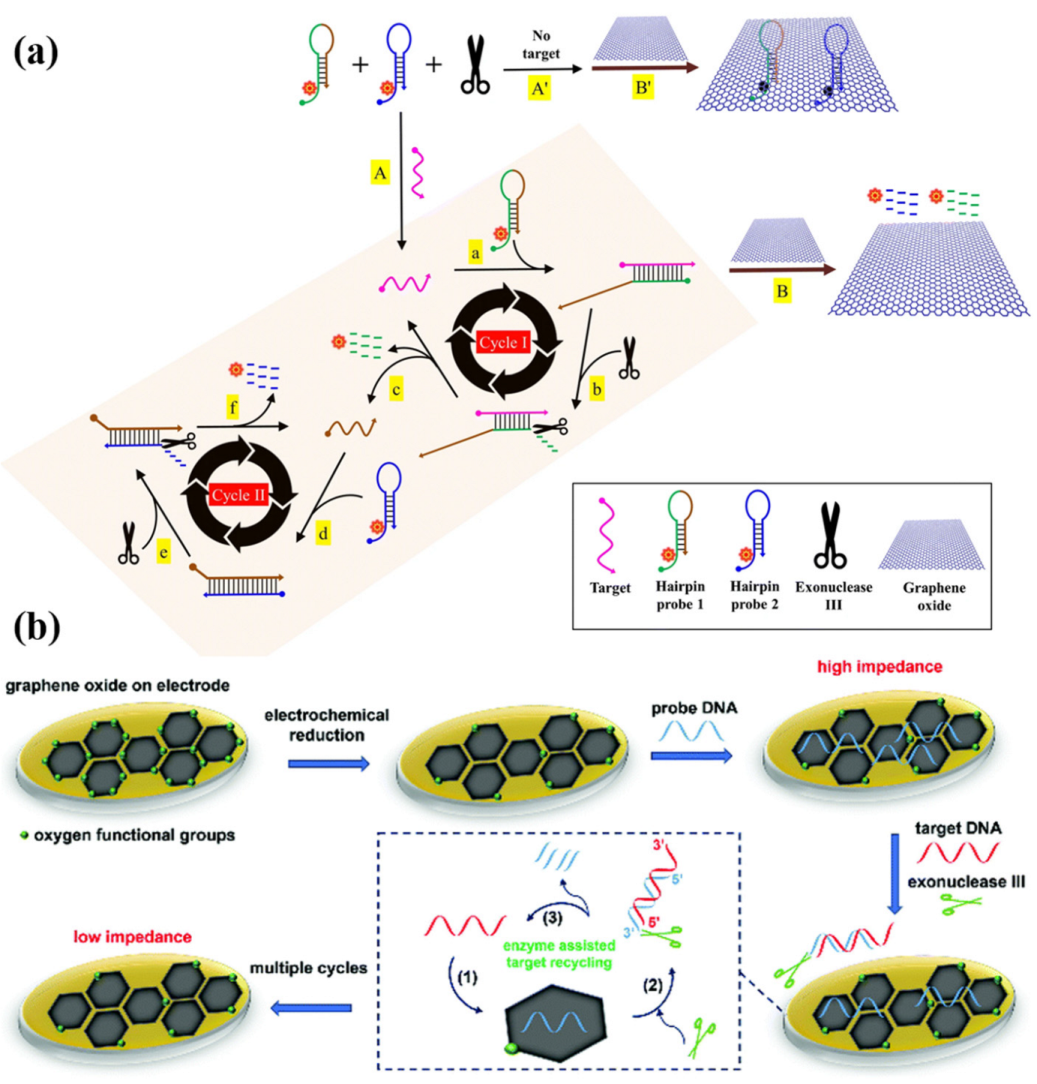
Porous Carbon
Carbon Nanotubes
2.1.2. Polymer and Bio-Nanomaterials
2.1.3. Other Nanomaterials
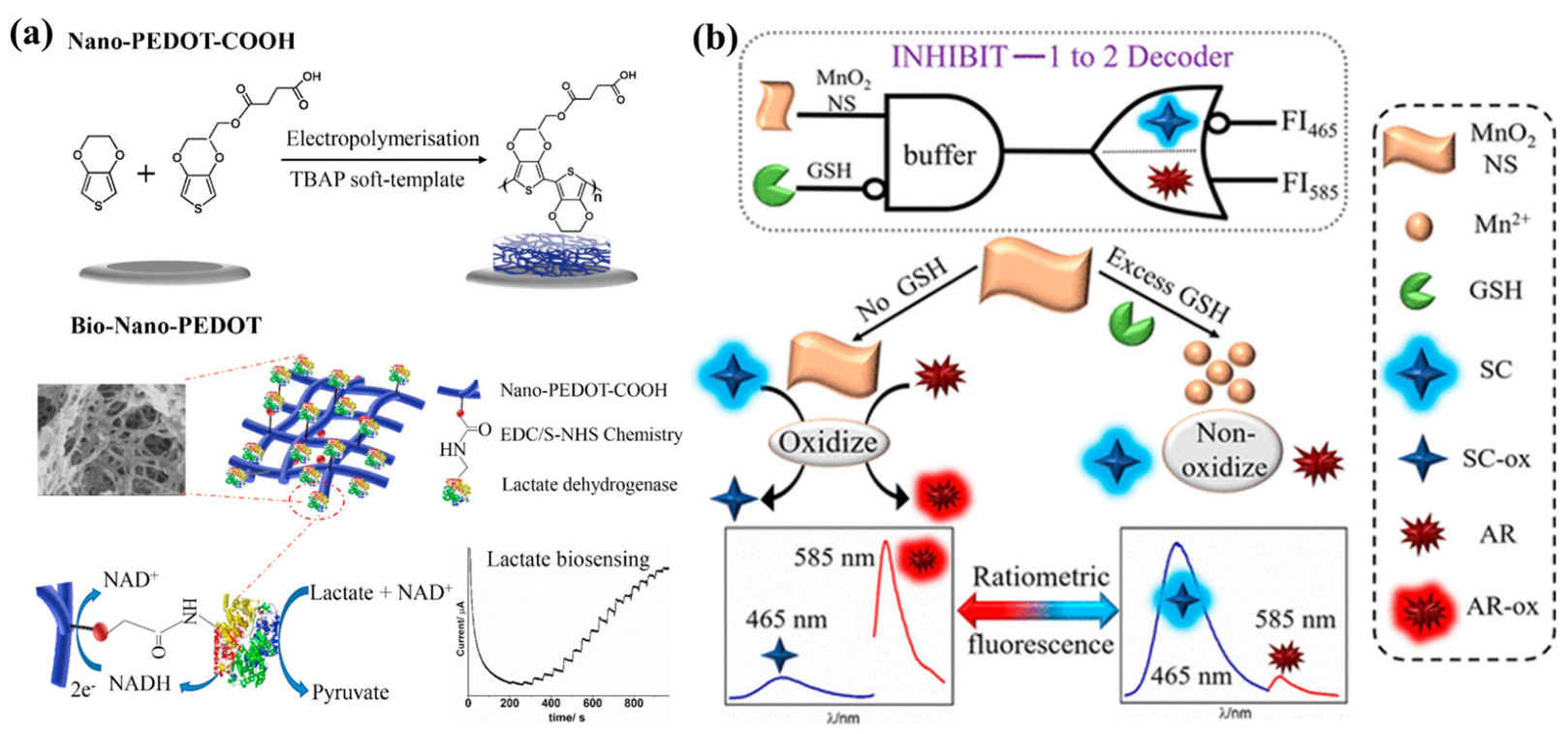
2.2. Small-Molecule-Mediated Interfacial Regulation
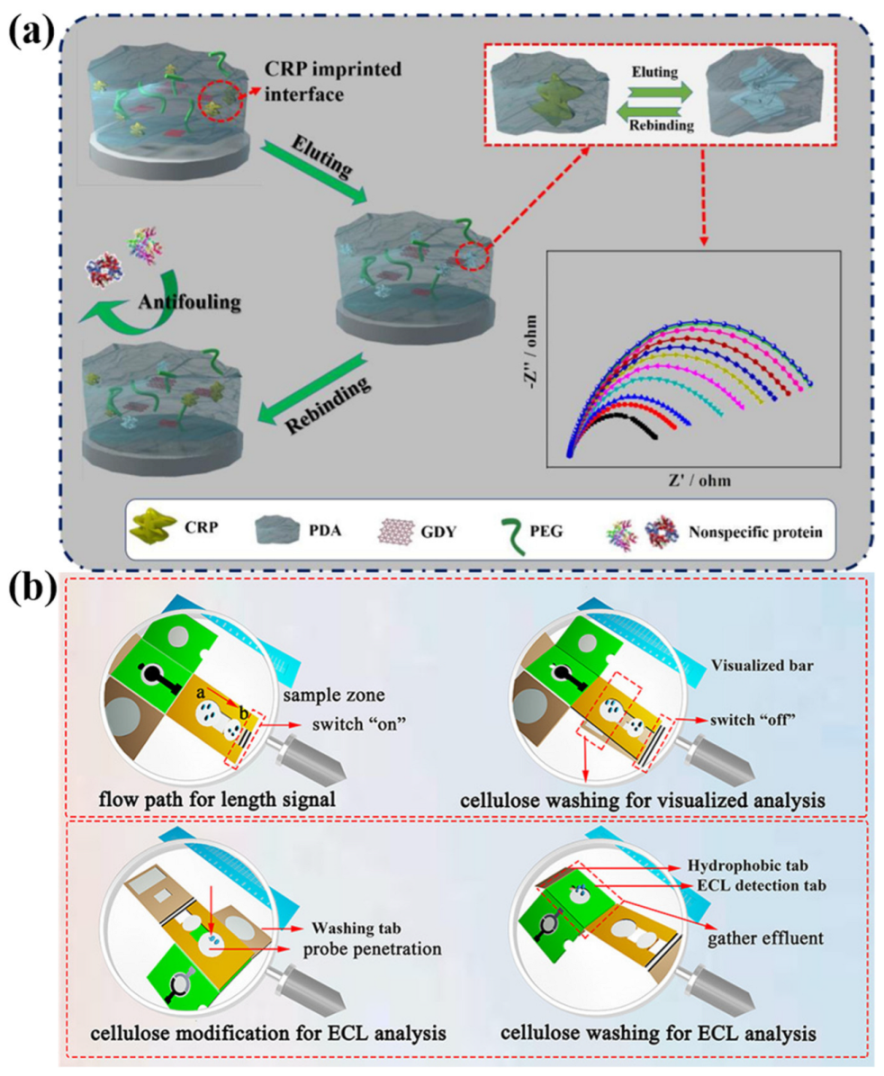
2.3. Biomacromolecule-Mediated Interfacial Regulation
2.3.1. Enzyme-Based Interfaces
2.3.2. DNA-Based Interfaces
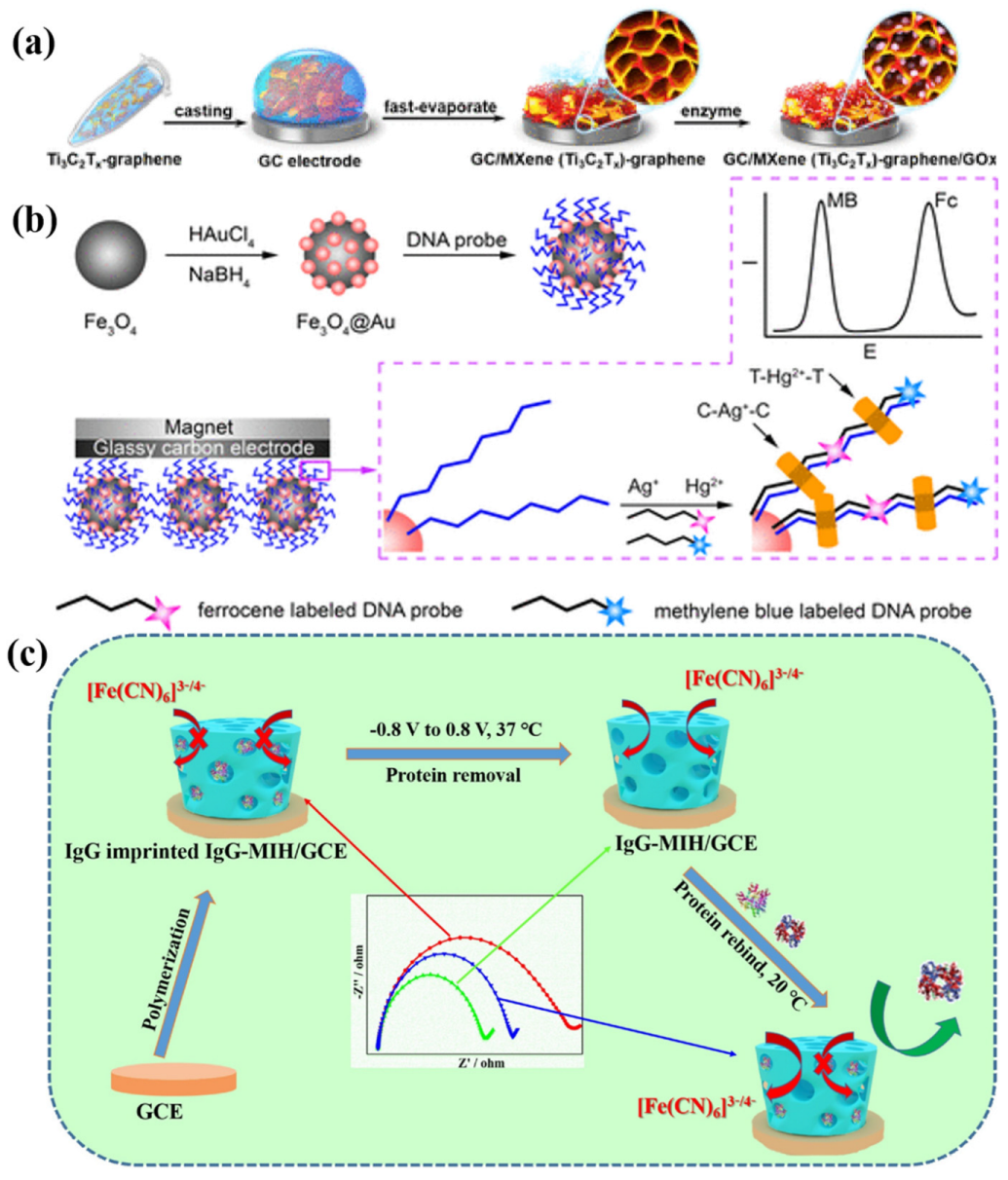
2.3.3. Protein-Based Interfaces
2.4. Cell-Based Interfaces
3. Electro-Click Chemistry for the Functionalization of Biosensor Interfaces
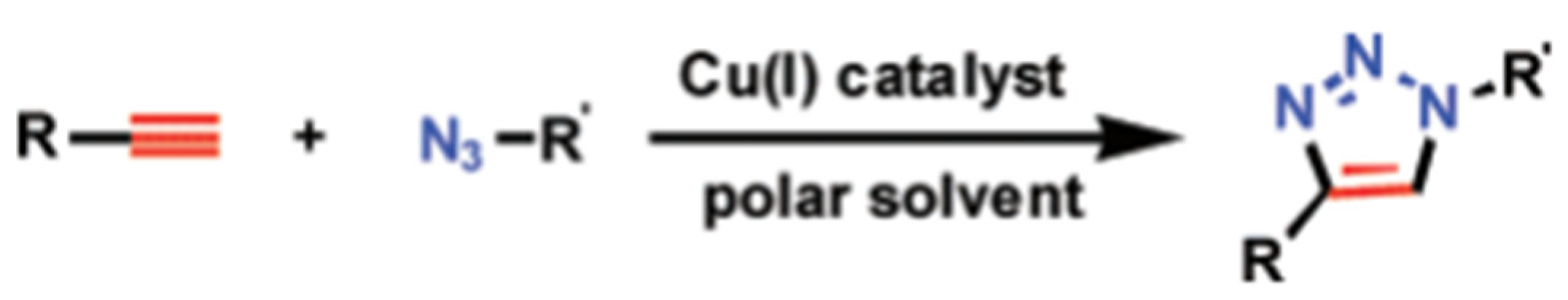
3.1. Traditional Click Technology
3.2. Electro-Click Technology
3.3. Electro-Click Strategies for the Functionalization of Biosensor Interfaces
3.3.1. Electro-Click Strategies for the Functionalization of Electrochemical Biosensor Interfaces
Electrografting
Electropolymerization
Electrodeposition
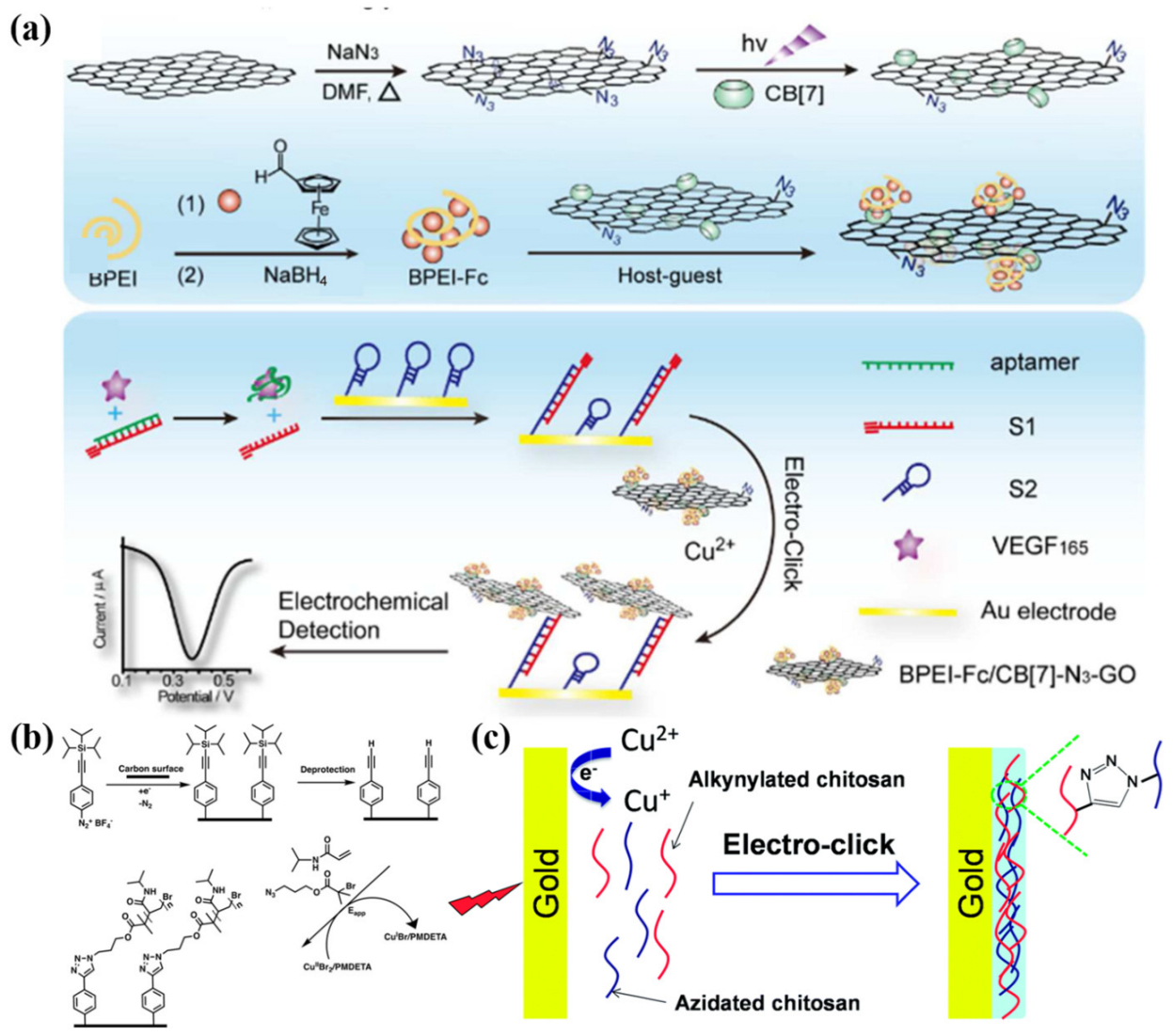
Bipolar Electrodes
Applications of Electro-Click Strategies in Electrochemical Biosensor Functionalization with Submicron Resolution
3.3.2. Electro-Click Strategies for the Functionalization of Optical Biosensor Interfaces
3.3.3. Limits of Electro-Click Strategies for the Functionalization of Biosensor Interfaces
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Acronym | Definition | Acronym | Definition |
| GO | Graphene oxide | ATPA | Adenosine triphosphate aptamer |
| Exo III | Exonuclease III | ATP | Adenosine triphosphate |
| HP1 and HP2 | Hairpin probes | DsDNA | Double-stranded DNA |
| PtNFS-GO | Flower-like Pt-graphene oxide | HDNA | Hairpin DNA |
| 8-OHdG | 8-hydroxy-2′-deoxyguanosine | SsDNA | Single-stranded DNA |
| rGO | Reduced graphene oxide | NPs | Nanoparticles |
| LOD | Low detection limit | RBD | Receptor-binding domain |
| Fe/N-C | N-doped porous-carbon-containing Fe | HSA | Human serum albumin |
| ECL | Electrochemiluminescence | SRB | Sulfate-reducing bacteria |
| SWCNTs | Single-walled carbon nanotubes | RBC | Red blood cell |
| MWCNTs | Multi-walled carbon nanotubes | PLNPs | Persistent luminescent nanophosphors |
| NIR | Near-infrared | MCF | Mesoporous carbon framework |
| PDAC | Pancreatic ductal adenocarcinoma | HEMA | Hydroxyethyl methacrylate |
| EDOT | 3,4-ethylenedioxythiophene | DEAEMA | 2-(diethylamino)ethyl methacrylate |
| PEDOT | Poly(3,4-ethylenedioxythiophene) | SPE | Gold screen-printed electrodes |
| Au@Pt | Au@Pt nanocrystals | AFB1 | Aflatoxin B1 |
| FNAs | Framework nucleic acids | OTA | Ochratoxin A |
| E. coli O157:H7 | Escherichia coli O157:H7 | NP | Nucleocapsid protein |
| AgNCs | Silver nanoclusters | IgG | Immunoglobulin G |
| SC | Scopoletin | CuAAC | Cu(I)-catalyzed alkyne–azide cycloaddition |
| AR | Amplex Red | CXCL7 | Chemokine (C-X-C motif) ligand 7 |
| GSH | Glutathione | GO-N3 | Azide-co-functionalized graphene oxide |
| PSA | Prostate-specific antigen | CDs | Carbon dots |
| PCa | Prostate cancer | CDs-DNA | Carbon-dot-labeled DNA |
| GDY | Graphdiyne | FRET | Fluorescence resonance energy transfer |
| C-MIPs | C-reactive-molecular-imprinted polymers | CEA | Carcinoembryonic antigen |
| HRP | Horseradish peroxidase | MspA | Mycobacterium smegmatis porin A |
| ROS | Reactive oxygen species | DBCO | Dibenzocyclooctyne |
| O2− | Superoxide radicals | PAA | Poly(acrylic acid) |
| Au/VO2 | Gold-loaded 2D VO2 nanobelts | PAAalk | Poly(acrylic acid) multilayers with alkynyl |
| RET | Resonance energy transfer | PAAaz | Poly(acrylic acid) multilayers with azide |
| MIT | Molecular-imprinting technology | BDD | Boron-doped diamond |
| HER-2 | Human epidermal growth factor receptor 2 | EP | Electropolymerization |
| HOCl | Hypochlorous acid | CV | Cyclic voltammetry |
| OONO− | Peroxynitrite | SI-eATRP | Surface-initiated atom-transfer radical polymerization |
| GOx | Glucose oxidase | PNIPAM | Poly(N-isopropylacrylamide) |
| MG | Mxene–graphene | BPEs | Bipolar electrodes |
| CHI | Chitosan | PEDOT | Poly(3,4-ethylenedioxythiophene) |
| Co3O4 @ MCF | Cobalt oxide-loaded mesoporous carbon framework | BEMD | Bipolar electrolytic micelle disruption |
| AChE | Acetylcholinesterase | 3T | 2,2′:3′,2″-terthiophene |
| pp | Plasma-polymerized | PET | Poly(ethylene terephthalate) |
| FDNA | Framework DNA | BODIPY | 4,4-Difluoro-8-(4-trimethylsilylethynylphenyl)-1,3,5,7-tetramethyl-2,6-diethyl-4-bora-3a,4a-diaza-s-indacene |
References
- Walther, B.K.; Dinu, C.Z.; Guldi, D.M.; Sergeyev, V.G.; Creager, S.E.; Cooke, J.P.; Guiseppi-Elie, A. Nanobiosensing with graphene and carbon quantum dots: Recent advances. Mater. Today 2020, 39, 23–46. [Google Scholar]
- Singh, S.; Kumar, V.; Dhanjal, D.S.; Datta, S.; Prasad, R.; Singh, J. Biological biosensors for monitoring and diagnosis. In Microbial Biotechnology; Springer: Berlin/Heidelberg, Germany, 2020; pp. 317–335. [Google Scholar]
- Cheng, Z.; Wei, J.; Gu, L.; Zou, L.; Wang, T.; Chen, L.; Li, Y.; Yang, Y.; Li, P. DNAzyme-based biosensors for mercury (Ⅱ) detection: Rational construction, advances and perspectives. J. Hazard. Mater. 2022, 431, 128606. [Google Scholar]
- Gosai, A.; Khondakar, K.R.; Ma, X.; Ali, M.A. Application of Functionalized Graphene Oxide Based Biosensors for Health Monitoring: Simple Graphene Derivatives to 3D Printed Platforms. Biosensors 2021, 11, 384. [Google Scholar] [PubMed]
- Bandodkar, A.J.; Wang, J. Non-invasive wearable electrochemical sensors: A review. Trends Biotechnol. 2014, 32, 363–371. [Google Scholar]
- Zhao, Q.; Zhang, Q.; Sun, Y.; Liu, Y.; Lu, H.; Fan, X.; Wang, H.; Zhang, Y.; Wang, H. Design synthesis of a controllable flower-like Pt-graphene oxide architecture through electrostatic self-assembly for DNA damage biomarker 8-hydroxy-2′-deoxyguanosine biosensing research. Analyst 2018, 143, 3619–3627. [Google Scholar] [PubMed]
- Levien, M.; Farka, Z.; Pastucha, M.; Skládal, P.; Nasri, Z.; Weltmann, K.-D.; Fricke, K. Functional plasma-polymerized hydrogel coatings for electrochemical biosensing. Appl. Surf. Sci. 2022, 584, 152511. [Google Scholar]
- Kolb, H.C.; Finn, M.G.; Sharpless, K.B. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew. Chem. Int. Ed. 2001, 40, 2004–2021. [Google Scholar]
- Hein, J.E.; Fokin, V.V. Copper-catalyzed azide–alkyne cycloaddition (CuAAC) and beyond: New reactivity of copper (I) acetylides. Chem. Soc. Rev. 2010, 39, 1302–1315. [Google Scholar]
- Deng, Y.; Shavandi, A.; Okoro, O.V.; Nie, L. Alginate modification via click chemistry for biomedical applications. Carbohydr. Polym. 2021, 270, 118360. [Google Scholar]
