Fluorescent Protein-Based Metal Biosensors
Abstract
1. Introduction
2. Spectroscopic Properties of Metal-Induced FP
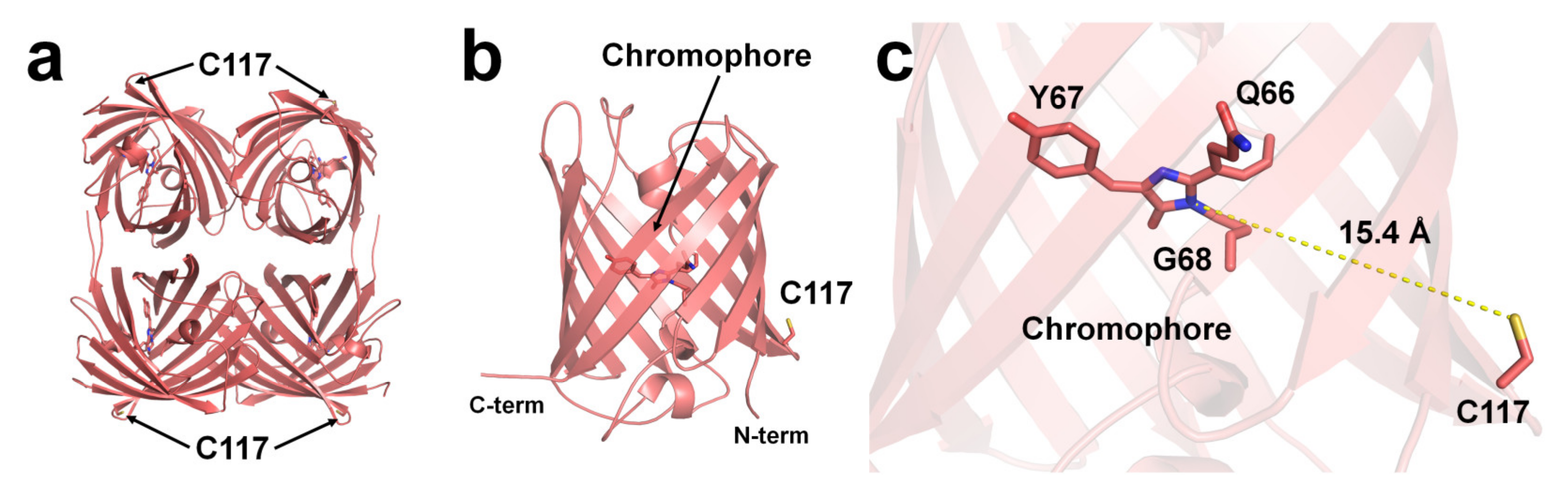
2.1. DsRed
2.2. BFPms1
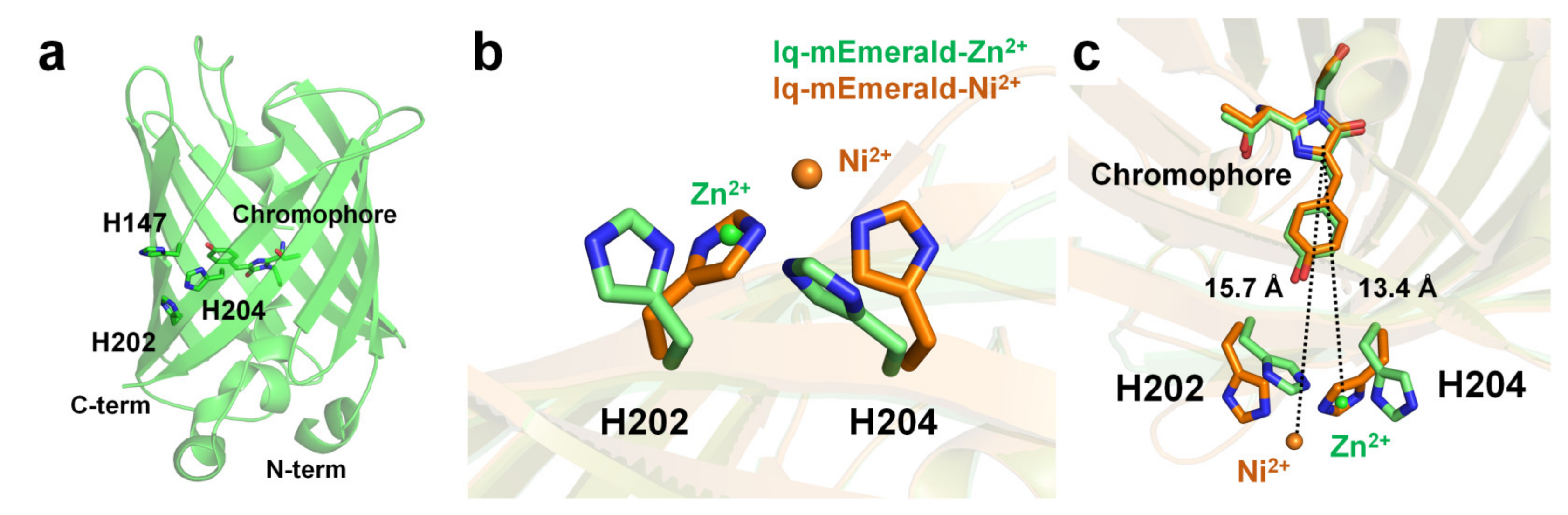
2.3. iq-mEmerald
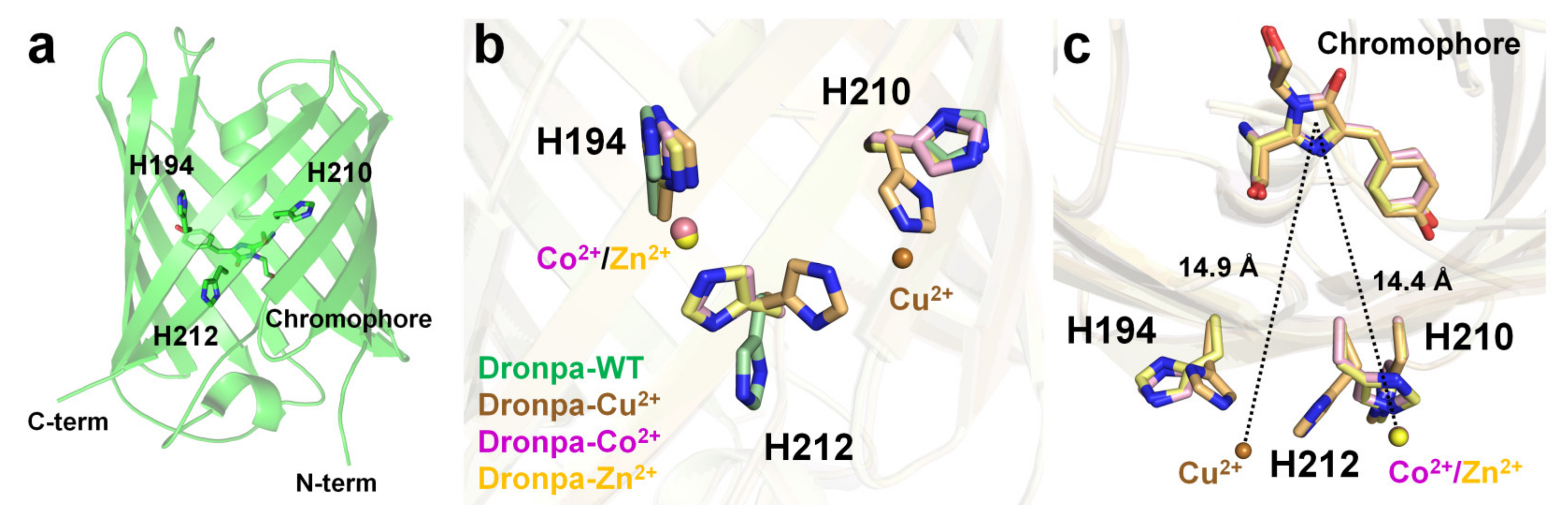
2.4. Dronpa
2.5. AmCyan
2.6. mOrange2
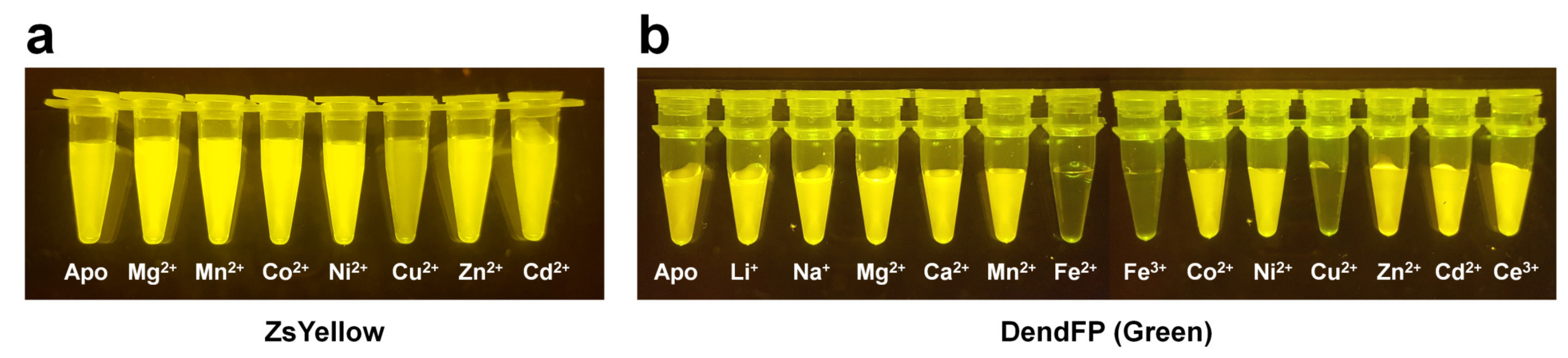
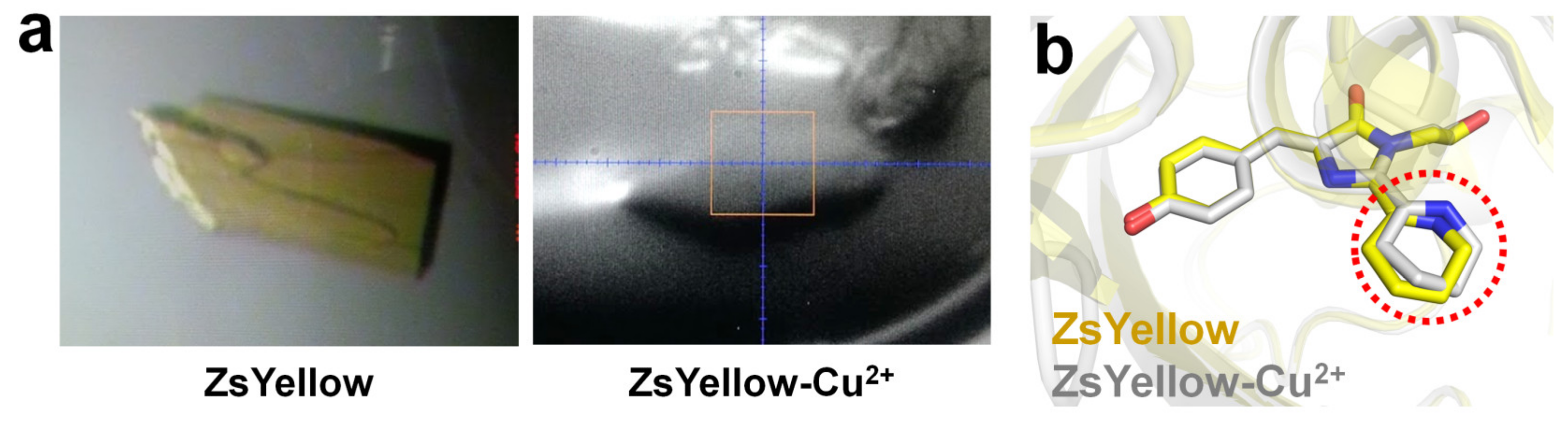
2.7. ZsYellow
2.8. ZsGreen
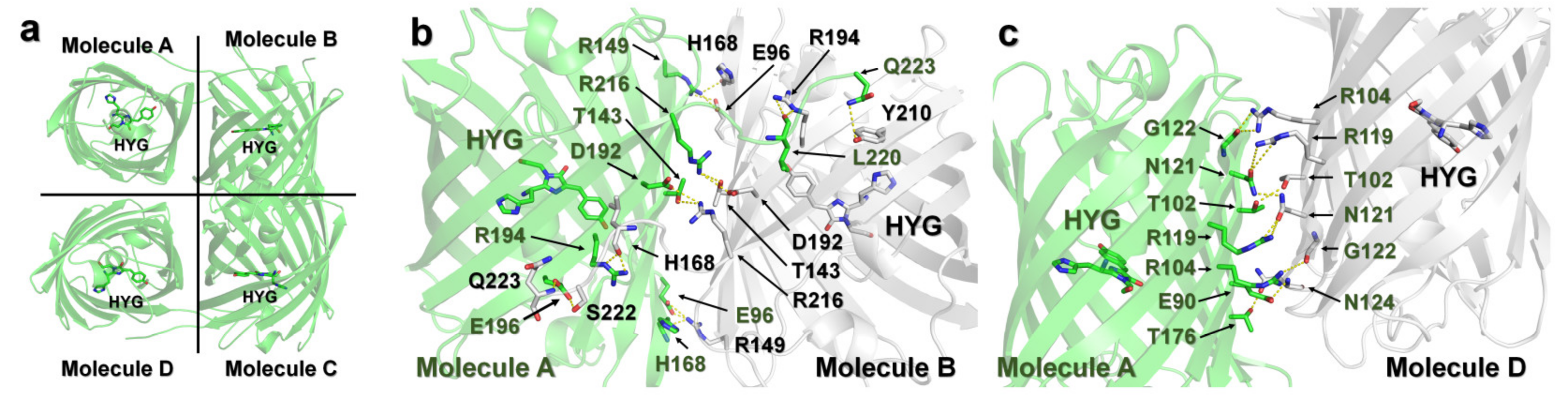
2.9. DendFP
3. Discussion
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Carter, K.P.; Young, A.M.; Palmer, A.E. Fluorescent Sensors for Measuring Metal Ions in Living Systems. Chem. Rev. 2014, 114, 4564–4601. [Google Scholar] [CrossRef] [PubMed]
- Moustakas, M. The Role of Metal Ions in Biology, Biochemistry and Medicine. Materials 2021, 14, 549. [Google Scholar] [CrossRef] [PubMed]
- Prasad, A.S. Zinc deficiency. BMJ 2003, 326, 409–410. [Google Scholar] [CrossRef] [PubMed]
- Theil, E.C. Iron Homeostasis and Nutritional Iron Deficiency. J. Nutr. 2011, 141, 724S–728S. [Google Scholar] [CrossRef]
- Tang, Z.; Wang, H.Q.; Chen, J.; Chang, J.D.; Zhao, F.J. Molecular mechanisms underlying the toxicity and detoxification of trace metals and metalloids in plants. J. Integr. Plant Biol. 2023, 65, 570–593. [Google Scholar] [CrossRef]
- Avila, D.S.; Puntel, R.L.; Aschner, M. Manganese in Health and Disease. In Interrelations between Essential Metal Ions and Human Diseases; Metal Ions in Life Sciences; Springer: Berlin/Heidelberg, Germany, 2013; pp. 199–227. [Google Scholar]
