Abstract
Molecular recognition based on non-covalent interactions between two or more molecules plays a crucial role in biological systems. Specific biological molecule recognition has been widely applied in biotechnology, clinical diagnosis, and treatment. The efficiency and affinity of molecular recognition are greatly determined by the spatial conformation of biomolecules. The designability of DNA nanotechnology makes possible the precise programming of the spatial conformation of biomolecules including valency and spacing, further achieving spatial pattern recognition regulation between biomolecules. This review summarizes recent achievements with DNA-based molecular spatial pattern recognition systems, the important factors affecting spatial pattern recognition, and their applications in biosensing, bioimaging, and targeted therapy. The future challenges in and development of this field are discussed and prospected. This review will provide valuable guidance for the creation of new DNA tools to enhance the efficiency and specificity of biomolecular recognition.
1. Introduction
In living organisms, molecular recognition based on the specific interaction between molecules is a crucial foundation for maintaining life, as mutual recognition and interaction between biomolecules are essential for processes such as cell signaling, metabolism, cell proliferation, and differentiation [1]. For example, cell-surface receptor molecules can interact with signaling molecules, triggering intracellular signal transduction to regulate cellular physiology and metabolism [2,3]. Enzymes can recognize their substrate molecules, catalyzing the conversion of the substrate [4,5]. In the field of biotechnology, specific molecular recognition also has a wide range of applications in the diagnostic and biosensing fields. For example, the recognition of specific proteins on the surface of tumor cells can facilitate early diagnosis and targeted treatment of tumors [6,7]. Biosensors leverage molecular recognition principles to identify particular molecules, harnessing the specificity of enzymes to detect their corresponding substrates [8,9].
In biomolecular recognition, non-covalent interactions such as hydrogen bonds, ionic interactions, and van der Waals forces between molecules in the binding site are usually involved. These non-covalent interactions usually occur based on the spatial conformation of biomolecules [10]. For example, proteins on the cell surface cluster into a specific pattern and interact with ligands through specific structural domains, triggering downstream biological processes such as immune response and cell adhesion. Therefore, the conformation of biomolecules plays an important role in biomolecular recognition. Recently, by linking biomolecules to organic/inorganic or biological scaffolds, researchers have constructed ex vivo molecular recognition systems. Various spatial conformations were constructed by adjusting the valence and spacing of biomolecules on the scaffold [11]. Efficient biomolecular spatial pattern recognition and relevant applications, including targeted diagnosis, can ultimately be achieved.
In addition to polymers [12], nanoparticles [13], and dendritic polymers [14], nucleic acids have emerged as novel scaffold materials for biomolecule coupling. With the development of DNA chemical synthesis technology, various chemical groups, including fluorescent dyes, thiols, and biotin, can be modified on DNA chains. Directional coupling and the precise spatial arrangement of various biomolecules can be achieved. In addition, DNA molecules have excellent biocompatibility and biological stability. In particular, DNA technology enables a class of one-dimensional to three-dimensional nanostructures formed by the precise Watson–Crick base pairing of DNA [15]. DNA nanostructures exhibit unparalleled monodispersity and atomic-level precision when compared to other nanoparticles [16]. Excellent chemical addressability, precise assembly, and highly ordered conformation make DNA nanostructure the perfect template for a variety of nanoscale molecules, such as small molecules, proteins, liposomes, and nanoparticles [17,18]. Thus, biomolecular spatial pattern recognition can be easily programmed by DNA [19]. This review provides a comprehensive overview of DNA-based molecular spatial pattern recognition systems and DNA biomolecule conjugation chemistry. We emphasize the crucial factors influencing spatial pattern recognition and explore their applications (Scheme 1). Furthermore, we examine the challenges encountered in the advancement of DNA-programmed biomolecular spatial pattern recognition and propose future directions for this field.
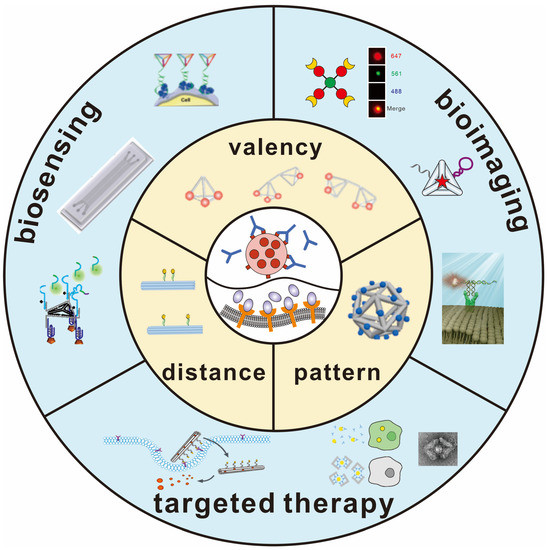
Scheme 1.
Schematic illustration of DNA-programmed biomolecular spatial pattern recognition.
2. Biomolecular Systems Based on Spatial Pattern Recognition
Specific recognition between antibodies and antigens is a common molecular recognition in biological systems and is typically used to recognize and clear foreign substances or abnormal cells in living organisms (Figure 1a). Antibodies, which are proteins generated by the immune system, possess the ability to identify and attach themselves to particular antigen molecules, creating a complex interaction with the antigen. The recognition and binding between antibodies and antigens are highly specific, with only antigens that precisely match the antibody being capable of recognition and binding [20]. In addition, antigen molecules on the surface of tumor cells or viruses are often multivalent and arranged into specific graphic patterns. For example, the number and arrangement of spikes on the surface of enveloped viruses differ among various viral species, leading to distinct characteristics in terms of infection mechanisms and behaviors. Therefore, multivalent antibodies are often designed to match the spatial conformation of antigens to enhance the affinity of antigens and antibodies through molecular spatial pattern recognition in antibody-based virus neutralization and tumor therapy [21]. For example, the conformation of complementary-determining regions (CDRs) of natural antibodies can be reconstructed on gold nanoparticles to achieve a high binding affinity with target antigens [22].
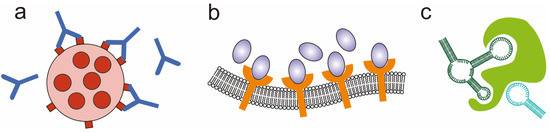
Figure 1.
Biomolecular spatial pattern recognition based on antibody–antigen (a), receptor–ligand (b), and aptamer–target (c).
Receptor–ligand recognition is another ubiquitous molecular recognition in vivo (Figure 1b). Receptors are proteins on the cell membrane that can bind to specific ligands, triggering biological reactions inside the cell. Ligand molecules often include metal ions, nucleic acids, proteins, etc. [23]. The recognition between receptor and ligand molecules is highly selective. Ligands with specific chemical and structural features bind to specific receptors. This selectivity ensures the specificity and effectiveness of biological reactions [24]. Since the receptor molecules often cluster on the cell membrane, spatial pattern recognition between receptor and ligand has been applied in the studies of receptor-mediated biological processes. Palma’s group proposed a biomimetic nanoscale array fabrication strategy based on triangular origami. By assembling integrin-specific binding ligands and epidermal growth factor (EGF) in a multivalent form in patterned nanoscale arrays, they studied receptor–ligand recognition in the spread of cancer cells with nanoscale spatial resolution and single-molecule control. They demonstrated that the synergistic effect of integrin and EGF in the spread of melanoma cells is related to the number and ratio of the two ligands [25]. Dutta et al. designed a six-helix DNA origami tension probe by customizing different numbers of ligands on DNA origami. They used single-molecule force spectroscopy to measure the tension signal of platelets and found that the total tension signal of the platelets increases with the number of modified ligands on the probe [26].
Nucleic acid aptamers, characterized by unique secondary and tertiary structures, are single-strand nucleic acid oligomers that possess the capability to selectively bind to specific target molecules (Figure 1c). These aptamers are commonly referred to as chemical antibodies and offer several advantages over traditional antibodies due to their high specificity [27]. First, the molecular weights of nucleic acid aptamers are relatively small and their structures are simple. Therefore, they penetrate cell membranes more easily and can bind to target molecules within the cell. Second, aptamers are generated from an in vitro process known as the systematic evolution of ligands by exponential enrichment (SELEX), which is more economical and can be prepared on a large scale. Last, nucleic acid aptamers can maintain their structural stability over a wide range of temperatures and pH. Their resistance to degradation by proteases makes them well-suited for utilization in intricate biological systems [28]. In addition, aptamers can be easily modified with different functional groups. Therefore, nucleic acid aptamers have been increasingly used in the research and application of biomolecular recognition [29]. Significantly, drawing inspiration from natural multivalent interactions, researchers have ventured into exploring multivalent aptamers that exhibit enhanced binding affinity, heightened specificity, and extended circulation duration compared to monomeric aptamers [30]. The rational conformational design of multivalent aptamers is key to their target recognition. For example, human thrombin aptamers with defined distances and orientations were established to study the effect of the aptamer geometry on binding properties. It has been proven that optimization of the spatial pattern of the aptamers can improve the affinity of binding efficiently [31].
3. Approaches for the Construction of DNA-Based Multivalent Biomolecules
When establishing a DNA-based biomolecular recognition system, the conjugation of DNA and biomolecules is the first and most important step. There are several highly specific and efficient methods for modifying biomolecules on DNA, including biotin–avidin interactions, and click chemistry reactions. The approaches are mainly divided into non-covalent conjugation and covalent conjugation based on the mechanism of the reaction (Table 1) [32,33].
3.1. Non-Covalent Conjugation
The non-covalent conjugation between DNA and biomolecules is realized through the interaction between the connecting molecules on DNA and ligands such as biotin–avidin, Ni2+–NTA–Histag, etc.
The non-covalent interaction between biotin and avidin is a highly specific and high-affinity binding interaction, commonly employed in the conjugation of DNA with biomolecules. With the maturation of solid-phase DNA synthesis technology, biotin-modified oligonucleotides have been commercialized. The common strategy is to conjugate biotin-modified DNA with biomolecules modified with tetrameric streptavidin protein under mild conditions. Using this strategy, different ligands have been ligated to DNA nanostructures [34,35,36]. However, this method has some drawbacks. For example, the large size of tetrameric streptavidin may cause steric hindrance, which will affect the efficiency of ligation. The structure of the tetramer may lead to uncertain stoichiometry between DNA and biomolecules. These problems may be solved by using monomeric avidin, but the binding affinity would be weakened accordingly.