- Shi, W.; Tang, F.; Ao, J.; Yu, Q.; Liu, J.; Tang, Y.; Jiang, B.; Ren, X.; Huang, H.; Yang, W. Manipulating the Click Reactivity of Dibenzoazacyclooctynes: From Azide Click Component to Caged Acylation Reagent by Silver Catalysis. Angew. Chem. Int. Ed. 2020, 132, 20112–20116. [Google Scholar]
- Yoon, H.Y.; Lee, D.; Lim, D.K.; Koo, H.; Kim, K. Copper-Free Click Chemistry: Applications in Drug Delivery, Cell Tracking, and Tissue Engineering. Adv. Mater. 2022, 34, 2107192. [Google Scholar]
- Kondengadan, S.M.; Bansal, S.; Yang, C.; Liu, D.; Fultz, Z.; Wang, B. Click chemistry and drug delivery: A bird’s-eye view. Acta Pharm. Sin. B 2022, 13, 1990–2016. [Google Scholar] [CrossRef]
- Fitzgerald, P.R.; Paegel, B.M. DNA-encoded chemistry: Drug discovery from a few good reactions. Chem. Rev. 2020, 121, 7155–7177. [Google Scholar] [CrossRef]
- Kim, E.; Koo, H. Biomedical applications of copper-free click chemistry: In vitro, in vivo, and ex vivo. Chem. Sci. 2019, 10, 7835–7851. [Google Scholar] [CrossRef]
- Rydzek, G.; Ji, Q.; Li, M.; Schaaf, P.; Hill, J.P.; Boulmedais, F.; Ariga, K. Electrochemical nanoarchitectonics and layer-by-layer assembly: From basics to future. Nano Today 2015, 10, 138–167. [Google Scholar] [CrossRef]
- Deo, K.A.; Jaiswal, M.K.; Abasi, S.; Lokhande, G.; Bhunia, S.; Nguyen, T.-U.; Namkoong, M.; Darvesh, K.; Guiseppi-Elie, A.; Tian, L. Nanoengineered Ink for Designing 3D Printable Flexible Bioelectronics. ACS Nano 2022, 16, 8798–8811. [Google Scholar] [CrossRef]
- Zhang, W.; Zhu, S.; Luque, R.; Han, S.; Hu, L.; Xu, G. Recent development of carbon electrode materials and their bioanalytical and environmental applications. Chem. Soc. Rev. 2016, 45, 715–752. [Google Scholar]
- Lopez, A.; Liu, J. Covalent and Noncovalent Functionalization of Graphene Oxide with DNA for Smart Sensing. Adv. Intell. Syst. 2020, 2, 2000123. [Google Scholar] [CrossRef]
- Georgakilas, V.; Tiwari, J.N.; Kemp, K.C.; Perman, J.A.; Bourlinos, A.B.; Kim, K.S.; Zboril, R. Noncovalent functionalization of graphene and graphene oxide for energy materials, biosensing, catalytic, and biomedical applications. Chem. Rev. 2016, 116, 5464–5519. [Google Scholar]
- Zhang, M.; Li, Y.; Su, Z.; Wei, G. Recent advances in the synthesis and applications of graphene–polymer nanocomposites. Polym. Chem. 2015, 6, 6107–6124. [Google Scholar] [CrossRef]
- Liu, B.; Salgado, S.; Maheshwari, V.; Liu, J. DNA adsorbed on graphene and graphene oxide: Fundamental interactions, desorption and applications. Curr. Opin. Colloid. Interface Sci. 2016, 26, 41–49. [Google Scholar]
- Szunerits, S.; Boukherroub, R. Graphene-based bioelectrochemistry and bioelectronics: A concept for the future? Curr. Opin. Electrochem. 2018, 12, 141–147. [Google Scholar] [CrossRef]
- Vivaldi, F.M.; Dallinger, A.; Bonini, A.; Poma, N.; Sembranti, L.; Biagini, D.; Salvo, P.; Greco, F.; Di Francesco, F. Three-dimensional (3D) laser-induced graphene: Structure, properties, and application to chemical sensing. ACS Appl. Mater. Interfaces 2021, 13, 30245–30260. [Google Scholar] [CrossRef]
- Wei, T.; Dai, Z.; Lin, Y.; Du, D. Electrochemical Immunoassays Based on Graphene: A Review. Electroanalysis 2016, 28, 4–12. [Google Scholar] [CrossRef]
- Perrozzi, F.; Prezioso, S.; Ottaviano, L. Graphene oxide: From fundamentals to applications. J. Phys. Condens. Matter 2014, 27, 013002. [Google Scholar] [CrossRef]
- Gao, L.; Lian, C.; Zhou, Y.; Yan, L.; Li, Q.; Zhang, C.; Chen, L.; Chen, K. Graphene oxide-DNA based sensors. Biosens. Bioelectron. 2014, 60, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Iwe, I.; Li, Z.; Huang, J. Graphene oxide and enzyme-assisted dual-cycling amplification method for sensitive fluorometric determination of DNA. Mikrochim. Acta 2019, 186, 716. [Google Scholar] [CrossRef]
- Wei, T.; Chen, Y.; Tu, W.; Lan, Y.; Dai, Z. A phosphomolybdic acid anion probe-based label-free, stable and simple electrochemical biosensing platform. Chem. Commun. 2014, 50, 9357–9360. [Google Scholar] [CrossRef]
- Shamsipur, M.; Molaei, K.; Molaabasi, F.; Hosseinkhani, S.; Taherpour, A.; Sarparast, M.; Moosavifard, S.E.; Barati, A. Aptamer-based fluorescent biosensing of adenosine triphosphate and cytochrome c via aggregation-induced emission enhancement on novel label-free DNA-capped silver nanoclusters/graphene oxide nanohybrids. ACS Appl. Mater. Interfaces 2019, 11, 46077–46089. [Google Scholar] [CrossRef] [PubMed]
- Asefifeyzabadi, N.; Holland, T.E.; Sivakumar, P.; Talapatra, S.; Senanayake, I.M.; Goodson, B.M.; Shamsi, M.H. Sequence-Independent DNA Adsorption on Few-Layered Oxygen-Functionalized Graphene Electrodes: An Electrochemical Study for Biosensing Application. Biosensors 2021, 11, 273. [Google Scholar] [CrossRef] [PubMed]
- Pena-Bahamonde, J.; Nguyen, H.N.; Fanourakis, S.K.; Rodrigues, D.F. Recent advances in graphene-based biosensor technology with applications in life sciences. J. Nanobiotechnology 2018, 16, 75. [Google Scholar] [PubMed]
- Qiu, L.; Li, D.; Cheng, H.M. Structural Control of Graphene-Based Materials for Unprecedented Performance. ACS Nano 2018, 12, 5085–5092. [Google Scholar] [CrossRef]
- Cao, S.H.; Li, L.H.; Wei, W.Y.; Feng, Y.; Jiang, W.L.; Wang, J.L.; Zhang, X.P.; Cai, S.H.; Chen, Z. A label-free and ultrasensitive DNA impedimetric sensor with enzymatic and electrical dual-amplification. Analyst 2019, 144, 4175–4179. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Hu, C.; Jahan, S.; Yuan, B.; Saleh, M.S.; Ju, E.; Gao, S.J.; Panat, R. Sensing of COVID-19 Antibodies in Seconds via Aerosol Jet Nanoprinted Reduced-Graphene-Oxide-Coated 3D Electrodes. Adv. Mater. 2021, 33, 2006647. [Google Scholar] [CrossRef]
- Maduraiveeran, G.; Jin, W. Nanomaterials based electrochemical sensor and biosensor platforms for environmental applications. Trends Environ. Anal. 2017, 13, 10–23. [Google Scholar]
- Guan, H.; Zhong, T.; He, H.; Zhao, T.; Xing, L.; Zhang, Y.; Xue, X. A self-powered wearable sweat-evaporation-biosensing analyzer for building sports big data. Nano Energy 2019, 59, 754–761. [Google Scholar] [CrossRef]
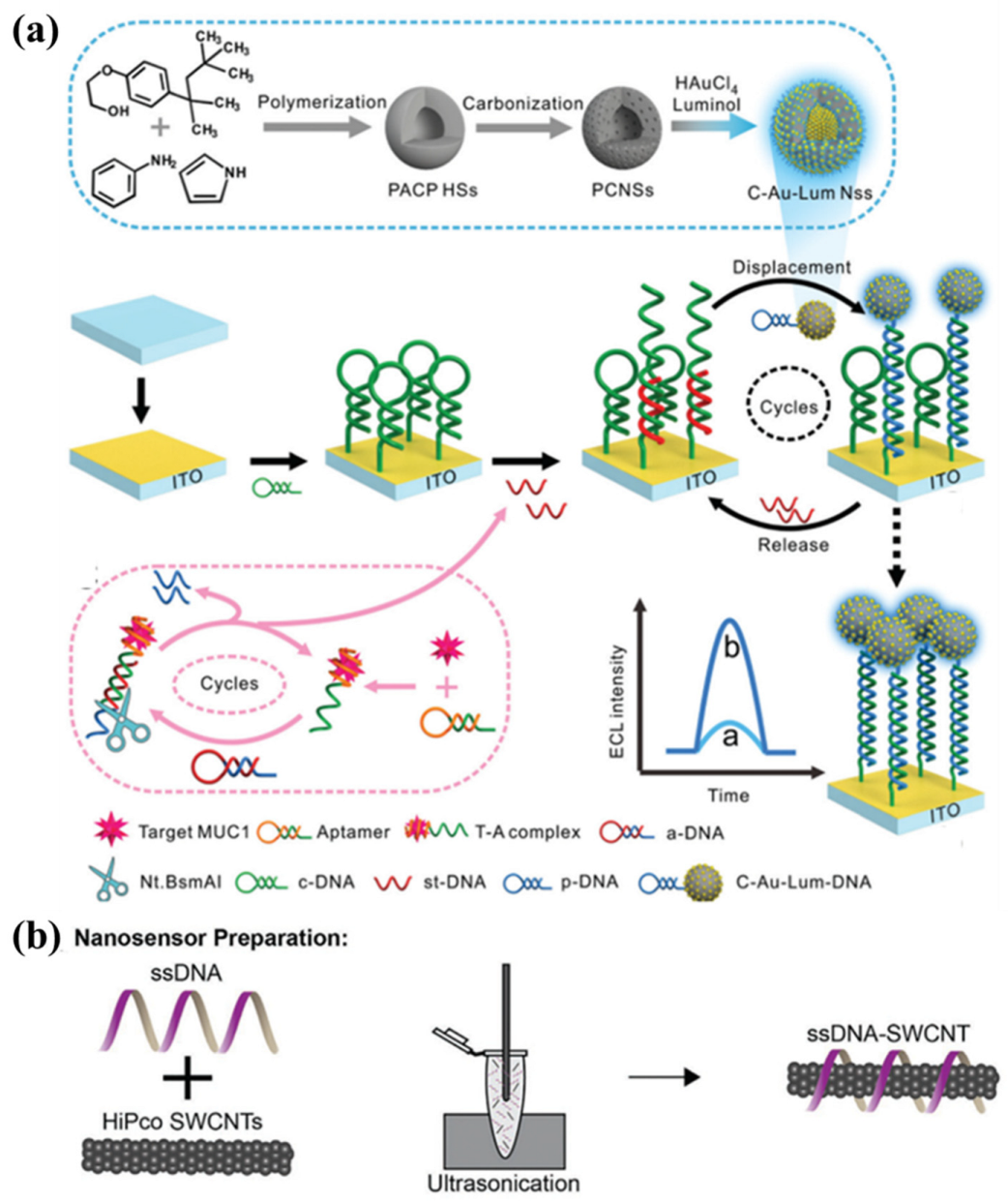
- Gao, J.W.; Chen, M.M.; Wen, W.; Zhang, X.; Wang, S.; Huang, W.H. Au-Luminol-decorated porous carbon nanospheres for the electrochemiluminescence biosensing of MUC1. Nanoscale 2019, 11, 16860–16867. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Tan, B.; Dang, X.; Zhao, H. Enhanced Electrochemiluminescence Detection for Hydrogen Peroxide Using Peroxidase-Mimetic Fe/N-Doped Porous Carbon. J. Electrochem. Soc. 2019, 166, 1594–1601. [Google Scholar] [CrossRef]