- Zheng, X.; Cheng, W.; Ji, C.; Zhang, J.; Yin, M. Detection of metal ions in biological systems: A review. Rev. Anal. Chem. 2020, 39, 231–246. [Google Scholar] [CrossRef]
- Mitra, S.; Chakraborty, A.J.; Tareq, A.M.; Emran, T.B.; Nainu, F.; Khusro, A.; Idris, A.M.; Khandaker, M.U.; Osman, H.; Alhumaydhi, F.A.; et al. Impact of heavy metals on the environment and human health: Novel therapeutic insights to counter the toxicity. J. King Saud Univ. Sci. 2022, 34, 101865. [Google Scholar] [CrossRef]
- Yu, X.; Strub, M.P.; Barnard, T.J.; Noinaj, N.; Piszczek, G.; Buchanan, S.K.; Taraska, J.W. An engineered palette of metal ion quenchable fluorescent proteins. PLoS ONE 2014, 9, e95808. [Google Scholar] [CrossRef]
- Ghaedi, M.; Ahmadi, F.; Shokrollahi, A. Simultaneous preconcentration and determination of copper, nickel, cobalt and lead ions content by flame atomic absorption spectrometry. J. Hazard. Mater. 2007, 142, 272–278. [Google Scholar] [CrossRef]
- Ashenef, A. Essential and toxic metals in tea (Camellia sinensis) imported and produced in Ethiopia. Food Addit. Contam. B Surveill. 2013, 7, 30–36. [Google Scholar] [CrossRef]
- Williams, M.; Luo, W.; McWhirter, K.; Ikegbu, O.; Talbot, P. Chemical Elements, Flavor Chemicals, and Nicotine in Unused and Used Electronic Cigarettes Aged 5–10 Years and Effects of pH. Int. J. Environ. Health Res. 2022, 19, 16931. [Google Scholar] [CrossRef]