Ni2+–NTA–Histag non-covalent interaction is another commonly used linking strategy. In the presence of Ni2+ ions, molecules labeled with multiple histidine residues (Histags) are bound to NTA-labeled DNA to construct DNA–biomolecule complexes [37]. This interaction is reversible in the presence of strong chelators such as ethylenediaminetetraacetic acid (EDTA). As the protein labeled with multiple histidine residues can be easily fused with the target protein through protein engineering, this method is more commonly used for the coupling of protein and DNA [38].
In addition, non-covalent interactions between proteins are also used to construct DNA–protein complexes. Protein A/G is a protein ligand that binds specifically to the Fc region of antibodies. The binding between protein A/G and the Fc region of antibodies is achieved through a combination of electrostatic interactions, hydrophobic interactions, and hydrogen bonding. These forces enable the formation of a stable complex between protein A/G and the Fc region of antibodies. A ProA/G-dRep fusion protein was developed for the conjugation of DNA and antibody with precise stoichiometry. In addition, protein-binding cyclic peptides can also bind to certain proteins through non-covalent interactions. Gothelf’s group selected the FC-III, a 13-amino-acid cyclic peptide binder of the human immunoglobulin G (Ig G) Fc domain, to direct DNA protein conjugation. Quantitative conversion of DNA is achieved at low stoichiometries and the reaction can be performed in complex biological matrixes, such as cell lysates [39]. Although the non-covalent interaction between proteins can precisely control the ratio of DNA to protein in conjugation, it is not universally applicable to other biomolecules because such interactions occur mostly in antibodies. Some DNA binding proteins that bind to the DNA of specific sequences non-covalently, such as Zinc-finger protein and transcription activator-like effector nucleases (TALEN), also have potential in DNA biomolecule conjugation [40,41].
3.2. Covalent Conjugation
Compared with non-covalent conjugation, covalent conjugation provides a much stronger binding between DNA and biomolecules, further improving the stability of DNA–biomolecule complexes and promoting their applications in physiological environments.
The most commonly used strategy of covalent conjugation is to use the native functional group on the biomolecules. For example, covalent reactions occur between the amine group in lysine and different chemical groups modified on DNA, such as carboxyl. The thiol group is also an active group for covalent reactions. In addition to direct reactions such as acid–base condensation reactions, some heterobifunctional crosslinking agents, such as succinimide 3 (-2-pyridyl dithionyl) -propionate (SPDP) and sulfonyl succinimide 4- (n-maleimide methyl) cyclohexane-1-carboxylate (Sulfo-SMCC) were also introduced in the conjugation of two different chemical groups [42,43]. The use of a native chemical group of biomolecules for conjugation is simple but lacks selectivity. When more than one functional group used for ligation exists in biomolecules, it is difficult to control the valency and direction of the coupled DNA. Therefore, click chemistry with high selectivity, fast reaction rates, and few side reactions has been more and more applied in the conjugation of DNA and biomolecules [44,45]. Cu(I)-catalyzed azide–alkyne cycloaddition (CuAAC), the first widely accepted click reaction, was first proposed by Sharpless in 2001 [46]. Alkyne-labeled DNA strands have been conjugated with the genetically incorporated azide group on glycoproteins or enzymes to form DNA–biomolecule complexes. However, the requirement for copper ions as a catalyst limits its application in vivo. Later, a copper-free click reaction was proposed by Bertozzi and widely used for biomolecule conjugation [47]. One of the most commonly used reactions is strain-promoted alkyne–azide cycloaddition (SPAAC), which conjugates azide and dibenzocyclooctyne (DBCO). Knappe and colleagues developed a versatile DNA origami functionalization platform based on SPAAC, realizing the in situ conjugation of carbohydrates, small molecules, peptides, polymers, and proteins to DNA scaffolds [48].
Another method for covalent linkage is to introduce a tag protein on the biomolecules, which provides a self-catalytic reaction site for reaction with modified DNA. The commonly used tag proteins are O6-alkylguanine-DNA-alkyltransferase (SNAP-tag) [49] and haloalkane dehalogenase (Halo-tag) [50]. SNAP-tag transfers the benzyl group on O6-benzyl-guanine-modified DNA to cysteine, while phenylalanine in Halo-tag undergoes displacement reaction with chlorine atoms on DNA, forming a covalent linkage between the ligand and DNA. Since these two reactions are orthogonal, multiple biomolecules can be simultaneously modified on DNA using different tags [51].

Table 1.
Summary of approaches for the construction of DNA-based multivalent biomolecules.
Table 1.
Summary of approaches for the construction of DNA-based multivalent biomolecules.
| Type | Modification | Reference | |
|---|---|---|---|
| Non-covalent | Biotin–avidin interaction | Biotinylated protein and avidin-modified DNA | [34] |
| Ni2+–NTA–Histag interaction | Protein bearing histidine clusters and NTA-modified DNA | [38] | |
| Protein–protein interaction | Protein A/G or protein binding peptide-modified DNA | [39] | |
| Protein–DNA interaction | Protein bearing Zinc-finger protein/TALEN | [41] | |
| Covalent | Heterobifunctional crosslinking | Amine and thiol modification | [43] |
| Click chemistry | Azide and alkyne modification | [48] | |
| Tag-protein-mediated conjugation | Protein fused with SNAP-/Halo-tag | [51] | |
4. Factors Affecting Spatial Pattern Recognition
After conjugating DNA with biomolecules, the spatial conformation of these biomolecules can be arranged precisely according to design. The interaction between receptor and ligand molecules with different spatial conformations can be studied with the help of many techniques including X-ray crystallography, nuclear magnetic resonance, and surface plasmon resonance. So far, researchers have investigated several factors that influence spatial pattern molecular recognition by leveraging the programmability of DNA. By optimizing these factors, they have successfully achieved efficient molecular recognition.
4.1. Valency
Spatial pattern recognition often involves multivalent interactions between multiple biomolecules. In nature, multivalent interactions are characterized by the binding of multiple ligands on one biological entity with multiple receptors on another, exhibiting characteristics that are not present in monovalent interactions [52,53]. In contrast to weak monovalent binding, multivalent interactions substantially augment the molecular-level binding between receptors and ligands. For example, the multivalent interaction between a virus and its host cell allows the virus to stably adhere to the cell surface, achieving efficient invasion [54]. Immunoglobulin M (IgM) is the first defense for the body against foreign pathogens, typically binding to antigens in the form of pentamers, and subsequently activating the complement response [55]. Hence, through chemical synthesis that mimics endogenous multivalent biomolecular arrays, researchers can investigate the pivotal role of multivalent interactions in molecular spatial pattern recognition. Moreover, it becomes possible to construct novel multivalent biomolecular patterns that can alter the efficiency of molecular recognition through mechanisms beyond the capabilities of natural substances [56].
Low-valence multivalent biomolecules can be constructed by the site-specific conjugation of molecules on single- or double-strand DNA, which is also called DNA functionalization or modification. These multivalent biomolecules, such as proteins, small molecules, and nucleic acids, can achieve the selective recognition and quantitative measurement of the target molecules. In addition to single- and double-strand DNA, DNA nanostructures are more widely used in the construction of multivalent biomolecules. DNA nanostructures are self-assembled from multiple DNA strands with high precision and predictability, with one-dimensional to three-dimensional sizes and geometries [15,16]. For example, the DNA tetrahedron is a three-dimensional framework DNA nanostructure, usually formed by the hybridization of four DNA single strands [57]. Small molecules, peptides, nucleic acids, and other biomolecules can be ligated to the four DNA single strands through covalent or non-covalent connections, forming multivalent biomolecules with different valencies (primarily 1–4 valence) [58,59,60]. Li et al. connected 1–4 unmethylated CpG oligodeoxynucleotide sequences to the vertices of a DNA tetrahedron. The multivalent CpG specifically recognizes Toll-like receptor 9 (TLR9) on the surface of macrophages and successfully activates downstream immune regulatory functions [61] (Figure 2a). Other DNA nanostructures composed of a few single-strand DNA materials such as DNA G-quadruplex and DNA tile, can also be used as scaffolds for low-valence multivalent biomolecule construction. Liu et al. designed trivalent and tetravalent nucleic acid aptamers based on the J1 connection and 4 × 4 DNA tile structures, both of which showed strong affinity when binding to target cells. Based on this, two types of nucleic acid aptamers were linked to the end of a DNA tile dimer to construct an octavalent double-specific nucleic acid aptamer, which mediated the connection between the two types of cells [62].
Sometimes, it is necessary to construct tens or even hundreds of multivalent biomolecules for molecular recognition, where DNA origami serves as a good candidate for a multivalent scaffold. DNA origami is a type of bottom-up synthesized nanostructure ranging in size from tens of nanometers to sub-micrometers. A typical DNA origami structure contains about 200 staple strands with unique sequences and positions. By conjugating biological molecules to these staple strands, multivalent biomolecules with hundreds of valencies can, in principle, be constructed [63]. Song’s group precisely arranged 10 to 90 SARS-CoV-2 RBDs (receptor binding domains) on a ~74 nm DNA soccer-ball origami structure and studied the multivalent molecular recognition between RBD and ACE2 (angiotensin-converting enzyme 2) on the host cell. It was found that both the affinity and the rate of the DNA-based multivalent RBDs binding to the host cell increase with the RBD number [64] (Figure 2c). Furthermore, certain DNA nanostructures, such as DNA tetrahedra, can serve as fundamental units for constructing higher-order nanostructures. By attaching biomolecules to each DNA nanostructure monomer, multivalent molecular recognition can be achieved, thereby expanding the capabilities and applications of these DNA-based architectures. Yang’s group arranged sub-10 nm DNA tetrahedrons on a microfluidic chip and ligated nucleic acid aptamers (SYL3C) to them. The multivalent nucleic acid aptamer formed on the interface increased the affinity with human colon cancer cells (SW480) by about four times compared to free nucleic acid aptamers [65]. DNA nanostructures self-assembled via the hybridization chain reaction (HCR) or rolling circle amplification (RCA) can also serve as multivalent nanoscaffolds. Tan’s group designed and constructed several nanostructures with multivalent aptamers such as “nanocentipede” and “nanoflower”, which specifically bind to target cells with high affinity through multivalent recognition [66,67] (Figure 2b). Nonetheless, molecular recognition based on these structures can only be studied qualitatively due to the unpredictable nature of the reactions.
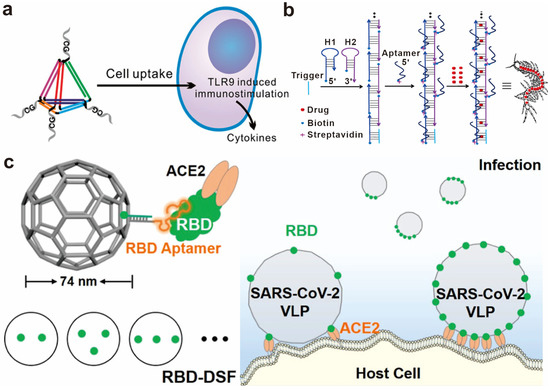
Figure 2.