- Safaee, M.M.; Gravely, M.; Roxbury, D. A Wearable Optical Microfibrous Biomaterial with Encapsulated Nanosensors Enables Wireless Monitoring of Oxidative Stress. Adv. Funct. Mater. 2021, 31, 2006254. [Google Scholar] [CrossRef]
- Saheb, A.; Janata, J.; Josowicz, M. Reference electrode for ionic liquids. Electroanalysis 2006, 18, 405–409. [Google Scholar] [CrossRef]
- Iijima, S.; Ichihashi, T. Single-shell carbon nanotubes of 1-nm diameter. Nature 1993, 363, 603–605. [Google Scholar]
- Iijima, S. Helical microtubules of graphitic carbon. Nature 1991, 354, 56–58. [Google Scholar] [CrossRef]
- Gooding, J.J. Nanostructuring electrodes with carbon nanotubes: A review on electrochemistry and applications for sensing. Electrochim. Acta 2005, 50, 3049–3060. [Google Scholar]
- Lew, T.T.S.; Koman, V.B.; Silmore, K.S.; Seo, J.S.; Gordiichuk, P.; Kwak, S.Y.; Park, M.; Ang, M.C.; Khong, D.T.; Lee, M.A.; et al. Real-time detection of wound-induced H2O2 signalling waves in plants with optical nanosensors. Nat. Plants 2020, 6, 404–415. [Google Scholar]
- Bhattacharya, S.; Gong, X.; Wang, E.; Dutta, S.K.; Caplette, J.R.; Son, M.; Nguyen, F.T.; Strano, M.S.; Mukhopadhyay, D. DNA-SWCNT Biosensors Allow Real-Time Monitoring of Therapeutic Responses in Pancreatic Ductal Adenocarcinoma. Cancer Res. 2019, 79, 4515–4523. [Google Scholar] [PubMed]
- Harvey, J.D.; Williams, R.M.; Tully, K.M.; Baker, H.A.; Shamay, Y.; Heller, D.A. An in Vivo Nanosensor Measures Compartmental Doxorubicin Exposure. Nano Lett. 2019, 19, 4343–4354. [Google Scholar]
- Salem, D.P.; Gong, X.; Liu, A.T.; Akombi, K.; Strano, M.S. Immobilization and Function of nIR-Fluorescent Carbon Nanotube Sensors on Paper Substrates for Fluidic Manipulation. Anal. Chem. 2020, 92, 916–923. [Google Scholar]
- Meng, L.; Turner, A.P.F.; Mak, W.C. Tunable 3D nanofibrous and bio-functionalised PEDOT network explored as a conducting polymer-based biosensor. Biosens. Bioelectron. 2020, 159, 112181. [Google Scholar]
- Zhao, Z.; Chen, H.; Cheng, Y.; Huang, Z.; Wei, X.; Feng, J.; Cheng, J.; Mugo, S.M.; Jaffrezic-Renault, N.; Guo, Z. Electrochemical aptasensor based on electrodeposited poly (3, 4-ethylenedioxythiophene)-graphene oxide coupled with Au@ Pt nanocrystals for the detection of 17β-estradiol. Microchim. Acta 2022, 189, 178. [Google Scholar]
- Zhu, F.; Bian, X.; Zhang, H.; Wen, Y.; Chen, Q.; Yan, Y.; Li, L.; Liu, G.; Yan, J. Controllable design of a nano-bio aptasensing interface based on tetrahedral framework nucleic acids in an integrated microfluidic platform. Biosens. Bioelectron. 2021, 176, 112943. [Google Scholar]
- Jia, Y.; Yi, X.; Li, Z.; Zhang, L.; Yu, B.; Zhang, J.; Wang, X.; Jia, X. Recent advance in biosensing applications based on two-dimensional transition metal oxide nanomaterials. Talanta 2020, 219, 121308. [Google Scholar]
- Fan, Y.; Liu, S.; Yi, Y.; Rong, H.; Zhang, J. Catalytic Nanomaterials toward Atomic Levels for Biomedical Applications: From Metal Clusters to Single-Atom Catalysts. ACS Nano 2021, 15, 2005–2037. [Google Scholar]
- Zhou, W.; Saran, R.; Liu, J. Metal sensing by DNA. Chem. Rev. 2017, 117, 8272–8325. [Google Scholar]
- Fan, D.; Shang, C.; Gu, W.; Wang, E.; Dong, S. Introducing Ratiometric Fluorescence to MnO2 Nanosheet-Based Biosensing: A Simple, Label-Free Ratiometric Fluorescent Sensor Programmed by Cascade Logic Circuit for Ultrasensitive GSH Detection. ACS Appl. Mater. Interfaces 2017, 9, 25870–25877. [Google Scholar]
- Yadav, V.; Roy, S.; Singh, P.; Khan, Z.; Jaiswal, A. 2D MoS(2)-Based Nanomaterials for Therapeutic, Bioimaging, and Biosensing Applications. Small 2019, 15, 1803706. [Google Scholar]
- Yan, R.; Lu, N.; Han, S.; Lu, Z.; Xiao, Y.; Zhao, Z.; Zhang, M. Simultaneous detection of dual biomarkers using hierarchical MoS2 nanostructuring and nano-signal amplification-based electrochemical aptasensor toward accurate diagnosis of prostate cancer. Biosens. Bioelectron. 2022, 197, 113797. [Google Scholar]
- Han, H.-H.; Tian, H.; Zang, Y.; Sedgwick, A.C.; Li, J.; Sessler, J.L.; He, X.-P.; James, T.D. Small-molecule fluorescence-based probes for interrogating major organ diseases. Chem. Soc. Rev. 2021, 50, 9391–9429. [Google Scholar]
- Cui, M.; Che, Z.; Gong, Y.; Li, T.; Hu, W.; Wang, S. A graphdiyne-based protein molecularly imprinted biosensor for highly sensitive human C-reactive protein detection in human serum. Chem. Eng. J. 2022, 431, 133455. [Google Scholar]
- Salimian, R.; Kékedy-Nagy, L.; Ferapontova, E.E. Specific Picomolar Detection of a Breast Cancer Biomarker HER-2/neu Protein in Serum: Electrocatalytically Amplified Electroanalysis by the Aptamer/PEG-Modified Electrode. ChemElectroChem 2017, 4, 872–879. [Google Scholar]
- Zhang, L.; Xiao, X.; Xu, Y.; Chen, D.; Chen, J.; Ma, Y.; Dai, Z.; Zou, X. Electrochemical assay for continuous monitoring of dynamic DNA methylation process. Biosens. Bioelectron. 2018, 100, 184–191. [Google Scholar] [PubMed]
- Tian, X.; Murfin, L.C.; Wu, L.; Lewis, S.E.; James, T.D. Fluorescent small organic probes for biosensing. Chem. Sci. 2021, 12, 3406–3426. [Google Scholar]
- Ma, X.; Gao, W.; Du, F.; Yuan, F.; Yu, J.; Guan, Y.; Sojic, N.; Xu, G. Rational Design of Electrochemiluminescent Devices. Acc. Chem. Res. 2021, 54, 2936–2945. [Google Scholar] [PubMed]
- Ma, C.; Cao, Y.; Gou, X.; Zhu, J.J. Recent Progress in Electrochemiluminescence Sensing and Imaging. Anal. Chem. 2020, 92, 431–454. [Google Scholar] [PubMed]
- Zhang, Y.; Xu, J.; Zhou, S.; Zhu, L.; Lv, X.; Zhang, J.; Zhang, L.; Zhu, P.; Yu, J. DNAzyme-Triggered Visual and Ratiometric Electrochemiluminescence Dual-Readout Assay for Pb(II) Based on an Assembled Paper Device. Anal. Chem. 2020, 92, 3874–3881. [Google Scholar]
- Guo, J.; Xie, M.; Du, P.; Liu, Y.; Lu, X. Signal Amplification Strategy Using Atomically Gold-Supported VO(2) Nanobelts as a Co-reaction Accelerator for Ultrasensitive Electrochemiluminescent Sensor Construction Based on the Resonance Energy Transfer Platform. Anal. Chem. 2021, 93, 10619–10626. [Google Scholar]
- Wu, K.; Zheng, Y.; Chen, R.; Zhou, Z.; Liu, S.; Shen, Y.; Zhang, Y. Advances in electrochemiluminescence luminophores based on small organic molecules for biosensing. Biosens. Bioelectron. 2023, 223, 115031. [Google Scholar]