- Amerikanou, C.; Karavoltsos, S.; Gioxari, A.; Tagkouli, D.; Sakellari, A.; Papada, E.; Kalogeropoulos, N.; Forbes, A.; Kaliora, A.C. Clinical and inflammatory biomarkers of inflammatory bowel diseases are linked to plasma trace elements and toxic metals; new insights into an old concept. Front. Nutr. 2022, 9, 997356. [Google Scholar] [CrossRef]
- Kim, H.H.; Lee, G.H.; Pyo, G.J.; Kwon, E.S.; Myung, K.B.; Cheong, S.H. Nickel, cobalt, and chromium in nail sticker and tip products in Korea. Contact Derm. 2023. ahead of print. [Google Scholar] [CrossRef]
- Nazir, N.U.A.; Abbas, S.R. Identification of phenol 2,2-methylene bis, 6 [1,1-D] as breath biomarker of hepatocellular carcinoma (HCC) patients and its electrochemical sensing: E-nose biosensor for HCC. Anal. Chim. Acta 2023, 1242, 340752. [Google Scholar] [CrossRef]
- Patel, M.; Bisht, N.; Prabhakar, P.; Sen, R.K.; Kumar, P.; Dwivedi, N.; Ashiq, M.; Mondal, D.P.; Srivastava, A.K.; Dhand, C. Ternary nanocomposite-based smart sensor: Reduced graphene oxide/polydopamine/alanine nanocomposite for simultaneous electrochemical detection of Cd2+, Pb2+, Fe2+, and Cu2+ ions. Environ. Res. 2023, 221, 115317. [Google Scholar] [CrossRef]
- Loa, J.D.A.; Cruz-Rodríguez, I.A.; Rojas-Avelizapa, N.G. Colorimetric Detection of Metals Using CdS-NPs Synthesized by an Organic Extract of Aspergillus niger. Appl. Biochem. Biotechnol. 2023. ahead of print. [Google Scholar] [CrossRef]
- Wang, X.; Liu, S.; Zhou, J.; Zhang, S.; Hou, C.; Huo, D. Colorimetric detection of Cu2+ based on the inhibition strategy for etching reaction of AgNCs. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2023, 289, 122229. [Google Scholar] [CrossRef]