Biomolecular spatial recognition based on valency regulation. (a) Schematic showing of the assembly of a CpG-bearing DNA tetrahedron and its immunostimulatory effect. Reprinted with permission from [61], copyright 2011, American Chemical Society. (b) The nanocentipede consists of multivalent aptamers as a function of targeting moieties to target cells. Reprinted with permission from [66], copyright 2016, American Chemical Society. (c) Schematic illustration for the assembly of RBDs on a DSF and the interaction with the host cell. Reprinted with permission from [64], copyright 2022, American Chemical Society.
In general, a higher number of valence states in biomolecules provides an advantage in molecular recognition due to the multivalent synergistic effect. Nevertheless, several studies have indicated the presence of a valence threshold in specific molecular recognition systems, wherein the affinity no longer improves beyond a certain number of valence states, despite an increase in their quantity. Consequently, it becomes desirable, from both economic and practical perspectives, to determine the precise threshold of valence states for optimal molecular recognition. In the DNA-based multivalent biomolecular systems mentioned above, the structures formed through HCR and RCA result in a substantial amplification of valence states due to the repetitive units of the structure. However, the lack of control in their reactions hinders the assurance of precise and uniform valence states. Conversely, multivalent biomolecules based on DNA tetrahedra and DNA origami provide accurate manipulation of valence states, offering greater advantages in research on biomolecular recognition that relies on spatial conformation.
4.2. Distance
The distances between molecules play a crucial role in molecular recognition, particularly in spatial pattern recognition, as they provide valuable information about specific interaction patterns. Biomolecules, in particular, can exhibit even distribution on the cell membrane or localize in microdomains due to the fluidity of the phospholipid bilayer. Consequently, the distances between biomolecules can vary depending on the cellular state [68]. Understanding and analyzing these distances is essential for deciphering molecular recognition processes and spatial patterns [69]. DNA nanostructures enable precise localization of biomolecules. Among them, DNA origami with strong designability, span scales from nanometers to hundreds of nanometers, perfectly matching the scale range of molecular arrangements on the cell surface. In addition, the remarkable spatial addressability of DNA origami allows for the precise anchoring of biomolecules at the nanometer scale. These capabilities enable researchers to investigate biomolecular networks with controlled distances, facilitating in-depth studies in distance-dependent molecular recognition.
In 2008, Yan’s group first utilized the spatial addressing ability of self-assembled DNA nanoscaffolds to construct bivalent aptamers with different distance intervals for recognizing thrombin protein and visualized this interaction at the single-molecule level. The bivalent aptamers with the highest thrombin binding activity were found to have a distance of 5.8 nm between the two aptamers [70] (Figure 3a). Similarly, Shaw et al. modified DNA origami with the Eph receptor tyrosine kinase ligand ephrin-A5, and constructed a series of “nano-rulers” with different ligand spacing to study the role of distances between ephrin-A5 in Eph receptor recognition and receptor-mediated signal transduction [71]. Furthermore, various biomolecules such as caspase-9 monomers and cell-binding ligand RGD have been strategically positioned on diverse DNA nanostructures. These investigations aim to elucidate the impact of spacing on the molecular recognition capacity and the effectiveness of downstream signal transduction [72,73].
An antibody molecule typically consists of two antigen-binding fragments (Fab), with each Fab region capable of binding to an antigen. Consequently, a single antibody molecule can simultaneously bind to two identical or different antigens. This bivalent binding mechanism plays a vital role in enhancing the antibody’s affinity and specificity, thereby increasing its effectiveness in immune responses. As a result, the distance between antigens becomes crucial in immune reactions. By precisely controlling the spacing of antigen molecules, it is possible to optimize antigen–antibody recognition and interactions, enhance the efficacy and selectivity of immune responses, and provide guidance and principles for the design and optimization of molecular vaccines. For example, Shaw et al. used DNA origami to precisely control the distance between antigens and characterized the binding of antibodies with identical antigen-binding domains. They found that the antibodies bound to two antigens at distances of 3–17 nm, and the binding affinity varied with the distance between the antigens, with the highest affinity observed for antigens at a distance of approximately 16 nm [74]. Fan’s group constructed artificial antigen epitopes of 3–20 nm by anchoring antigens onto triangular DNA origami and used high-speed atomic force microscopy to image the antigen–antibody interaction at the single-molecule level, providing dynamic evidence for the antigen-binding process of IgG from monovalent to bivalent [75] (Figure 3b). In addition to antibody binding, antigens can also activate B-cell receptor (BCR) signaling pathways to activate antigen-specific B cells. To further explore the effect of antigen spacing on IgM–BCR activation, Bathe’s group conjugated the clinical vaccine immunogen eOD-GT8 (an HIV-1 glycoprotein-120 external domain) to the surface of icosahedral and six-helix bundle DNA origami at different numbers and distances. It was shown that the activation of B cells was maximized with an antigen spacing of 25–30 nm. This work provides optimization principles for the design of molecular vaccines based on B-cell immune responses [76] (Figure 3c).
Overall, the influence of spacing on biomolecular recognition varies according to the specific recognition system. It is necessary to investigate the optimal distance within specific types of recognition biomolecules and environments to attain the highest efficiency in molecular recognition, which can ultimately be applied in biomedical fields such as disease diagnosis, vaccine design, and other related areas.
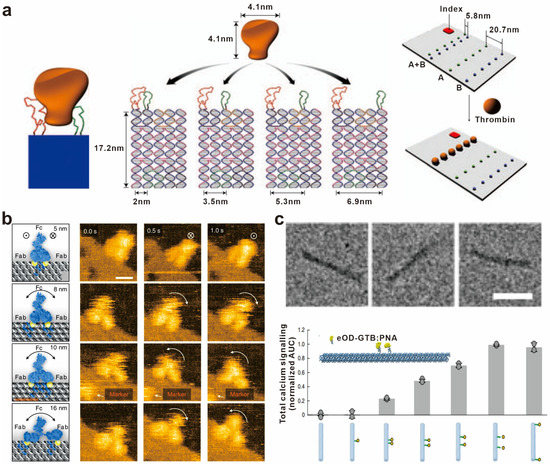
Figure 3.
Biomolecular spatial recognition based on distance regulation. (a) Schematic showing a rigid DNA tile (blue) spatially separating two aptamers (red and green) at a controlled distance for bivalent binding. Reprinted with permission from [70], copyright 2008, Springer Nature. (b) Snapshot HS-AFM images of single IgGs captured by DNA origami epitopes with designed distances of 5, 8, 10, and 16 nm, respectively. Reprinted with permission from [75], copyright 2020, Springer Nature. (c) TEM images of six-helix bundle DNA origami and the total calcium signaling in B cells stimulated with DNA-eOD-GT8 dimers with inter-antigen distances between 7 nm and 80 nm. Reprinted with permission from [76], copyright 2020, Springer Nature.
4.3. Pattern Arrangement
Molecular recognition in the biological system is based on specific interactions between ligands and receptors, which often occur on the cell membrane and exhibit topological cluster characteristics. As an illustration, surface antigens found on viruses and bacteria often exhibit a specific spatial topology [77]. Moreover, the response of T-cell receptors (TCRs) to antigens is intricately linked to the precise positioning of the antigen in three-dimensional space [78]. By orienting multiple biomolecules into a certain geometric pattern, multivalent ligands with various topological structures can be formed and specifically recognize target molecules. DNA nanostructures, ranging from one-dimensional to three-dimensional architectures, offer unparalleled advantages over other materials when it comes to the precise spatial arrangement of molecules. Through DNA structure design and modification site engineering, biomolecular patterns of diverse shapes can be constructed. Moreover, by precisely controlling the spatial arrangement of multivalent ligands on DNA, the affinity with target molecules can be significantly enhanced.
The algebraic topology of biomolecules can be arranged on a DNA framework to match the receptor patterns on the cell surface. Fan’s group topologically rearranged 1–3 aptamers on a DNA tetrahedral framework including point, line, and surface configurations, targeting overexpressed epithelial cell adhesion molecule (EpCAM) on tumor cell membranes. The multivalent ligands distributed in a surface pattern showed the strongest ability to recruit receptor aggregation on the cell membrane, with a 19-fold increase in affinity compared to free aptamers [79] (Figure 4a). By utilizing DNA tetrahedral dimers, researchers were able to broaden the topological configurations of the aptamers. This expansion resulted in altered molecular recognition properties and binding strength by inducing receptor aggregation on the cell membrane [80]. Shen and colleagues built a suite of nuclear pore complex (NPC) mimics by programmably arranging multiple nucleoporin proteins on DNA origami. They designed two octagonal-shaped DNA origami channels to mimic the NPC’s eightfold rotational symmetry. Different numbers of nucleoporin proteins formed different topologies in the structure and showed different binding affinity to the capsid of human immunodeficiency virus 1 (HIV-1), which provides a mechanistic insight for elucidating how viruses enter the nucleus [81] (Figure 4b).
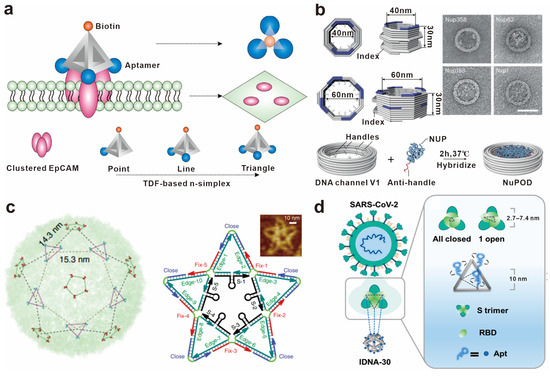
Figure 4.
Biomolecular spatial recognition based on pattern arrangement. (a) Scheme of the DNA-tetrahedra-based n-simplexes and recruitment-binding-induced EpCAM cluster on the cell membrane. Reprinted with permission from [79], copyright 2019, American Chemical Society. (b) Design of DNA origami channels as mimics of the NPC scaffold and the attachment of nup-DNA conjugates to a DNA origami channel by DNA hybridization. Reprinted with permission from [81], copyright 2023, Springer Nature. (c) Distribution of DENV ED3 clusters and the structure of the star-shaped DNA nanostructure carrying 10 envelope protein domain III (ED3) aptamers. Reprinted with permission from [82], copyright 2019, Springer Nature. (d) Spatially matched IDNA-30 against SARS-CoV-2. Reprinted with permission from [83], copyright 2022, American Chemical Society.