- Wu, X.; Li, Z.; Yang, L.; Han, J.; Han, S. A self-referenced nanodosimeter for reaction based ratiometric imaging of hypochlorous acid in living cells. Chem. Sci. 2013, 4, 460–467. [Google Scholar]
- Jia, X.; Chen, Q.; Yang, Y.; Tang, Y.; Wang, R.; Xu, Y.; Zhu, W.; Qian, X. FRET-Based Mito-Specific Fluorescent Probe for Ratiometric Detection and Imaging of Endogenous Peroxynitrite: Dyad of Cy3 and Cy5. J. Am. Chem. Soc. 2016, 138, 10778–10781. [Google Scholar] [PubMed]
- Gu, K.; Xu, Y.; Li, H.; Guo, Z.; Zhu, S.; Zhu, S.; Shi, P.; James, T.D.; Tian, H.; Zhu, W.H. Real-Time Tracking and In Vivo Visualization of beta-Galactosidase Activity in Colorectal Tumor with a Ratiometric Near-Infrared Fluorescent Probe. J. Am. Chem. Soc. 2016, 138, 5334–5340. [Google Scholar] [PubMed]
- Murfin, L.C.; Weber, M.; Park, S.J.; Kim, W.T.; Lopez-Alled, C.M.; McMullin, C.L.; Pradaux-Caggiano, F.; Lyall, C.L.; Kociok-Kohn, G.; Wenk, J.; et al. Azulene-Derived Fluorescent Probe for Bioimaging: Detection of Reactive Oxygen and Nitrogen Species by Two-Photon Microscopy. J. Am. Chem. Soc. 2019, 141, 19389–19396. [Google Scholar]
- Chen, H.; Tang, Y.; Ren, M.; Lin, W. Single near-infrared fluorescent probe with high- and low-sensitivity sites for sensing different concentration ranges of biological thiols with distinct modes of fluorescence signals. Chem. Sci. 2016, 7, 1896–1903. [Google Scholar]
- Jiao, Y.; Yin, J.; He, H.; Peng, X.; Gao, Q.; Duan, C. Conformationally Induced Off-On Cell Membrane Chemosensor Targeting Receptor Protein-Tyrosine Kinases for in Vivo and in Vitro Fluorescence Imaging of Cancers. J. Am. Chem. Soc. 2018, 140, 5882–5885. [Google Scholar] [CrossRef]
- Long, L.; Huang, M.; Wang, N.; Wu, Y.; Wang, K.; Gong, A.; Zhang, Z.; Sessler, J.L. A Mitochondria-Specific Fluorescent Probe for Visualizing Endogenous Hydrogen Cyanide Fluctuations in Neurons. J. Am. Chem. Soc. 2018, 140, 1870–1875. [Google Scholar] [CrossRef] [PubMed]
- Sassolas, A.; Blum, L.J.; Leca-Bouvier, B.D. Immobilization strategies to develop enzymatic biosensors. Biotechnol. Adv. 2012, 30, 489–511. [Google Scholar] [PubMed]
- Gu, H.; Xing, Y.; Xiong, P.; Tang, H.; Li, C.; Chen, S.; Zeng, R.; Han, K.; Shi, G. Three-Dimensional Porous Ti3C2Tx MXene–Graphene Hybrid Films for Glucose Biosensing. ACS Appl. Nano Mater. 2019, 2, 6537–6545. [Google Scholar] [CrossRef]
- Dhanjai; Balla, P.; Sinha, A.; Wu, L.; Lu, X.; Tan, D.; Chen, J. Co3O4 nanoparticles supported mesoporous carbon framework interface for glucose biosensing. Talanta 2019, 203, 112–121. [Google Scholar] [CrossRef]
- Yao, C.; Ou, J.; Tang, J.; Yang, D. DNA Supramolecular Assembly on Micro/Nanointerfaces for Bioanalysis. Acc. Chem. Res. 2022, 55, 2043–2054. [Google Scholar] [CrossRef]
- Li, F.; Li, Q.; Zuo, X.; Fan, C. DNA framework-engineered electrochemical biosensors. Sci. China Life Sci. 2020, 63, 1130–1141. [Google Scholar] [PubMed]
- Su, S.; Ma, J.; Xu, Y.; Pan, H.; Zhu, D.; Chao, J.; Weng, L.; Wang, L. Electrochemical Analysis of Target-Induced Hairpin-Mediated Aptamer Sensors. ACS Appl. Mater. Interfaces 2020, 12, 48133–48139. [Google Scholar] [CrossRef]
- Li, M.; Lv, M.; Wang, L.; Fan, C.; Zuo, X. Engineering electrochemical interface for biomolecular sensing. Curr. Opin. Electrochem. 2019, 14, 71–80. [Google Scholar]
- Zhu, C.; Liu, D.; Li, Y.; Ma, S.; Wang, M.; You, T. Hairpin DNA assisted dual-ratiometric electrochemical aptasensor with high reliability and anti-interference ability for simultaneous detection of aflatoxin B1 and ochratoxin A. Biosens. Bioelectron. 2021, 174, 112654. [Google Scholar]
- Miao, P.; Tang, Y.; Wang, L. DNA Modified Fe3O4@Au Magnetic Nanoparticles as Selective Probes for Simultaneous Detection of Heavy Metal Ions. ACS Appl. Mater. Interfaces 2017, 9, 3940–3947. [Google Scholar] [CrossRef]
- Han, C.; Li, W.; Li, Q.; Xing, W.; Luo, H.; Ji, H.; Fang, X.; Luo, Z.; Zhang, L. CRISPR/Cas12a-Derived electrochemical aptasensor for ultrasensitive detection of COVID-19 nucleocapsid protein. Biosens. Bioelectron. 2022, 200, 113922. [Google Scholar]
- Su, J.; Liu, W.; Chen, S.; Deng, W.; Dou, Y.; Zhao, Z.; Li, J.; Li, Z.; Yin, H.; Ding, X.; et al. A Carbon-Based DNA Framework Nano-Bio Interface for Biosensing with High Sensitivity and a High Signal-to-Noise Ratio. ACS Sens. 2020, 5, 3979–3987. [Google Scholar] [CrossRef]
- Mao, X.; Mao, D.; Chen, T.; Jalalah, M.; Al-Assiri, M.S.; Harraz, F.A.; Zhu, X.; Li, G. DNA Hydrogel-Based Three-Dimensional Electron Transporter and Its Application in Electrochemical Biosensing. ACS Appl. Mater. Interfaces 2020, 12, 36851–36859. [Google Scholar] [CrossRef]
- Cui, M.; Gong, Y.; Du, M.; Wang, K.; Li, T.; Zhu, X.; Wang, S.; Luo, X. An antifouling electrochemical biosensor based on a protein imprinted hydrogel for human immunoglobulin G recognition in complex biological media. Sens. Actuators B Chem. 2021, 337, 129820. [Google Scholar] [CrossRef]
- Berezovski, M.; Krylov, S.N. Using DNA-binding proteins as an analytical tool. J. Am. Chem. Soc. 2003, 125, 13451–13454. [Google Scholar] [CrossRef]
- Campuzano, S.; Pedrero, M.; Pingarrón, J.M. Viral protein-based bioanalytical tools for small RNA biosensing. Trends Analyt. Chem. 2016, 79, 335–343. [Google Scholar]
- Quijano-Rubio, A.; Yeh, H.W.; Park, J.; Lee, H.; Langan, R.A.; Boyken, S.E.; Lajoie, M.J.; Cao, L.; Chow, C.M.; Miranda, M.C.; et al. De novo design of modular and tunable protein biosensors. Nature 2021, 591, 482–487. [Google Scholar]
- Wei, Y.; Zeng, Q.; Hu, Q.; Wang, M.; Tao, J.; Wang, L. Self-cleaned electrochemical protein imprinting biosensor basing on a thermo-responsive memory hydrogel. Biosens. Bioelectron. 2018, 99, 136–141. [Google Scholar] [CrossRef]
- Wei, Y.; Zeng, Q.; Huang, J.; Guo, X.; Wang, L.; Wang, L. Preparation of Gas-Responsive Imprinting Hydrogel and Their Gas-Driven Switchable Affinity for Target Protein Recognition. ACS Appl. Mater. Interfaces 2020, 12, 24363–24369. [Google Scholar] [CrossRef]
- Kerry, R.G.; Ukhurebor, K.E.; Kumari, S.; Maurya, G.K.; Patra, S.; Panigrahi, B.; Majhi, S.; Rout, J.R.; del Pilar Rodriguez-Torres, M.; Das, G. A comprehensive review on the applications of nano-biosensor-based approaches for non-communicable and communicable disease detection. Biomater. Sci. 2021, 9, 3576–3602. [Google Scholar]
- Qi, P.; Wan, Y.; Zhang, D. Impedimetric biosensor based on cell-mediated bioimprinted films for bacterial detection. Biosens. Bioelectron. 2013, 39, 282–288. [Google Scholar]