- Lü, B.; Chen, Y.; Li, P.; Wang, B.; Müllen, K.; Yin, M. Stable radical anions generated from a porous perylenediimide metal-organic framework for boosting near-infrared photothermal conversion. Nat. Commun. 2019, 10, 767. [Google Scholar] [CrossRef]
- Forzani, E.S.; Zhang, H.; Chen, W.; Tao, N. Detection of Heavy Metal Ions in Drinking Water Using a High-Resolution Differential Surface Plasmon Resonance Sensor. Environ. Sci. Technol. 2004, 39, 1257–1262. [Google Scholar] [CrossRef]
- Malitesta, C.; Guascito, M.R. Heavy metal determination by biosensors based on enzyme immobilised by electropolymerisation. Biosens. Bioelectron. 2005, 20, 1643–1647. [Google Scholar] [CrossRef]
- Zhu, X.; Xu, L.; Lou, Y.; Yu, H.; Li, X.; Blake, D.A.; Liu, F. Preparation of Specific Monoclonal Antibodies (MAbs) against Heavy Metals: MAbs That Recognize Chelated Cadmium Ions. J. Agric. Food Chem. 2007, 55, 7648–7653. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-S.; Han, M.S.; Mirkin, C.A. Colorimetric Detection of Mercuric Ion (Hg2+) in Aqueous Media using DNA-Functionalized Gold Nanoparticles. Angew. Chem. Int. Ed. 2007, 46, 4093–4096. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Shi, L.; Wang, E.; Dong, S. Silver-Ion-Mediated DNAzyme Switch for the Ultrasensitive and Selective Colorimetric Detection of Aqueous Ag+ and Cysteine. Chem. Eur. J. 2009, 15, 3347–3350. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, M.; Riedel, K.; Adler, K.; Kunze, G. Amperometric measurement of copper ions with a deputy substrate using a novel Saccharomyces cerevisiae sensor. Biosens. Bioelectron. 2000, 15, 211–219. [Google Scholar] [CrossRef]