Some biomolecules combine into unique shapes and configurations for the specific recognition of target molecules. A noteworthy example is the diverse shapes of surface antigens found on viruses. These epitope configurations serve as specific markers for the viruses and play a vital role in enabling the immune system to recognize and mount an attack against the viral pathogens. The precise design of DNA nanostructures can be used to achieve a highly matched spatial conformation and achieve precise regulation of molecular recognition. A star-shaped DNA nanostructure carrying 10 envelope protein domain III (ED3) aptamers was developed to precisely match the trivalent and pentavalent ED3 epitope on the dengue virus surface, which can be used for virus detection and inhibition [82] (Figure 4c). The aptamers against the SARS-CoV-2 S protein were assembled into a trimeric complex on a DNA nanocage to match the pattern of the S protein trimer in the spatial arrangement, resulting in significantly higher affinity to the S protein trimer [83] (Figure 4d).
Precisely modulating the spatial arrangement of multivalent biomolecules to match the patterns of the target can maximize the strength of multivalent interactions and further enhance the affinity between biomolecules. However, the precise spatial arrangement of biomolecules on DNA nanostructures to achieve spatial pattern recognition can only be accomplished when the exact distribution of target molecules is known. For target molecules with unknown or imprecise distribution, a relatively flexible DNA scaffold may be constructed to enable dynamic and adaptive matching of biomolecules, thereby enhancing the affinity of biomolecular recognition.
5. Application of DNA-Based Spatial Pattern Recognition
DNA scaffolds have been utilized to precisely attach diverse biomolecules, enabling spatially controlled molecular recognition characterized by specificity and strong affinity. Since molecular recognition is crucial in many biomedical fields, such as immunology and drug development, emerging studies have focused on developing DNA-programmed multivalent biomolecules for biomedical applications, such as biosensing, bioimaging, and targeted therapy.
5.1. Biosensing
Based on the interactions between biosensors and the target substance, biosensing technology finds extensive application in the detection and analysis of target molecules within biological systems. Efforts have been made to improve the efficiency and sensitivity of the biosensor, such as developing new construction materials, optimizing the biorecognition element, and introducing a signal amplification strategy [84,85,86,87]. Among them, DNA-based biomolecular recognition can be conducive to highly sensitive biosensors in detecting cancer biomarkers [88]. The overexpression of distinct membrane proteins in tumor cells, which can be specifically recognized by corresponding ligands, plays a vital role in the early diagnosis of cancer. As an illustration, in liquid biopsy, various membrane proteins, such as EpCAM, which are overexpressed on the surface of circulating tumor cells (CTCs), serve as novel biomarkers, offering a broader understanding of the primary tumor tissue for precision medicine. Yang’s group combined microfluidic chips and DNA-tetrahedron-conjugated EpCAM aptamers to ensure highly ordered and perpendicular ligand orientation at the nanoscale, avoiding the non-specific binding and crowding effects of traditional microfluidic interfaces (Figure 5a). The chip they developed improved the capture efficiency of circulating tumor cells (CTCs) by nearly 60% compared to chips with monovalent ligand modification. Additionally, 83% cell release can be achieved by DNase Ι treatment, with a cell survival rate of 91% [65]. In addition, the spatial topology of the EpCAM aptamer was optimized with DNA tetrahedrons, and a capture efficiency of up to 97% for MCF-7 cells injected into whole blood was achieved, which was higher than the antibody capture method (~50%). CTCs from the whole blood samples of cancer patients were successfully captured, with 3–10 CTCs per milliliter for non-metastatic patients and 38–44 CTCs per milliliter for metastatic patients [79] (Figure 5b). In addition to DNA tetrahedra, DNA-origami-based multivalent aptamers were also developed, which match the spatial distribution of target protein clusters and detect CTCs with high affinity [89]. In comparison to DNA tetrahedra, DNA origami employed in CTC detection offers more binding sites. This enables the conjugation of recognition molecules with higher valence and different types. As a result, the affinity for binding with CTC is significantly enhanced. The binding equilibrium constant Kd is reduced from 7 nM (for DNA tetrahedra) to 260 pM (for DNA origami). However, the smaller size and greater flexibility of DNA tetrahedra make them more amenable to be modified at the interface. By combining with a microfluidic system, the thermodynamics of molecular recognition at the modified interface were greatly improved. The strategy can be used in a wide variety of areas, including non-invasive testing, and the interfacial regulation of cellular behavior.
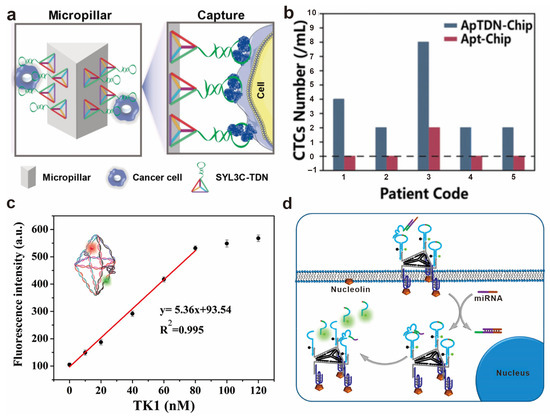
Figure 5.
Application of DNA-based spatial pattern recognition in biosensing. (a) Engineered multivalent aptamers on the microfluidic interface for CTCs capture. (b) CTCs capture performance of the chip with multivalent aptamers for clinical samples. Reprinted with permission from [65], copyright 2020, John Wiley and Sons. (c) TK1 mRNA detection curve of a DNA-octahedron-based fluorescence nanoprobe. Reprinted with permission from [90], copyright 2018, American Chemical Society. (d) DNAzyme walking and amplification imaging of miRNAs within living cells. Reprinted with permission from [91], copyright 2019, American Chemical Society.In addition to biosensors that directly detect molecules on the cell surface, high-affinity molecular recognition also mediates the entry of biosensors into cells for the efficient detection of intracellular biomolecules. After binding to specific molecules on the cell surface, DNA-based multivalent biomolecules can enter cells via endocytosis and detect target molecules inside cells. Lu et al. constructed a DNA octahedron with a divalent AS1411 aptamer, which can simultaneously detect and image two tumor-related mRNAs in living cells, distinguishing target cancer cells from normal cells. The fluorescence signal of the octahedron with the divalent AS1411 aptamer was stronger than that of the octahedron without aptamers, indicating that the DNA-based biosensor can be more effectively taken up by cancer cells through modification with the AS1411 aptamer [90] (c). Xue et al. integrated a DNAzyme walker into a triangular DNA scaffold functionalized with multivalent aptamers for highly sensitive detection of cell miRNA. The movement of the DNAzyme walker was activated by the target miRNA and produced signal amplification [91] (d).
Despite its polyanionic nature, DNA can cross the negatively charged membrane to enter living cells by assembling into specific nanostructures. It was reported that DNA nanostructures approach the membrane primarily with their corners to minimize electrostatic repulsion and their binding affinity and cellular internalization frequency depended on the corner angle of the structures [92,93]. Therefore, three-dimensional DNA nanostructures with corner structures, such as tetrahedra and octahedra, offer greater advantages for intracellular sensing applications than two-dimensional planar structures such as triangles and squares.
5.2. Bioimaging
Molecular recognition has a wide range of applications in biological imaging, including cell imaging, tumor imaging, and molecular localization. Fluorescent probes can be used to detect specific receptors on the surface of tumor cells, assisting in tumor diagnosis and treatment. For example, EpCAM aptamers with different numbers and directions were assembled on a DNA framework to construct a series of probes for the imaging of tumor cells. The directional-yet-flexible probe exhibited adaptability to the receptor distribution on cell surfaces and high affinity against target tumor cells and has the potential of becoming an excellent imaging probe for EpCAM-positive tumors [94] (Figure 6a). Multicolor probes were constructed by arranging different fluorescent molecules and aptamers on DNA tetrahedra. Molecular recognition facilitated the achievement of multiplexed cell imaging and classification of various tumor cells by harnessing the programmability of fractal DNA frameworks [95]. In terms of tumor imaging in vivo, Ding’s research group has employed rational design to create DNA-based probes that feature precisely organized tumor-targeting components along with imaging molecules such as fluorescent groups and magnetic resonance contrast agents. Through this approach, they have successfully achieved specific and non-invasive tumor imaging in mouse models using DNA nanostructures [96,97] (Figure 6b).
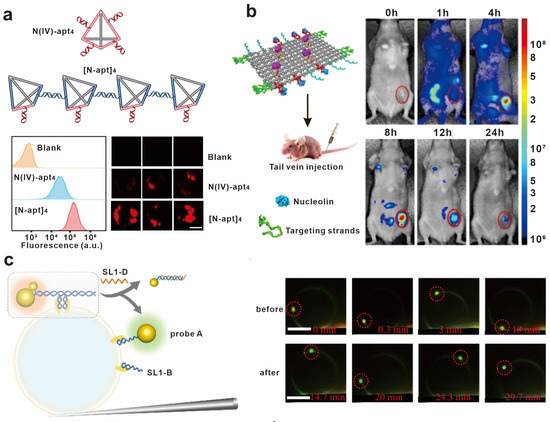
Figure 6.
Application of DNA-based spatial pattern recognition in bioimaging. (a) MCF-7 imaging with tetravalent aptamers and linear aptamer oligomer. Reprinted with permission from [94], copyright 2022, John Wiley and Sons. (b) Imaging of a mouse bearing a human breast tumor before and after intravenous injection of a Cy5.5-labeled nanorobot. Reprinted with permission from [96], copyright 2018, Springer Nature. (c) Schematic illustration of DNA-programmed single-molecule manipulation on giant plasma membrane vesicle and representative images of the vesicle over time before and after the DNA strand displacement. Reprinted with permission from [98], copyright 2023, American Chemical Society.
The biology and chemistry of cells can also be studied by probes based on biomolecular pattern recognition. For example, introducing imaging molecules into specific cellular structures can help in understanding the distribution and dynamic changes of molecules within cells. Xu’s group developed DNA-programmed plasma rulers for imaging the dimerization of the RTK on the cell membrane. The binding of aptamers to cell-surface receptors was regulated by DNA, further mediating the dimerization and dissociation of the receptors. Real-time imaging of RTK dimerization and dissociation processes was realized by detecting the scattered signal caused by the plasma coupling effect of gold nanoparticles. This method provides a more persistent and higher resolution tool for studying protein oligomerization, transport, and dynamics processes, which is of great significance for the in-depth interpretation and precise regulation of cellular signal transduction [98] (Figure 6c).