- Jiang, H.; Jiang, D.; Liu, X.; Yang, J. A self-driven PET chip-based imprinted electrochemical sensor for the fast detection of Salmonella. Sens. Actuators B Chem. 2021, 349, 130785. [Google Scholar] [CrossRef]
- Benson, V.S.S.a.D.E. Protein Design Provides Lead(II) Ion Biosensors for Imaging Molecular Fluxes around Red Blood Cells. Biochemistry 2009, 48, 462–470. [Google Scholar]
- Chen, C.; Liu, Y.; Gu, H.-Y. Cellular biosensor based on red blood cells immobilized on Fe3O4 Core/Au Shell nanoparticles for hydrogen peroxide electroanalysis. Microchim. Acta 2010, 171, 371–376. [Google Scholar] [CrossRef]
- Liu, J.M.; Zhang, D.D.; Fang, G.Z.; Wang, S. Erythrocyte membrane bioinspired near-infrared persistent luminescence nanocarriers for in vivo long-circulating bioimaging and drug delivery. Biomaterials 2018, 165, 39–47. [Google Scholar] [CrossRef]
- Rostovtsev, V.V.; Green, L.G.; Fokin, V.V.; Sharpless, K.B. A stepwise huisgen cycloaddition process: Copper (I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. 2002, 114, 2708–2711. [Google Scholar]
- Rodionov, V.O.; Presolski, S.I.; Díaz Díaz, D.; Fokin, V.V.; Finn, M. Ligand-accelerated Cu-catalyzed azide− alkyne cycloaddition: A mechanistic report. J. Am. Chem. Soc. 2007, 129, 12705–12712. [Google Scholar] [CrossRef]
- Chan, T.R.; Hilgraf, R.; Sharpless, K.B.; Fokin, V.V. Polytriazoles as copper (I)-stabilizing ligands in catalysis. Org. Lett. 2004, 6, 2853–2855. [Google Scholar] [CrossRef] [PubMed]
- Yanez-Sedeno, P.; Gonzalez-Cortes, A.; Campuzano, S.; Pingarron, J.M. Copper(I)-Catalyzed Click Chemistry as a Tool for the Functionalization of Nanomaterials and the Preparation of Electrochemical (Bio)Sensors. Sensors 2019, 19, 2379. [Google Scholar] [CrossRef]
- Nicosia, C.; Huskens, J. Reactive self-assembled monolayers: From surface functionalization to gradient formation. Mater. Horiz. 2014, 1, 32–45. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, Y.; Chen, J.; Yang, Y.; Li, G. Bioinspired Artificial “Clickase” for the Catalytic Click Immunoassay of Foodborne Pathogens. Anal. Chem. 2021, 93, 3217–3225. [Google Scholar] [CrossRef]
- Guerrero, S.; Cadano, D.; Agüí, L.; Barderas, R.; Campuzano, S.; Yáñez-Sedeño, P.; Pingarrón, J.M. Click chemistry-assisted antibodies immobilization for immunosensing of CXCL7 chemokine in serum. J. Electroanal. Chem. 2019, 837, 246–253. [Google Scholar] [CrossRef]
- Fomo, G.; Nwaji, N.; Nyokong, T. Low symmetric metallophthalocyanine modified electrode via click chemistry for simultaneous detection of heavy metals. J. Electroanal. Chem. 2018, 813, 58–66. [Google Scholar]
- Genzer, J.; Bhat, R.R. Surface-bound soft matter gradients. Langmuir 2008, 24, 2294–2317. [Google Scholar] [PubMed]
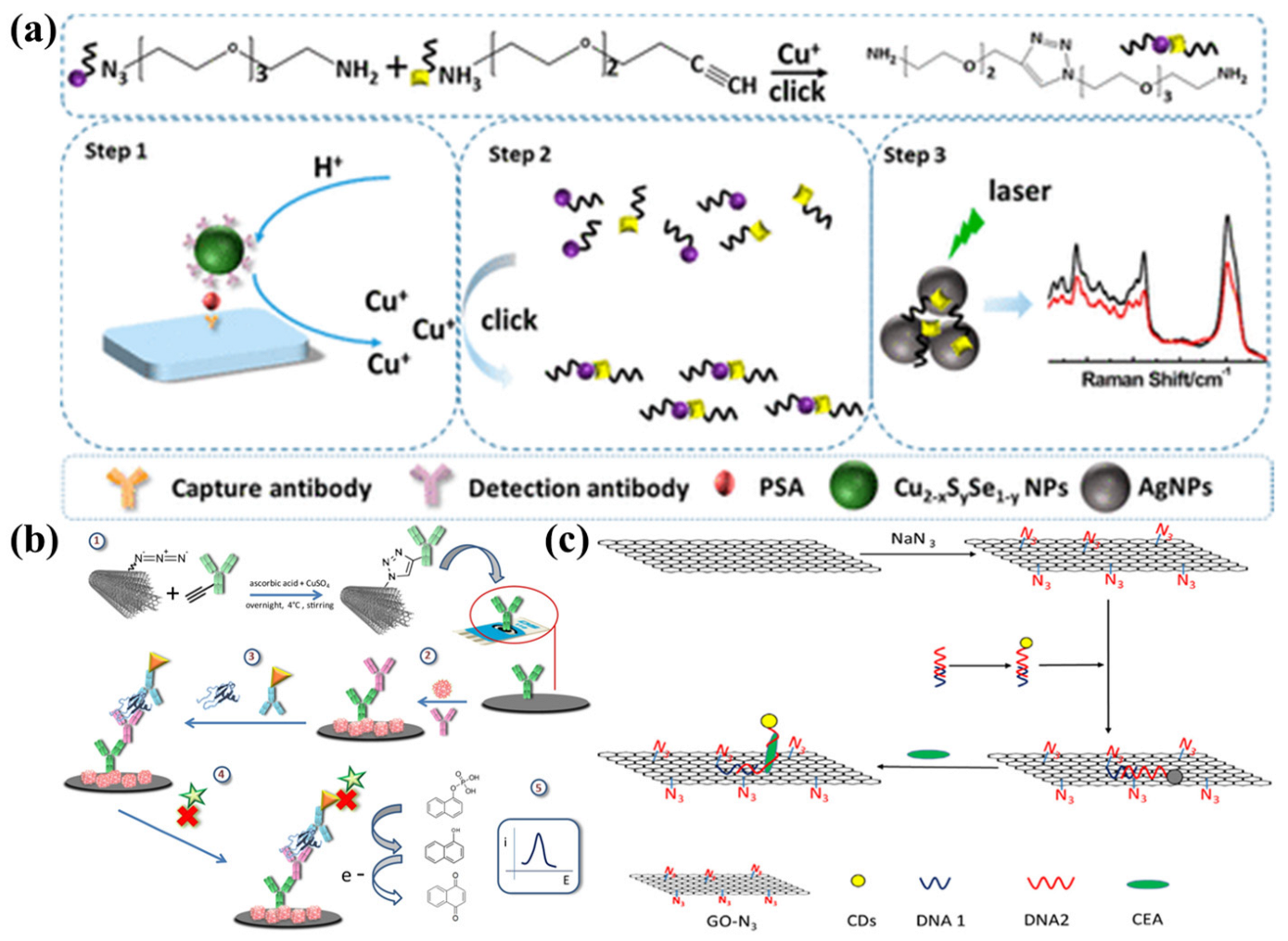
- Yang, L.; Gao, M.X.; Zou, H.Y.; Li, Y.F.; Huang, C.Z. Plasmonic Cu2-xSySe1-y Nanoparticles Catalyzed Click Chemistry Reaction for SERS Immunoassay of Cancer Biomarker. Anal. Chem. 2018, 90, 11728–11733. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Yang, Y.; Wu, S.; Huang, Y.; Li, J.; Xiang, Y.; Li, G. In Situ Reduction of Porous Copper Metal-Organic Frameworks for Three-Dimensional Catalytic Click Immunoassay. Anal. Chem. 2020, 92, 2972–2978. [Google Scholar] [CrossRef]
- Xiang, W.; Zhang, Z.; Weng, W.; Wu, B.; Cheng, J.; Shi, L.; Sun, H.; Gao, L.; Shi, K. Highly sensitive detection of carcinoembryonic antigen using copper-free click chemistry on the surface of azide cofunctionalized graphene oxide. Anal. Chim. Acta 2020, 1127, 156–162. [Google Scholar] [CrossRef]
- Lutz, J.F. Copper-free azide-alkyne cycloadditions: New insights and perspectives. Angew. Chem. Int. Ed. 2008, 47, 2182–2184. [Google Scholar] [CrossRef]
- Agard, N.J.; Prescher, J.A.; Bertozzi, C.R. A strain-promoted [3+2] azide− alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 2004, 126, 15046–15047. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, K.; Zhang, S.; Zheng, X.; Cui, T.; Yang, X.; Liu, Y.; Lu, D.; Wang, Y.; Tian, X.; et al. Site-Specific Introduction of Bioorthogonal Handles to Nanopores by Genetic Code Expansion. Angew. Chem. Int. Ed. 2023, 62, 202216115. [Google Scholar] [CrossRef] [PubMed]
- Toulemon, D.; Pichon, B.P.; Leuvrey, C.d.; Zafeiratos, S.; Papaefthimiou, V.; Cattoën, X.; Bégin-Colin, S. Fast assembling of magnetic iron oxide nanoparticles by microwave-assisted copper (I) catalyzed alkyne–azide cycloaddition (CuAAC). Chem. Mater. 2013, 25, 2849–2854. [Google Scholar] [CrossRef]
- Cintas, P.; Barge, A.; Tagliapietra, S.; Boffa, L.; Cravotto, G. Alkyne–azide click reaction catalyzed by metallic copper under ultrasound. Nat. Prot. 2010, 5, 607–616. [Google Scholar] [CrossRef]
- Tasdelen, M.A.; Yagci, Y. Light-induced click reactions. Angew. Chem. Int. Ed. 2013, 52, 5930–5938. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Li, M.; Zhang, R.; Dong, M.; Ling, C. Double electrochemical covalent coupling method based on click chemistry and diazonium chemistry for the fabrication of sensitive amperometric immunosensor. Anal. Chim. Acta 2013, 792, 28–34. [Google Scholar] [CrossRef]
- Devaraj, N.K.; Dinolfo, P.H.; Chidsey, C.E.; Collman, J.P. Selective functionalization of independently addressed microelectrodes by electrochemical activation and deactivation of a coupling catalyst. J. Am. Chem. Soc. 2006, 128, 1794–1795. [Google Scholar] [CrossRef]
- Lesniewski, A.; Matyjewicz, J.; Palys, B.; Niedziolka-Jonsson, J. Electroassisted click chemistry immobilisation of gold nanoparticles on a solid substrate. Electrochem. Commun. 2015, 53, 20–23. [Google Scholar] [CrossRef]
- Villalba, M.; Bossi, M.; Pozo, M.D.; Calvo, E.J. Palladium Nanoparticles Embedded in a Layer-by-Layer Nanoreactor Built with Poly(Acrylic Acid) Using “Electro-Click Chemistry”. Langmuir 2016, 32, 6836–6842. [Google Scholar] [CrossRef]
- Yamamoto, T.; Akahori, M.; Natsui, K.; Saitoh, T.; Einaga, Y. Controlled decoration of boron-doped diamond electrodes by electrochemical click reaction (e−CLICK). Carbon 2018, 130, 350–354. [Google Scholar] [CrossRef]
- Rydzek, G.; Jierry, L.; Parat, A.; Thomann, J.S.; Voegel, J.C.; Senger, B.; Hemmerle, J.; Ponche, A.; Frisch, B.; Schaaf, P.; et al. Electrochemically triggered assembly of films: A one-pot morphogen-driven buildup. Angew. Chem. Int. Ed. 2011, 50, 4374–4377. [Google Scholar] [CrossRef]
- Hu, L.; Zhao, P.; Deng, H.; Xiao, L.; Qin, C.; Du, Y.; Shi, X. Electrical signal guided click coating of chitosan hydrogel on conductive surface. RSC Adv. 2014, 4, 13477. [Google Scholar] [CrossRef]