- Tsien, R.Y. The green fluorescent protein. Annu. Rev. Biochem. 1998, 67, 509–544. [Google Scholar] [CrossRef]
- Xu, Y.; Hwang, K.Y.; Nam, K.H. Spectral and structural analysis of large Stokes shift fluorescent protein dKeima570. J. Microbiol. 2018, 56, 822–827. [Google Scholar] [CrossRef]
- Kim, S.E.; Hwang, K.Y.; Nam, K.H. Spectral and structural analysis of a red fluorescent protein from Acropora digitifera. Protein Sci. 2019, 28, 375–381. [Google Scholar] [CrossRef]
- Lee, Y.R.; Park, J.-H.; Hahm, S.-H.; Kang, L.-W.; Chung, J.H.; Nam, K.-H.; Hwang, K.Y.; Kwon, I.C.; Han, Y.S. Development of Bimolecular Fluorescence Complementation Using Dronpa for Visualization of Protein–Protein Interactions in Cells. Mol. Imaging Biol. 2010, 12, 468–478. [Google Scholar] [CrossRef]
- Bajar, B.T.; Wang, E.S.; Zhang, S.; Lin, M.Z.; Chu, J. A Guide to Fluorescent Protein FRET Pairs. Sensors 2016, 16, 1488. [Google Scholar] [CrossRef]
- Liao, J.; Madahar, V.; Dang, R.; Jiang, L. Quantitative FRET (qFRET) Technology for the Determination of Protein–Protein Interaction Affinity in Solution. Molecules 2021, 26, 6339. [Google Scholar] [CrossRef]
- Skruzny, M.; Pohl, E.; Abella, M. FRET Microscopy in Yeast. Biosensors 2019, 9, 122. [Google Scholar] [CrossRef]
- Baird, G.S.; Zacharias, D.A.; Tsien, R.Y. Circular permutation and receptor insertion within green fluorescent proteins. Proc. Natl. Acad. Sci. USA 1999, 96, 11241–11246. [Google Scholar] [CrossRef]