Overall, in biological imaging systems, DNA origami has an advantage in multimodal imaging due to its numerous modification sites, by simultaneous modifications such as fluorescence and radiolabeling. Conversely, DNA double strands and DNA tetrahedra are more suitable for dynamic imaging due to their structural flexibility.
5.3. Targeted Therapy
One of the main challenges in the treatment of diseases, especially tumors, is the lack of effective and precise targeted delivery systems. Molecular recognition can be used to identify target molecules that are related to the occurrence of diseases. Consequently, there has been extensive research on utilizing modified DNA as drug carriers for targeted therapy, aiming at specific molecules.
Commonly used chemotherapy drugs such as doxorubicin (DOX) can intercalate into DNA base pairs. Nucleotide drugs such as siRNA and antisense oligodeoxynucleotide (ASO) can be hybridized with DNA by Watson–Crick complementary base pairing [99]. After being conjugated with ligands for recognition, DNA specifically delivers these drugs into target cells, without toxic side effects on normal cells. Lin’s group ligated the nucleic acid aptamer AS1411 to DNA tetrahedra (t-FNAs) to target the overexpressed nucleolin protein on tumor cells. Compared to tetrahedra without AS1411, AS1411–tFNA accumulated in the nuclei of MCF-7 cells. Through the additional loading of 5-fluorouracil (5-FU) onto t-FNAs, the successful induction of apoptosis in MCF-7 cells was achieved [100]. In addition to AS1411, other nucleic acid aptamers such as Gint4.T and GMT8 (which specifically recognize platelet-derived growth factor receptor beta and human glioblastoma cells U87MG, respectively) were also ligated to t-FNAs, and the loaded chemotherapeutic drug paclitaxel was target-released in tumor cells [101]. Ge and colleagues reported a DNA origami nanostructure modified with targeting ligand DUPA as an antibody–drug conjugate (ADC) analog for the targeted therapy of prostate cancer. The platform efficiently delivers DOX to prostate-specific membrane antigen (PSMA)-positive cells, and the therapeutic effect is dependent on the number of ligands connected [102] (Figure 7a). Compared to other organic and inorganic nanomaterials, DNA nanostructures demonstrate significant advantages in the targeted delivery of drugs because they can provide large payload capacity and improve cellular internalization with high specific recognition. However, the expression of surface biomarkers on cancer cells is heterogeneous, which causes the inaccurate identification of cancer cells by a single biomarker. To address this issue, Ju’s research group developed a dual-receptor-mediated DNA nanocarrier for siRNA delivery. DNA is self-assembled into a double-lock structure with aptamers Sgc8c and Sgc4f that bind to two specific receptors on the cell surface. The delivery system is active only when both of the aptamers were recognized by cells, thus improving the accuracy of cell identification and greatly reducing the off-target toxicity [103].
Functional molecules can be simultaneously modified on DNA frameworks, providing a powerful platform for multimodal cancer therapy. Zhang et al. combined a DNA tetrahedron modified with a nucleic acid aptamer and a metal–organic framework (MOF) nanoparticle to design a hybrid nanocarrier for loading DOX. The chemotherapy drug was released in cancer cells rich in adenosine triphosphate (ATP) or vascular endothelial growth factor (VEGF). At the same time, a photosensitizer, Zn (II) protoporphyrin IX (Zn (II)-PPIX), was introduced to produce high-intensity fluorescence efficiently and selectively in tumor cells, inducing the production of reactive oxygen species (ROS) and conducting photodynamic therapy on malignant tumor cells [104]. A nanomachine for DNA logic gate operation across cell membranes was designed, realizing precise photodynamic therapy for solid tumors in vivo. This DNA nanomachine consists of upconversion nanoparticle cores (UCNPs), DNA assemblies, and sgc8 aptamer. The overexpressed PTK-7 protein on the cancer cell membrane and the high-expressed miRNA-21 inside the cancer cell serve as two signals trigging the production of intracellular ROS [105] (Figure 7b). A DNA nanorobot was constructed based on a rectangular DNA origami structure for delivering and releasing thrombin in tumors. Thrombin molecules were placed inside the lumen of the tubular DNA nanorobot and then closed by a predesigned anchor chain containing the nucleic acid aptamer AS1411. The nucleic acid aptamer on the outer surface can guide and trigger the release of thrombin, activating clotting at the tumor site and ultimately resulting in tumor necrosis and growth inhibition [96]. The DNA-modified biomolecules not only act as guides for targeted delivery but also function as triggers for drug release or ROS production, thereby significantly enhancing the spatiotemporal control of targeted therapy.
Tumor immunotherapy, as an emerging method for cancer therapy, activates the immune system for better recognition and attacking of cancer cells, inhibiting the growth and metastasis of tumors. Tumor-specific antigens specifically expressed by tumor cells can be recognized by the immune system as foreign and trigger an immune response to eliminate these tumor cells. For example, T cells acquire adaptive immunity through the binding of T-cell receptor (TCR) to the major histocompatibility complex (pMHC) on the surface of antigen-presenting cells (APCs). By designing a series of DNA nanoscale junctions (DNJs) based on DNA tetrahedra with different sizes, the intermembrane distance at the interface of APC and T cells was precisely controlled. It was demonstrated that the axial distance of the immunological synapse plays an important role in T-cell recognition and activation, providing a basis for T-cell immunology research [106]. Recently, researchers constructed a set of self-adjuvant carriers known as framework nucleic acids (FNAs), which possess regulated rigidity and size. These FNAs were utilized to investigate the impact of epitope spacing on the efficacy of peptide vaccines. When epitopes were assembled on FNAs of appropriate size, the recognition between the epitopes and B-cell receptors (BCRs) as well as the immunogenicity, could be efficiently enhanced [107] (Figure 7c). The recognition and activation of T cells and B cells with antigen epitopes can be regulated and optimized through the design of DNA nanostructures. However, current research focuses primarily on regulating distances and there has been a lack of investigations into patterned arrangement recognition for immune cell activation.
In addition to the targeted treatment of tumors, molecular recognition also plays an important role in targeted antiviral therapy. By interfering with the specific molecular interactions between viruses and host cells, the infection of host cells by viruses could be prevented. SARS-CoV-2 infection was inhibited by anchoring neutralizing aptamers on a DNA scaffold in a pattern that matches the spatial configuration of the viral S protein [83]. Dietz’s group conjugated antivirus antibodies to a DNA icosahedral shell in a modular fashion. Viruses were recognized and trapped in the shell, realizing the neutralization of hepatitis B virus and adeno-associated virus [108] (Figure 7d). The virus-trapping strategy mediated by DNA provides a novel approach to antiviral therapy. The use of DNA-based agents potentially circumvents neutralization, phagocytosis, and degradation by pathways of the innate and adaptive immune system targeting protein structures. However, nucleic-acid-specific reactions, such as the activation of pattern-recognition receptors recognizing DNA, may occur in vivo. Thus, assessing the potential adverse effects in organisms remains to be an important challenge in application.
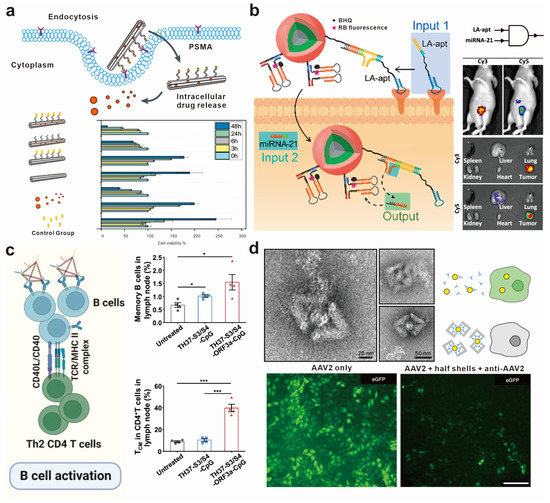
Figure 7.
Application of DNA-based spatial pattern recognition in targeted therapy. (a) Schematic illustration of the ligand-modified six-helical-bundle DNA origami as drug carriers and its working principle. Reprinted with permission from [102], copyright 2020, John Wiley and Sons. (b) Structural diagram of the transmembrane DNA computation and in vivo fluorescence imaging of mice based on different inputs. Reprinted with permission from [105], copyright 2021, American Chemical Society. (c) Immune responses induced by FNA-constructed COVID-19 epitope vaccines. The significance of difference was evaluated by p value; * p < 0.05, *** p < 0.001. Reprinted with permission from [107], copyright 2023, John Wiley and Sons. (d) Negative stain TEM images demonstrating the capture of AAV2 virus particles within antibody-modified DNA origami half shells and fluorescent microscopy images showing the anti-infective effect. Reprinted with permission from [108], copyright 2021, Springer Nature.
6. Conclusions and Perspectives
Advancements in DNA-programmed biomolecular spatial pattern recognition have led to the synthesis of multivalent biomolecules that demonstrate enhanced affinity to receptors, both in vivo and in vitro. These biomolecules exhibit exceptional performance in various applications, including biological detection and targeted therapy. However, DNA-based biomolecular recognition still faces many challenges in future applications: (1) Due to the small differences in the expression levels of some biomolecules in different cells, it is necessary to further optimize the spatial arrangement of biomolecules on DNA scaffolds to achieve precise control of recognition affinity and improve the sensitivity of molecular detection. (2) Presently, DNA-based multivalent ligands are capable of precisely targeting receptors with predetermined conformations on the cell surface. Nevertheless, the cell membrane exhibits mobility, and the distribution of receptors on its surface undergoes dynamic changes. Consequently, the development of adaptive multivalent biomolecules that leverage the adjustability of DNA to accurately match molecules on the cell membrane surface remains an ongoing challenge. (3) In the specific detection of membrane proteins on the cell surface, the cellular uptake of DNA scaffolds may affect the detection sensitivity. To address this issue, spatial pattern recognition can be designed on interfaces such as microfluidic chips and microbeads to eliminate the influence of cellular uptake. (4) Matrices are complex in the biological system and the biomolecular recognition in vivo can be affected by heterogeneity, steric hindrance, and so on. The factors that influence molecular spatial pattern recognition have not been fully elucidated. Computational simulations, such as the simulation of dissipative particle dynamics, may help the construction of a biomolecular recognition model.
Despite these challenges, the studies of DNA-programmed biomolecular spatial pattern recognition have provided compelling evidence for the power of this approach to reveal important physiological processes in cells and promoted the development of biosensing and drug delivery. We believe that with further developments in DNA nanotechnology, synthetic biology, microfluidics, and information technology, more accurate and adaptive control over biomolecular recognition will be realized, providing a powerful tool for the clinical diagnosis and treatment of diseases.