- Cheng, C.; Oueslati, R.; Wu, J.; Chen, J.; Eda, S. Capacitive DNA sensor for rapid and sensitive detection of whole genome human herpesvirus-1 dsDNA in serum. Electrophoresis 2017, 38, 1617–1623. [Google Scholar] [CrossRef]
- Castiello, F.R.; Porter, J.; Modarres, P.; Tabrizian, M. Interfacial capacitance immunosensing using interdigitated electrodes: The effect of insulation/immobilization chemistry. Phys. Chem. Chem. Phys. 2019, 21, 15787–15797. [Google Scholar] [CrossRef] [PubMed]
- Cesbron, M.; Levillain, E.; Breton, T.; Gautier, C. Click chemistry: A versatile method for tuning the composition of mixed organic layers obtained by reduction of diazonium cations. ACS Appl. Mater. Interfaces 2018, 10, 37779–37782. [Google Scholar] [CrossRef]
- Cernat, A.; Griveau, S.; Martin, P.; Lacroix, J.C.; Farcau, C.; Sandulescu, R.; Bedioui, F. Electrografted nanostructured platforms for click chemistry. Electrochem. Commun. 2012, 23, 141–144. [Google Scholar] [CrossRef]
- Fenoy, G.E.; Hasler, R.; Quartinello, F.; Marmisolle, W.A.; Lorenz, C.; Azzaroni, O.; Bauerle, P.; Knoll, W. “Clickable” Organic Electrochemical Transistors. JACS Au 2022, 2, 2778–2790. [Google Scholar] [CrossRef] [PubMed]
- Sciortino, F.; Rydzek, G.; Grasset, F.; Kahn, M.L.; Hill, J.P.; Chevance, S.; Gauffre, F.; Ariga, K. Electro-click construction of hybrid nanocapsule films with triggered delivery properties. Phys. Chem. Chem. Phys. 2018, 20, 2761–2770. [Google Scholar] [CrossRef]
- Guerrero, S.; Agui, L.; Yanez-Sedeno, P.; Pingarron, J.M. Design of electrochemical immunosensors using electro-click chemistry. Application to the detection of IL-1beta cytokine in saliva. Bioelectrochemistry 2020, 133, 107484. [Google Scholar] [CrossRef]
- Wei, T.; Dong, T.; Xing, H.; Liu, Y.; Dai, Z. Cucurbituril and Azide Cofunctionalized Graphene Oxide for Ultrasensitive Electro-Click Biosensing. Anal. Chem. 2017, 89, 12237–12243. [Google Scholar] [CrossRef] [PubMed]
- Levrie, K.; Jans, K.; Vos, R.; Ardakanian, N.; Verellen, N.; Van Hoof, C.; Lagae, L.; Stakenborg, T. Multiplexed site-specific electrode functionalization for multitarget biosensors. Bioelectrochemistry 2016, 112, 61–66. [Google Scholar] [CrossRef]
- Rydzek, G.; Terentyeva, T.G.; Pakdel, A.; Golberg, D.; Hill, J.P.; Ariga, K. Simultaneous electropolymerization and electro-click functionalization for highly versatile surface platforms. ACS Nano 2014, 8, 5240–5248. [Google Scholar] [CrossRef]
- Chmielarz, P.; Fantin, M.; Park, S.; Isse, A.A.; Gennaro, A.; Magenau, A.J.; Sobkowiak, A.; Matyjaszewski, K. Electrochemically mediated atom transfer radical polymerization (eATRP). Prog. Polym. Sci. 2017, 69, 47–78. [Google Scholar] [CrossRef]
- Wu, T.; Lankshear, E.R.; Downard, A.J. Simultaneous Electro-Click and Electrochemically Mediated Polymerization Reactions for One-Pot Grafting from a Controlled Density of Anchor Sites. ChemElectroChem 2019, 6, 5149–5154. [Google Scholar] [CrossRef]
- Kang, D.; Kim, T.W.; Kubota, S.R.; Cardiel, A.C.; Cha, H.G.; Choi, K.S. Electrochemical Synthesis of Photoelectrodes and Catalysts for Use in Solar Water Splitting. Chem. Rev. 2015, 115, 12839–12887. [Google Scholar] [CrossRef]
- Guiseppi-Elie, A. Electroconductive hydrogels: Synthesis, characterization and biomedical applications. Biomaterials 2010, 31, 2701–2716. [Google Scholar] [CrossRef]
- Nederberg, F.; Trang, V.; Pratt, R.C.; Kim, S.-H.; Colson, J.; Nelson, A.; Frank, C.W.; Hedrick, J.L.; Dubois, P.; Mespouille, L. Exploring the versatility of hydrogels derived from living organocatalytic ring-opening polymerization. Soft Matter 2010, 6, 2006. [Google Scholar] [CrossRef]
- Choi, E.J.; Shin, J.; Khaleel, Z.H.; Cha, I.; Yun, S.-H.; Cho, S.-W.; Song, C. Synthesis of electroconductive hydrogel films by an electro-controlled click reaction and their application to drug delivery systems. Polym. Chem. 2015, 6, 4473–4478. [Google Scholar] [CrossRef]
- Fosdick, S.E.; Knust, K.N.; Scida, K.; Crooks, R.M. Bipolar electrochemistry. Angew. Chem. Int. Ed. 2013, 52, 10438–10456. [Google Scholar] [CrossRef]
- Shida, N.; Zhou, Y.; Inagi, S. Bipolar Electrochemistry: A Powerful Tool for Electrifying Functional Material Synthesis. Acc. Chem. Res. 2019, 52, 2598–2608. [Google Scholar] [CrossRef] [PubMed]
- Shida, N.; Ishiguro, Y.; Atobe, M.; Fuchigami, T.; Inagi, S. Electro-Click Modification of Conducting Polymer Surface Using Cu(I) Species Generated on a Bipolar Electrode in a Gradient Manner. ACS Macro. Lett. 2012, 1, 656–659. [Google Scholar] [CrossRef]
- Zhou, Y.; Shida, N.; Tomita, I.; Inagi, S. Fabrication of Gradient and Patterned Organic Thin Films by Bipolar Electrolytic Micelle Disruption Using Redox-Active Surfactants. Angew. Chem. Int. Ed. 2021, 60, 14620–14629. [Google Scholar] [CrossRef]
- Khan, A.; Khan, A.A.; Asiri, A.M.; Rub, M.A.; Azum, N.; Rahman, M.M.; Khan, S.B.; Ghani, S.A. A new trend on biosensor for neurotransmitter choline/acetylcholine—An overview. Appl. Biochem. Biotechnol. 2013, 169, 1927–1939. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Mao, Z.; Tan, H.; Han, L.; Ren, T.; Gao, C. Gradient biomaterials and their influences on cell migration. Interface Focus 2012, 2, 337–355. [Google Scholar] [CrossRef] [PubMed]
- Rydzek, G.; Toulemon, D.; Garofalo, A.; Leuvrey, C.; Dayen, J.F.; Felder-Flesch, D.; Schaaf, P.; Jierry, L.; Begin-Colin, S.; Pichon, B.P.; et al. Selective Nanotrench Filling by One-Pot Electroclick Self-Constructed Nanoparticle Films. Small 2015, 11, 4638–4642. [Google Scholar] [CrossRef] [PubMed]
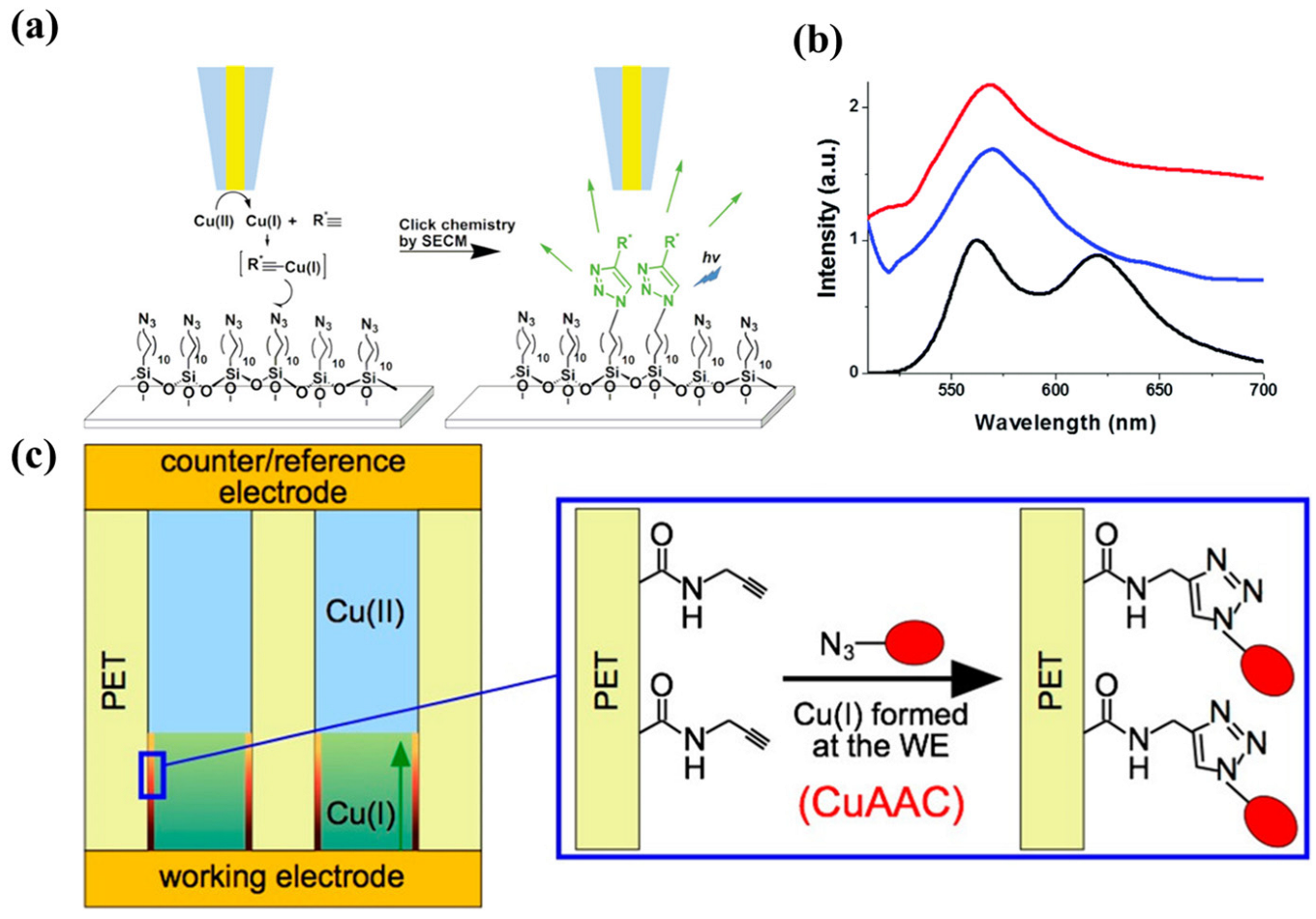
- Quinton, D.; Maringa, A.; Griveau, S.; Nyokong, T.; Bedioui, F. Surface patterning using scanning electrochemical microscopy to locally trigger a “click” chemistry reaction. Electrochem. Commun. 2013, 31, 112–115. [Google Scholar] [CrossRef]
- Krabbenborg, S.O.; Nicosia, C.; Chen, P.; Huskens, J. Reactivity mapping with electrochemical gradients for monitoring reactivity at surfaces in space and time. Nat. Commun. 2013, 4, 1667. [Google Scholar] [CrossRef]
- Nicosia, C.; Krabbenborg, S.O.; Chen, P.; Huskens, J. Shape-controlled fabrication of micron-scale surface chemical gradients via electrochemically activated copper(i) “click” chemistry. J. Mater. Chem. B 2013, 1, 5417–5428. [Google Scholar] [CrossRef]
- Hansen, T.S.; Daugaard, A.E.; Hvilsted, S.r.; Larsen, N.B. Spatially Selective Functionalization of Conducting Polymers by “Electroclick” Chemistry. Adv. Mat. 2009, 21, 4483–4486. [Google Scholar] [CrossRef]