Author Contributions
Conceptualization, L.W. and L.G.; writing-original draft preparation, Y.W. and L.R.; writing—review and editing, L.G.; visualization, H.P.; supervision, L.W.; funding acquisition, L.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key R&D Program of China 2021YFA1200900, the National Natural Science Foundation of China 22022410, 82050005, 12105352.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
There was no new data created or analyzed in this study. Data sharing does not apply to this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Persch, E.; Dumele, O.; Diederich, F. Molecular recognition in chemical and biological systems. Angew. Chem. Int. Ed. 2015, 54, 3290–3327. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef]
- Ruoslahti, E.; Pierschbacher, M.D. New Perspectives in Cell Adhesion: RGD and Integrins. Science 1987, 238, 491–497. [Google Scholar] [CrossRef]
- Ciferri, A. Critical issues in molecular recognition: The enzyme–substrate association. Soft Matter. 2021, 17, 8585–8589. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhao, Y. Molecular recognition of enzymes and modulation of enzymatic activity by nanoparticle conformational sensors. Chem. Commun. 2022, 58, 1732–1735. [Google Scholar] [CrossRef]
- Wu, X.; Chen, J.; Wu, M.; Zhao, J.X. Aptamers: Active targeting ligands for cancer diagnosis and therapy. Theranostics 2015, 5, 322–344. [Google Scholar] [CrossRef] [PubMed]
- Weigelt, B.; Peterse, J.L.; van’t Veer, L.J. Breast cancer metastasis: Markers and models. Nat. Rev. Cancer 2005, 5, 591–602. [Google Scholar] [CrossRef]
- Tong, P.; Zhang, L.; Xu, J.; Chen, H. Simply amplified electrochemical aptasensor of ochratoxin A based on exonuclease-catalyzed target recycling. Biosens. Bioelectron. 2011, 29, 97–101. [Google Scholar] [CrossRef]
- Lin, J.; Ju, H. Electrochemical and chemiluminescent immunosensors for tumor markers. Biosens. Bioelectron. 2005, 20, 1461–1470. [Google Scholar] [CrossRef]
- Jiao, Y.; Qiu, Y.; Zhang, L.; Liu, W.G.; Mao, H.; Chen, H.; Feng, Y.; Cai, K.; Shen, D.; Song, B.; et al. Electron-catalysed molecular recognition. Nature 2022, 603, 265–270. [Google Scholar] [CrossRef]
- Zhao, M.; Wu, T.; Xiao, X.; Liu, Y.; Su, X. New advances in molecular recognition based on biomolecular scaffolds. Anal. Bioanal. Chem. 2013, 405, 5679–5685. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Sefah, K.; Liu, H.; Wang, R.; Tan, W. DNA aptamer-micelle as an efficient detection/delivery vehicle toward cancer cells. Proc. Natl. Acad. Sci. USA 2010, 107, 5–10. [Google Scholar] [CrossRef]
- Sheng, W.; Chen, T.; Tan, W.; Fan, Z.H. Multivalent DNA Nanospheres for enhanced capture of cancer cells in microluidic devices. ACS Nano 2013, 7, 7067–7076. [Google Scholar] [CrossRef] [PubMed]
- Geng, Z.; Wang, L.; Liu, K.; Liu, J.; Tan, W. Enhancing anti-PD-1 Immunotherapy by Nanomicelles Self-Assembled from Multivalent Aptamer Drug Conjugates. Angew. Chem. Int. Ed. 2021, 60, 15459–15465. [Google Scholar] [CrossRef]
- Seeman, N.; Sleiman, H. DNA nanotechnology. Nat. Rev. Mater. 2018, 3, 17068. [Google Scholar] [CrossRef]
- Ge, Z.; Gu, H.; Li, Q.; Fan, C. Concept and Development of Framework Nucleic Acids. J. Am. Chem. Soc. 2018, 140, 17808–17819. [Google Scholar] [CrossRef]
- Ouyang, X.; Wang, M.; Guo, L.; Cui, C.; Liu, T.; Ren, Y.; Zhao, Y.; Ge, Z.; Guo, X.; Xie, G.; et al. DNA Nanoribbon-Templated Self-Assembly of Ultrasmall Fluorescent Copper Nanoclusters with Enhanced Luminescence. Angew. Chem. Int. Ed. 2020, 59, 11836–11844. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Liedl, T. DNA-Assembled Advanced Plasmonic Architectures. Chem. Rev. 2018, 118, 3032–3053. [Google Scholar] [CrossRef]
- Chen, S.; Xu, Z.; Yang, W.; Lin, X.; Li, J.; Li, J.; Yang, H. Logic-Gate-Actuated DNA-Controlled Receptor Assembly for the Programmable Modulation of Cellular Signal Transduction. Angew. Chem. Int. Ed. 2019, 58, 18186–18190. [Google Scholar] [CrossRef]
- Holliger, P.; Hudson, P.J. Engineered antibody fragments and the rise of single domains. Nat. Biotechnol. 2005, 23, 1126–1136. [Google Scholar] [CrossRef]
- Chiu, G.N.; Edwards, L.A.; Kapanen, A.I.; Malinen, M.M.; Dragowska, W.H.; Warburton, C.; Chikh, G.G.; Fang, K.Y.; Tan, S.; Sy, J.; et al. Modulation of cancer cell survival pathways using multivalent liposomal therapeutic antibody constructs. Mol. Cancer Ther. 2007, 6, 844–855. [Google Scholar] [CrossRef]
- Yan, G.; Wang, K.; Shao, Z.; Luo, L.; Song, Z.; Chen, J.; Jin, R.; Deng, X.; Wang, H.; Cao, Z.; et al. Artificial antibody created by conformational reconstruction of the complementary-determining region on gold nanoparticles. Proc. Natl. Acad. Sci. USA 2018, 115, E34–E43. [Google Scholar] [CrossRef] [PubMed]
- Kiessling, L.L.; Gestwicki, J.E.; Strong, L.E. Synthetic multivalent ligands in the exploration of cell-surface interactions. Curr. Opin. Chem. Biol. 2000, 4, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Patel, K.; Perez-Garrido, S.; Marshall, J.F.; Palma, M. DNA Origami Nanoarrays for Multivalent Investigations of Cancer Cell Spreading with Nanoscale Spatial Resolution and Single-Molecule Control. ACS Nano 2019, 13, 728–736. [Google Scholar] [CrossRef]
- Dutta, P.K.; Zhang, Y.; Blanchard, A.T.; Ge, C.; Rushdi, M.; Weiss, K.; Zhu, C.; Ke, Y.; Salaita, K. Programmable Multivalent DNA-Origami Tension Probes for Reporting Cellular Traction Forces. Nano Lett. 2018, 18, 4803–4811. [Google Scholar] [CrossRef]
- Tan, W.; Donovan, M.J.; Jiang, J. Aptamers from cell-based selection for bioanalytical applications. Chem. Rev. 2013, 113, 2842–2862. [Google Scholar] [CrossRef]
- Iliuk, A.B.; Hu, L.; Tao, W.A. Aptamer in bioanalytical applications. Anal. Chem. 2011, 83, 4440–4452. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhang, L.; Yuan, Q.; Tan, J. Current Advances in Aptamer-based Biomolecular Recognition and Biological Process Regulation. Chem. Res. Chin. Univ. 2022, 38, 847–855. [Google Scholar] [CrossRef]
- Lin, M.; Zhang, J.; Wan, H.; Yan, C.; Xia, F. Rationally Designed Multivalent Aptamers Targeting Cell Surface for Biomedical Applications. ACS Appl. Mater. Interfaces 2021, 13, 9369–9389. [Google Scholar] [CrossRef]
- Aghebat Rafat, A.; Sagredo, S.; Thalhammer, M.; Simmel, F.C. Barcoded DNA origami structures for multiplexed optimization and enrichment of DNA-based protein-binding cavities. Nat. Chem. 2020, 12, 852–859. [Google Scholar] [CrossRef] [PubMed]
- Kong, G.; Xiong, M.; Liu, L.; Hu, L.; Meng, H.M.; Ke, G.; Zhang, X.B.; Tan, W. DNA origami-based protein networks: From basic construction to emerging applications. Chem. Soc. Rev. 2021, 50, 1846–1873. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Kong, Y.; Zhao, S.; Xing, H. Engineering Functional DNA-Protein Conjugates for Biosensing, Biomedical, and Nanoassembly Applications. Top. Curr. Chem. 2020, 378, 41. [Google Scholar] [CrossRef]
- Mallik, L.; Dhakal, S.; Nichols, J.; Mahoney, J.; Dosey, A.; Jiang, S.; Sunahara, R.; Skiniotis, G.; Walter, N. Electron Microscopic Visualization of protein assemblies on flatted DNA origami. ACS Nano 2015, 9, 7133–7141. [Google Scholar] [CrossRef]
- Kuzuya, A.; Kimura, M.; Numajiri, K.; Koshi, N.; Ohnishi, T.; Okada, F.; Komiyama, M. Precisely Programmed and Robust 2D Streptavidin Nanoarrays by Using Periodical Nanometer-Scale Wells Embedded in DNA Origami Assembly. Chembiochem 2009, 10, 1811–1815. [Google Scholar] [CrossRef] [PubMed]
- Numajiri, K.; Kimura, M.; Kuzuya, A.; Komiyama, M. Stepwise and reversible nanopatterning of proteins on a DNA origami scaffold. Chem. Commun. 2010, 46, 5127–5129. [Google Scholar] [CrossRef]
- Shen, W.; Zhong, H.; Neff, D.; Norton, M. NTA Directed Protein Nanopatterning on DNA Origami Nanoconstructs. J. Am. Chem. Soc. 2009, 131, 6660–6661. [Google Scholar] [CrossRef]
- Ouyang, X.; De Stefano, M.; Krissanaprasit, A.; Bank Kodal, A.L.; Bech Rosen, C.; Liu, T.; Helmig, S.; Fan, C.; Gothelf, K.V. Docking of Antibodies into the Cavities of DNA Origami Structures. Angew. Chem. Int. Ed. 2017, 56, 14423–14427. [Google Scholar] [CrossRef]