- Cui, X.; Wei, T.; Hao, M.; Qi, Q.; Wang, H.; Dai, Z. Highly sensitive and selective colorimetric sensor for thiocyanate based on electrochemical oxidation-assisted complexation reaction with Gold nanostars etching. J. Hazard. Mater. 2020, 391, 122217. [Google Scholar] [CrossRef]
- Goll, M.; Ruff, A.; Muks, E.; Goerigk, F.; Omiecienski, B.; Ruff, I.; Gonzalez-Cano, R.C.; Lopez Navarrete, J.T.; Ruiz Delgado, M.C.; Ludwigs, S. Functionalized branched EDOT-terthiophene copolymer films by electropolymerization and post-polymerization “click”-reactions. Beilstein. J. Org. Chem. 2015, 11, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Saylan, Y.; Erdem, O.; Unal, S.; Denizli, A. An Alternative Medical Diagnosis Method: Biosensors for Virus Detection. Biosensors 2019, 9, 65. [Google Scholar] [CrossRef] [PubMed]
- Ku, S.-Y.; Wong, K.-T.; Bard, A.J. Surface patterning with fluorescent molecules using click chemistry directed by scanning electrochemical microscopy. J. Am. Chem. Soc. 2008, 130, 2392–2393. [Google Scholar] [CrossRef] [PubMed]
- Coceancigh, H.; Tran-Ba, K.H.; Siepser, N.; Baker, L.A.; Ito, T. Longitudinally Controlled Modification of Cylindrical and Conical Track-Etched Poly(ethylene terephthalate) Pores Using an Electrochemically Assisted Click Reaction. Langmuir 2017, 33, 11998–12006. [Google Scholar] [CrossRef]
- Rydzek, G.; Polavarapu, P.; Rios, C.; Tisserant, J.-N.; Voegel, J.-C.; Senger, B.; Lavalle, P.; Frisch, B.; Schaaf, P.; Boulmedais, F.; et al. Morphogen-driven self-construction of covalent films built from polyelectrolytes and homobifunctional spacers: Buildup and pH response. Soft Matter 2012, 8, 10336. [Google Scholar] [CrossRef]
- Chatterjee, A.K.; Chakraborty, R.; Basu, T. Mechanism of antibacterial activity of copper nanoparticles. Nanotechnology 2014, 25, 135101. [Google Scholar] [CrossRef]
- Yang, M.; Jalloh, A.S.; Wei, W.; Zhao, J.; Wu, P.; Chen, P.R. Biocompatible click chemistry enabled compartment-specific pH measurement inside E. coli. Nat. Commun. 2014, 5, 4981. [Google Scholar] [CrossRef]
- Cheng, X.; Liu, D.; Jin, Y.; Yang, M.; Xiang, J. Addressing Cu2+ interference for accurate aptamer-based biomarker determinations of Alzheimer’s disease. Anal. Sci. 2022, 38, 317–322. [Google Scholar] [CrossRef]
- Liu, G.; Gui, S.; Zhou, H.; Zeng, F.; Zhou, Y.; Ye, H. A strong adsorbent for Cu2+: Graphene oxide modified with triethanolamine. Dalton. Trans. 2014, 43, 6977–6980. [Google Scholar] [CrossRef]


| Functionalized Goal | Functionalized Strategy | Nanostructure | Principle | Effect | Reference |
|---|---|---|---|---|---|
| Fluorescence analysis detection of hemochromatosis protein gene | Direct adsorption | GO | π-stacking interactions | Ensures a very low background signal | [28] |
| Specific detection of biomarker-8-hydroxy-2′-deoxyguanosine | Layer–layer electrostatic self-assembly | GO | Electrostatic interaction | Improves the electrocatalytic performance of Pt nanoparticles | [6] |
| Detection of ultrasensitive target DNA | Direct adsorption | rGO | π-stacking interactions | Enlarges impedimetric signals | [34] |
| Detection of COVID-19 antibodies within seconds | Amidation reaction | rGO | Covalent bond | Enhances the transport of diffusing species in an electrochemical cell | [35] |
| Building of a self-powered wearable-lactate analyzer | Direct adsorption | Porous carbon | π-stacking interactions | Soaks up environmental thermal energy to generate electricity | [37] |
| Achievement of an ultrasensitive ECL biosensor | In situ reduction | Porous carbon | Electron transfer | Increases the mass transfer of reagents; accelerates the electron transport | [38] |
| Development of a highly sensitive and selective ECL * sensor based on the noble metal-free electrode | Pyrolytic process | Porous carbon | Breaking and polymerization of molecular bonds | Facilitates rapid electron transfer; improves the conductivity of the electrode | [39] |
| Real-time, in situ biochemical measurement of H2O2 in plants | DNA wrapping | SWNT | π-stacking interactions | An ideal probe for in vivo plant applications | [45] |
| Detection of H2O2 to determine the response to tumor therapy | DNA wrapping | SWNT | π-stacking interactions | Determines dynamic alteration of hydrogen peroxide in tumor; evaluates the effectiveness of chemotherapeutics | [46] |
| Continuous monitoring of ROS through a wearable diagnostic platform to prevent chronic and pathogenic infections | DNA wrapping | SWNT | π-stacking interactions | In situ measurements of peroxide in wounds | [40] |
| Use of implantable optical nanosensors to rapidly quantificate doxorubicin in living tissues | DNA wrapping | SWNT | π-stacking interactions | Quantifies doxorubicin exposure to tissues within living organisms | [47] |
| Fixing of near-infrared fluorescent SWCNT sensor on the paper substrate for sensing | DNA wrapping | SWNT | π-stacking interactions | Immobilizates SWCNTs onto paper substrates; distinguishes metal-ion contaminants | [48] |
| Fabrication of a Bio-Nano-PEDOT *-based biosensor for lactate detection | EDC/S–NHS chemistry | PEDOT-COOH | Chemical coupling | Low charge-transfer resistance; high transduction activity towards the co-enzyme NADH | [49] |
| Preparation of an electrochemical aptasensor electrodeposited of PEDOT *-GO coupled with Au@Pt | EDC/S–NHS chemistry | PEDOT-COOH | Chemical coupling | Fabricates a simple, label-free electrochemical aptasensor to detect estradiol | [50] |
| Development of an integrated “one-stop” microfluidic biosensor | Complementary base pairing | Framework nucleic acids | Hydrogen bond | One-stop detection of E.Coli O157:H7 with capture, release, enrichment, cell culture, and antimicrobial susceptibility testing | [51] |
| Detection of dual PCa * biomarkers, PSA and sarcosine | DNA wrapping | DNA-SiO2 | π-stacking interactions | Enhances the diagnostic performance of PCa * | [57] |
| Functionalized Goal | Functionalized Strategy | Small Molecule | Principle | Effect | Reference |
|---|---|---|---|---|---|
| Construction of a highly sensitive protein MIP biosensor | MIT * | Dopamine | Hydrogen bond, multipoint electrostatic attraction | Achieves the highly sensitive and selective recognition of human C-reactive protein | [59] |
| Development of an electrocatalytically amplified assay for analysis of HER-2 * | Reaction of Au with mercapto groups | Polyethylene glycol | Au-S bond | Achieves sensitive and specific sensing of cancer biomarkers in serum | [60] |