- Nielsen, T.B.; Thomsen, R.P.; Mortensen, M.R.; Kjems, J.; Nielsen, P.F.; Nielsen, T.E.; Kodal AL, B.; Clo, E.; Gothelf, K.V. Peptide-Directed DNA-Templated Protein Labelling for The Assembly of a Pseudo-IgM. Angew. Chem. Int. Ed. 2019, 58, 9068–9072. [Google Scholar] [CrossRef]
- Nakata, E.; Liew, F.F.; Uwatoko, C.; Kiyonaka, S.; Mori, Y.; Katsuda, Y.; Endo, M.; Sugiyama, H.; Morii, T. Zinc-finger proteins for site-specific protein positioning on DNA-origami structures. Angew. Chem. Int. Ed. 2012, 51, 2421–2424. [Google Scholar] [CrossRef]
- Praetorius, F.; Dietz, H. Self-assembly of genetically encoded DNA-protein hybrid nanoscale shapes. Science 2017, 355, eaam5488. [Google Scholar] [CrossRef] [PubMed]
- Fisher, P.D.E.; Shen, Q.; Akpinar, B.; Davis, L.K.; Chung, K.K.H.; Baddeley, D.; Saric, A.; Melia, T.J.; Hoogenboom, B.W.; Lin, C.; et al. A Programmable DNA Origami Platform for Organizing Intrinsically Disordered Nucleoporins within Nanopore Confinement. ACS Nano 2018, 12, 1508–1518. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Liu, M.; Liu, Y.; Woodbury, N.W.; Yan, H. Interenzyme substrate diffusion for an enzyme cascade organized on spatially addressable DNA nanostructures. J. Am. Chem. Soc. 2012, 134, 5516–5519. [Google Scholar] [CrossRef]
- El-Sagheer, A.H.; Brown, T. Click chemistry with DNA. Chem. Soc. Rev. 2010, 39, 1388–1405. [Google Scholar] [CrossRef]
- Tang, W.; Becker, M.L. “Click” reactions: A versatile toolbox for the synthesis of peptide-conjugates. Chem. Soc. Rev. 2014, 43, 7013–7039. [Google Scholar] [CrossRef]
- Kolb, H.C.; Finn, M.G.; Sharpless, K.B. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew. Chem. Int. Ed. 2001, 40, 2004–2021. [Google Scholar] [CrossRef]
- Laughlin, S.T.; Bertozzi, C.R. Metabolic labeling of glycans with azido sugars and subsequent glycan-profiling and visualization via Staudinger ligation. Nat. Protoc. 2007, 2, 2930–2944. [Google Scholar] [CrossRef]
- Knappe, G.A.; Wamhoff, E.C.; Read, B.J.; Irvine, D.J.; Bathe, M. In Situ Covalent Functionalization of DNA Origami Virus-like Particles. ACS Nano 2021, 15, 14316–14322. [Google Scholar] [CrossRef] [PubMed]
- Nakata, E.; Dinh, H.; Ngo, T.A.; Saimura, M.; Morii, T. A modular zinc finger adaptor accelerates the covalent linkage of proteins at specific locations on DNA nanoscaffolds. Chem. Commun. 2015, 51, 1016–1019. [Google Scholar] [CrossRef]
- Koßmann, K.J.; Ziegler, C.; Angelin, A.; Meyer, R.; Skoupi, M.; Rabe, K.S.; Niemeyer, C.M. A Rationally Designed Connector for Assembly of Protein-Functionalized DNA Nanostructures. Chembiochem 2016, 17, 1102–1106. [Google Scholar] [CrossRef]
- Saccà, B.; Meyer, R.; Erkelenz, M.; Kiko, K.; Arndt, A.; Schroeder, H.; Rabe, K.S.; Niemeyer, C.M. Orthogonal Protein Decoration of DNA Origami. Angew. Chem. Int. Ed. 2010, 49, 9378–9383. [Google Scholar] [CrossRef] [PubMed]
- Fasting, C.; Schalley, C.A.; Weber, M.; Seitz, O.; Hecht, S.; Koksch, B.; Dernedde, J.; Graf, C.; Knapp, E.-W.; Haag, R. Multivalency as a Chemical Organization and Action Principle. Angew. Chem. Int. Ed. 2012, 51, 10472–10498. [Google Scholar] [CrossRef] [PubMed]
- Mammen, M.; Choi, S.-K.; Whitesides, G.M. Polyvalent Interactions in Biological Systems: Implications for Design and Use of Multivalent Ligands and Inhibitors. Angew. Chem. Int. Ed. 1998, 37, 2754–2794. [Google Scholar] [CrossRef]
- Muller, M.; Lauster, D.; Wildenauer, H.H.K.; Herrmann, A.; Block, S. Mobility-Based Quantification of Multivalent Virus-Receptor Interactions: New Insights Into Influenza A Virus Binding Mode. Nano Lett. 2019, 19, 1875–1882. [Google Scholar] [CrossRef]
- Czajkowskya, D.M.; Shaoa, Z. The human IgM pentamer is a mushroom-shaped molecule with a flexural bias. Proc. Natl. Acad. Sci. USA 2009, 106, 14960–14965. [Google Scholar] [CrossRef]
- Hernandez-Lopez, R.A.; Yu, W.; Cabral, K.A.; Creasey, O.A.; Pazmino, M.D.P.L.; Tonai, Y.; De Guzman, A.; Mäkelä, A.; Saksela, K.; Gartner, Z.J.; et al. T cell circuits that sense antigen density with an ultrasensitive threshold. Science 2021, 371, 1166–1171. [Google Scholar] [CrossRef]
- Goodman, R.P.; Berry, R.M.; Turberfield, A.J. The single-step synthesis of a DNA tetrahedron. Chem. Commun. 2004, 12, 1372–1373. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Lytton-Jean, A.K.; Chen, Y.; Love, K.T.; Park, A.I.; Karagiannis, E.D.; Sehgal, A.; Querbes, W.; Zurenko, C.S.; Jayaraman, M.; et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotechnol. 2012, 7, 389–393. [Google Scholar] [CrossRef]
- Tian, T.; Xiao, D.; Zhang, T.; Li, Y.; Shi, S.; Zhong, W.; Gong, P.; Liu, Z.; Li, Q.; Lin, Y. A Framework Nucleic Acid Based Robotic Nanobee for Active Targeting Therapy. Adv. Funct. Mater. 2021, 31, 2007342. [Google Scholar] [CrossRef]
- Zhang, T.; Tian, T.; Lin, Y. Functionalizing Framework Nucleic-Acid-Based Nanostructures for Biomedical Application. Adv. Mater. 2022, 34, e2107820. [Google Scholar] [CrossRef]
- Li, J.; Pei, H.; Zhu, B.; Liang, L.; Wei, M.; He, Y.; Chen, N.; Li, D.; Huang, Q.; Fan, C. Self-Assembled Multivalent DNA Nanostructures for Noninvasive Intracellular Delivery of Immunostimulatory CpG Oligonucleotides. ACS Nano 2011, 5, 8783–8789. [Google Scholar] [CrossRef]
- Liu, X.; Yan, H.; Liu, Y.; Chang, Y. Targeted cell-cell interactions by DNA nanoscaffold-templated multivalent bispecific aptamers. Small 2011, 7, 1673–1682. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.; Fan, C.; Gothelf, K.V.; Li, J.; Lin, C.; Liu, L.; Liu, N.; Nijenhuis, M.A.D.; Saccà, B.; Simmel, F.C.; et al. DNA origami. Nat. Rev. Methods Prim. 2021, 1, 13. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, Y.; Chen, M.; Huang, Y.; Song, T.; Yang, C.; Yang, Y.; Song, Y. Elucidating the Effect of Nanoscale Receptor-Binding Domain Organization on SARS-CoV-2 Infection and Immunity Activation with DNA Origami. J. Am. Chem. Soc. 2022, 144, 21295–21303. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lin, B.; Wu, L.; Huang, M.; Li, X.; Zhang, H.; Song, J.; Wang, W.; Zhao, G.; Song, Y.; et al. DNA Nanolithography Enables a Highly Ordered Recognition Interface in a Microfluidic Chip for the Efficient Capture and Release of Circulating Tumor Cells. Angew. Chem. Int. Ed. 2020, 59, 14115–14119. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yang, X.; He, L.; Wang, K.; Wang, Q.; Huang, J.; Liu, J.; Wu, B.; Xu, C. Self-Assembled DNA Nanocentipede as Multivalent Drug Carrier for Targeted Delivery. ACS Appl. Mater. Interfaces 2016, 8, 25733–25740. [Google Scholar] [CrossRef]
- Zhu, G.; Hu, R.; Zhao, Z.; Chen, Z.; Zhang, X.; Tan, W. Noncanonical self-assembly of multifunctional DNA nanoflowers for biomedical applications. J. Am. Chem. Soc. 2013, 135, 16438–16445. [Google Scholar] [CrossRef]
- Simons, K.; Toomre, D. Lipid rafts and signal transduction. Nat. Rev. 2000, 1, 31–41. [Google Scholar] [CrossRef]
- de Castro, M.A.G.; Wildhagen, H.; Sograte-Idrissi, S.; Hitzing, C.; Binder, M.; Trepel, M.; Engels, N.; Opazo, F. Differential organization of tonic and chronic B cell antigen receptors in the plasma membrane. Nat. Commun. 2019, 10, 820. [Google Scholar] [CrossRef]
- Rinker, S.; Ke, Y.; Liu, Y.; Chhabra, R.; Yan, H. Self-assembled DNA nanostructures for distance-dependent multivalent ligand-protein binding. Nat. Nanotechnol. 2008, 3, 418–422. [Google Scholar] [CrossRef]
- Shaw, A.; Lundin, V.; Petrova, E.; Fördős, F.; Benson, E.; Al-Amin, R.A.; Herland, A.; Blokzijl, A.; Högberg, B.; Teixeira, A. Spatial control of membrane receptor function using ligand nanocalipers. Nat. Methods 2014, 11, 841–846. [Google Scholar] [CrossRef] [PubMed]
- Rosier, B.; Markvoort, A.J.; Gumi Audenis, B.; Roodhuizen, J.A.L.; Den Hamer, A.; Brunsveld, L.; De Greef, T.F.A. Proximity-induced caspase-9 activation on a DNA origami-based synthetic apoptosome. Nat. Catal. 2020, 3, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Deng, R.; Sun, Y.; Zhang, L.; Li, J. Reversible control of cell membrane receptor function using DNA nano-spring multivalent ligands. Chem. Sci. 2017, 8, 7098–7105. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.; Hoffecker, I.T.; Smyrlaki, I.; Rosa, J.; Grevys, A.; Bratlie, D.; Sandlie, I.; Michaelsen, T.E.; Andersen, J.T.; Högberg, B. Binding to nanopatterned antigens is dominated by the spatial tolerance of antibodies. Nat. Nanotechnol. 2019, 14, 184–190. [Google Scholar] [CrossRef]
- Zhang, P.; Liu, X.; Liu, P.; Wang, F.; Ariyama, H.; Ando, T.; Lin, J.; Wang, L.; Hu, J.; Li, B.; et al. Capturing transient antibody conformations with DNA origami epitopes. Nat. Commun. 2020, 11, 3114. [Google Scholar] [CrossRef]
- Veneziano, R.; Moyer, T.J.; Stone, M.B.; Wamhoff, E.-C.; Read, B.J.; Mukherjee, S.; Shepherd, T.R.; Das, J.; Schief, W.R.; Irvine, D.J.; et al. Role of nanoscale antigen organization on B-cell activation probed using DNA origami. Nat. Nanotechnol. 2020, 15, 716–723. [Google Scholar] [CrossRef]
- Harrison, S.C. Looking Inside Adenovirus. Science 2010, 329, 1026–1027. [Google Scholar] [CrossRef]
- Joglekar, A.V.; Li, G. T cell antigen discovery. Nat. Methods 2021, 18, 873–880. [Google Scholar] [CrossRef]
- Li, M.; Ding, H.; Lin, M.; Yin, F.; Song, L.; Mao, X.; Li, F.; Ge, Z.; Wang, L.; Zuo, X.; et al. DNA Framework-Programmed Cell Capture via Topology-Engineered Receptor-Ligand Interactions. J. Am. Chem. Soc. 2019, 141, 18910–18915. [Google Scholar] [CrossRef]
- Yin, F.; Li, M.; Mao, X.; Li, F.; Xiang, X.; Li, Q.; Wang, L.; Zuo, X.; Fan, C.; Zhu, Y. DNA Framework-Based Topological Cell Sorters. Angew. Chem. Int. Ed. 2020, 59, 10406–10410. [Google Scholar] [CrossRef]
- Shen, Q.; Feng, Q.; Wu, C.; Xiong, Q.; Tian, T.; Yuan, S.; Shi, J.; Bedwell, G.J.; Yang, R.; Aiken, C.; et al. Modeling HIV-1 nuclear entry with nucleoporin-gated DNA-origami channels. Nat. Struct. Mol. Biol. 2023, 30, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Kwon, P.S.; Ren, S.; Kwon, S.J.; Kizer, M.E.; Kuo, L.; Xie, M.; Zhu, D.; Zhou, F.; Zhang, F.; Kim, D.; et al. Designer DNA architecture offers precise and multivalent spatial pattern-recognition for viral sensing and inhibition. Nat. Chem. 2020, 12, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xu, Y.; Huang, Y.; Sun, M.; Liu, S.; Wan, S.; Chen, H.; Yang, C.; Yang, Y.; Song, Y. Spatially Patterned Neutralizing Icosahedral DNA Nanocage for Efficient SARS-CoV-2 Blocking. J. Am. Chem. Soc. 2022, 144, 13146–13153. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Liu, S.; Ju, H.; Chen, H. Direct electron transfer and enzymatic activity of hemoglobin in a hexagonal mesoporous silica matrix. Biosens. Bioelectron. 2004, 19, 861–867. [Google Scholar] [CrossRef]
- Liu, S.; Dai, Z.; Chen, H.; Ju, H. Immobilization of hemoglobin on zirconium dioxide nanoparticles for preparation of a novel hydrogen peroxide biosensor. Biosens. Bioelectron. 2004, 19, 963–969. [Google Scholar] [CrossRef]
- Dong, H.; Gao, W.; Yan, F.; Ji, H.; Ju, H. Fluorescence resonance energy transfer between quantum dots and graphene oxide for sensing biomolecules. Anal. Chem. 2010, 82, 5511–5517. [Google Scholar] [CrossRef]
- Lei, J.; Ju, H. Signal amplification using functional nanomaterials for biosensing. Chem. Soc. Rev. 2012, 41, 2122–2134. [Google Scholar] [CrossRef]
- Feng, Q.M.; Guo, Y.H.; Xu, J.; Chen, H. A surface-confined DNA assembly amplification strategy on DNA nanostructural scaffold for electrochemiluminescence biosensing. Biosens. Bioelectron. 2018, 100, 571–576. [Google Scholar] [CrossRef]
- Mao, M.; Lin, Z.; Chen, L.; Zou, Z.; Zhang, J.; Dou, Q.; Wu, J.; Chen, J.; Wu, M.; Niu, L.; et al. Modular DNA-Origami-Based Nanoarrays Enhance Cell Binding Affinity through the “Lock-and-Key” Interaction. J. Am. Chem. Soc. 2023, 145, 5447–5455. [Google Scholar] [CrossRef]
- Zhong, L.; Cai, S.; Huang, Y.; Yin, L.; Yang, Y.; Lu, C.; Yang, H. DNA Octahedron-Based Fluorescence Nanoprobe for Dual Tumor-Related mRNAs Detection and Imaging. Anal. Chem. 2018, 90, 12059–12066. [Google Scholar] [CrossRef]
- Xue, C.; Zhang, S.; Li, C.; Yu, X.; Ouyang, C.; Lu, Y.; Wu, Z.S. Y-Shaped Backbone-Rigidified Triangular DNA Scaffold-Directed Stepwise Movement of a DNAzyme Walker for Sensitive MicroRNA Imaging within Living Cells. Anal. Chem. 2019, 91, 15678–15685. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Li, J.; Chen, N.; Hu, X.; Yang, X.; Guo, L.; Li, Q.; Zuo, X.; Wang, L.; Ma, Y.; et al. DNA nanostructure-programmed like-charge attraction at the cell-membrane interface. ACS Cent. Sci. 2018, 4, 1344–1351. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Fang, S.; Ji, B.; Li, M.; Song, J.; Qiu, L.; Tan, W. DNA nanostructure-programmed cell entry via corner angle-mediated molecular interaction with membrane receptors. Nano Lett. 2021, 21, 6946–6951. [Google Scholar] [CrossRef]
- Guo, L.; Zhang, Y.; Wang, Y.; Xie, M.; Dai, J.; Qu, Z.; Zhou, M.; Cao, S.; Shi, J.; Wang, L.; et al. Directing Multivalent Aptamer-Receptor Binding on the Cell Surface with Programmable Atom-Like Nanoparticles. Angew. Chem. Int. Ed. 2022, 61, e202117168. [Google Scholar]
- Xie, M.; Guo, L.; Xing, S.; Cao, S.; Zhao, Z.; Liang, K.; Li, J.; Luo, S.; Zhang, Y.; Wang, L. Cell imaging with multi-color DNA framework probes. Chem. Commun. 2021, 57, 11318–11321. [Google Scholar] [CrossRef]
- Li, S.; Jiang, Q.; Liu, S.; Zhang, Y.; Tian, Y.; Song, C.; Wang, J.; Zou, Y.; Anderson, G.J.; Han, J.Y.; et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 2018, 36, 258–264. [Google Scholar] [CrossRef]
- Han, L.; Wang, Y.; Tang, W.; Liu, J.; Ding, B. Bioimaging Based on Nucleic Acid Nanostructures. Chem. Res. Chin. Univ. 2021, 37, 823–828. [Google Scholar] [CrossRef]
- Wang, J.; Song, J.; Zhang, X.; Wang, S.M.; Kang, B.; Li, X.L.; Chen, H.; Xu, J. DNA-Programed Plasmon Rulers Decrypt Single-Receptor Dimerization on Cell Membrane. J. Am. Chem. Soc. 2023, 145, 1273–1284. [Google Scholar] [CrossRef]
- Xie, S.; Sun, W.; Fu, T.; Liu, X.; Chen, P.; Qiu, L.; Qu, F.; Tan, W. Aptamer-Based Targeted Delivery of Functional Nucleic Acids. J. Am. Chem. Soc. 2023, 145, 7677–7691. [Google Scholar] [CrossRef]
- Li, Q.; Zhao, D.; Shao, X.; Lin, S.; Xie, X.; Liu, M.; Ma, W.; Shi, S.; Lin, Y. Aptamer-Modified Tetrahedral DNA Nanostructure for Tumor-Targeted Drug Delivery. ACS Appl. Mater. Interfaces 2017, 9, 36695–36701. [Google Scholar] [CrossRef]
- Shi, S.; Fu, W.; Lin, S.; Tian, T.; Li, S.; Shao, X.; Zhang, Y.; Zhang, T.; Tang, Z.; Zhou, Y.; et al. Targeted and effective glioblastoma therapy via aptamer-modified tetrahedral framework nucleic acid-paclitaxel nanoconjugates that can pass the blood brain barrier. Nanomedicine 2019, 21, 102061. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Guo, L.; Wu, G.; Li, J.; Sun, Y.; Hou, Y.; Shi, J.; Song, S.; Wang, L.; Fan, C.; et al. DNA Origami-Enabled Engineering of Ligand-Drug Conjugates for Targeted Drug Delivery. Small 2020, 16, e1904857. [Google Scholar] [CrossRef] [PubMed]
- Ren, K.; Liu, Y.; Wu, J.; Zhang, Y.; Zhu, J.; Yang, M.; Ju, H. A DNA dual lock-and-key strategy for cell-subtype-specific siRNA delivery. Nat. Commun. 2016, 7, 13580. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Fischer, A.; Ouyang, Y.; Wang, J.; Sohn, Y.S.; Nechushtai, R.; Pikarsky, E.; Fan, C.; Willner, I. Aptamer-modified DNA tetrahedra-gated metal–organic framework nanoparticle carriers for enhanced chemotherapy or photodynamic therapy. Chem. Sci. 2021, 12, 14473–14483. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, W.; Fang, Y.; Zhang, X.; Liu, Y.; Ju, H. Activating a DNA Nanomachine via Computation across Cancer Cell Membranes for Precise Therapy of Solid Tumors. J. Am. Chem. Soc. 2021, 143, 15233–15242. [Google Scholar] [CrossRef]
- Du, Y.; Lyu, Y.; Lin, J.; Ma, C.; Zhang, Q.; Zhang, Y.; Qiu, L.; Tan, W. Membrane-anchored DNA nanojunctions enable closer antigen-presenting cell-T-cell contact in elevated T-cell receptor triggering. Nat. Nanotechnol. 2023. [Google Scholar] [CrossRef]
- Shen, F.; Xiong, Z.; Wu, Y.; Peng, R.; Wang, Y.; Sun, L.; Fan, C.; Liu, Z. Precise Epitope Organization with Self-adjuvant Framework Nucleic Acid for Efficient COVID-19 Peptide Vaccine Construction. Angew. Chem. Int. Ed. 2023, 62, e202301147. [Google Scholar] [CrossRef]
- Sigl, C.; Willner, E.M.; Engelen, W.; Kretzmann, J.A.; Sachenbacher, K.; Liedl, A.; Kolbe, F.; Wilsch, F.; Aghvami, S.A.; Protzer, U.; et al. Programmable icosahedral shell system for virus trapping. Nat. Mater. 2021, 20, 1281–1289. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).