| Preparation of an electrochemical sensor via the co-assembling of DNA probe and 6-mercapto-1-hexanol onto a gold electrode | Reaction of Au with mercapto groups | 6-mercapto-1-hexanol | Au-S bond | Analyzes dynamic DNA methylation process | [61] |
| Integration of visual readings and ratiometric ECL analysis on paper substrates | Direct adsorption | Luminol | Au-N bond, electrostatic interaction | Obtains accurate monitoring performance for H2O2 | [65] |
| Preparation of a new luminol-ROS ECL system to detect GSH * | Direct adsorption | Luminol | Au-N bond, electrostatic interaction | Greatly promotes the ECL emission | [66] |
| Forster resonance energy transfer based ratiometric imaging of lysosomal HOCl * | Redox reaction | Non-fluorescent spirothioether unit | Electron transfer | Exhibits distinct biochemical properties for facile monitoring of HOCl via conventional flow cytometry | [68] |
| Exhibition of high detection sensitivity to OONO− * | Chemical reaction | Cyanine 3, cyanine 5 | Condensation reaction | Produces a ratiometric fluorescence signal | [69] |
| Development of “smart” noninvasive bioimaging probes for trapping specific enzyme activities | Substitution reaction | β-galactosidase | Nucleophilic substitution | Real-time fluorescence quantification; capture of in vivo and in situ β-GAL activity | [70] |
| Use of new fluorophore (azulene) to prepare an effective two-photon fluorescent probe | Inversion of internal charge transfer | Boronate | Electron transfer | Detects reactive oxygen species; has good cell penetration | [71] |
| Development of fluorescent probes that can show different modes of fluorescence signals fo distinct concentrations | Substitution reaction | 2,4-dinitrobenzenesulfonate, the chloro group | Nucleophilic substitution | Shows the signal in the low-concentration range of thiols and the ratio response to high-concentration thiols | [72] |
| A conformationally induced “off–on” tyrosine kinase cell membrane fluorescent sensor | Linker group connectivity | Sunitinib, pyrene | Hydrogen bond | Enables fluorescence microscopy imaging of receptor protein tyrosine kinases in the cell membranes of living cells | [73] |
| Design of a mitochondria-specific coumarin pyrrolidinium-derived fluorescence probe | Linker group connectivity | 7-diethylamino-coumarin moiety | Hydrogen bond | Allows real-time ratio imaging of HCN in living cells | [74] |
| Functionalized Goal | Functionalized Strategy | Biomacro-Molecule | Principle | Effect | Reference |
|---|---|---|---|---|---|
| Construction of a 3D porous Ti3C2Tx MG * hybrid film for the determination of glucose in serum | Membrane encapsulation | GOx | Wrapped | Enhances the stable fixation and retention of GOx in the membrane | [76] |
| Preparation of advanced functional nanostructures based on Co3O4@MCF * | Physical adsorption | GOx | Hydrogen bond | Highly selective detection of glucose | [77] |
| Study of the bio-interaction properties of PP hydrogel composed of HEMA * and DEAEMA * on SPE | Covalent binding | GOx | Covalent bond | Optimizes the response of the sensor via different biomolecular ratios | [7] |
| Construction of a targeted induced hairpin-mediated biosensing interface | Reaction of Au with mercapto groups | Hairpin DNA | Au-S bond | Enhances the effect of probe DNA on the detection performance of ATPA * and ATP * | [80] |
| Development of a dual-ratiometric electrochemical apta-sensing strategy for the simultaneous detection of AFB1 * and OTA * | Complementary base pairing | ssDNA | Hydrogen bond | Greatly improves the assembly and recognition efficiency of the sensing interface | [82] |
| Development of an electrochemical biosensor based on DNA-modified Fe3O4 @ Au magnetic NPs for the detection of trace heavy-metal ions | Reaction of Au with mercapto groups | DNA | Au-S bond | Detects heavy-metal ions with no obvious interference at the same time; maintains the high sensitivity | [83] |
| Proposition of an electrochemical aptamer sensor based on CRISPR/CAS12a | Reaction of Au with mercapto groups | Polyadenine DNA | Au-S bond | Detects COVID-19 NPs * rapidly and is ultrasensitive | [84] |
| Preparation of an electrochemical biosensor by using a specific DNA skeleton-DNA tetrahedron | Covalent binding | DNA tetrahedron | Covalent bond | Detects a variety of bioactive molecules with high signal-to-noise ratio, sensitivity, and specificity. | [85] |
| Construction of an electrochemical biosensor by introducing DNAzyme with peroxidase-like activity into the junction of hydrogel | Embedding binding | DNAzyme | π–π conjugation, hydrophobic interaction | Overcomes the limitation of two-dimensional electrode, obtains long-distance catalytic signal of DNAzyme | [86] |
| Construction of a specific and long-acting antifouling biosensor interface based on protein-imprinted hydrogel | Complex | Template protein IgG * | Multiple-point electrostatic interaction, hydrogen bond | Detects target immunoglobulins in complex biological samples | [87] |
| Preparation of a new gas-sensitive-imprinted hydrogel via free-radical polymerization | Free-radical polymerization method | π bond is broken and start of polymerization reaction | Human serum albumin (HSA)-template proteins | Shows unique self-recognition characteristics to HSA protein | [92] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.; Xie, Y.; Zhu, W.; Wei, T. Recent Advances in Functionalization Strategies for Biosensor Interfaces, Especially the Emerging Electro-Click: A Review. Chemosensors 2023, 11, 481. https://doi.org/10.3390/chemosensors11090481
Wang F, Xie Y, Zhu W, Wei T. Recent Advances in Functionalization Strategies for Biosensor Interfaces, Especially the Emerging Electro-Click: A Review. Chemosensors. 2023; 11(9):481. https://doi.org/10.3390/chemosensors11090481
Chicago/Turabian StyleWang, Feiyu, Yiwen Xie, Weijie Zhu, and Tianxiang Wei. 2023. "Recent Advances in Functionalization Strategies for Biosensor Interfaces, Especially the Emerging Electro-Click: A Review" Chemosensors 11, no. 9: 481. https://doi.org/10.3390/chemosensors11090481
APA StyleWang, F., Xie, Y., Zhu, W., & Wei, T. (2023). Recent Advances in Functionalization Strategies for Biosensor Interfaces, Especially the Emerging Electro-Click: A Review. Chemosensors, 11(9), 481. https://doi.org/10.3390/chemosensors11090481