- Galietta, L.J.V.; Haggie, P.M.; Verkman, A.S. Green fluorescent protein-based halide indicators with improved chloride and iodide affinities. FEBS Lett. 2001, 499, 220–224. [Google Scholar] [CrossRef]
- Barondeau, D.P.; Kassmann, C.J.; Tainer, J.A.; Getzoff, E.D. Structural chemistry of a green fluorescent protein Zn biosensor. J. Am. Chem. Soc. 2002, 124, 3522–3524. [Google Scholar] [CrossRef]
- Bizzarri, R.; Arcangeli, C.; Arosio, D.; Ricci, F.; Faraci, P.; Cardarelli, F.; Beltram, F. Development of a Novel GFP-based Ratiometric Excitation and Emission pH Indicator for Intracellular Studies. Biophys. J. 2006, 90, 3300–3314. [Google Scholar] [CrossRef]
- Frommer, W.B.; Davidson, M.W.; Campbell, R.E. Genetically encoded biosensors based on engineered fluorescent proteins. Chem. Soc. Rev. 2009, 38, 2833–2841. [Google Scholar] [CrossRef]
- Tantama, M.; Hung, Y.P.; Yellen, G. Optogenetic reporters. In Optogenetics: Tools for Controlling and Monitoring Neuronal Activity; Progress in Brain Research; Elsevier: Amsterdam, The Netherlands, 2012; pp. 235–263. [Google Scholar]
- Akerboom, J.; Carreras Calderón, N.; Tian, L.; Wabnig, S.; Prigge, M.; Tolö, J.; Gordus, A.; Orger, M.B.; Severi, K.E.; Macklin, J.J.; et al. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front. Mol. Neurosci. 2013, 6, 2. [Google Scholar] [CrossRef]
- Broyles, C.; Robinson, P.; Daniels, M. Fluorescent, Bioluminescent, and Optogenetic Approaches to Study Excitable Physiology in the Single Cardiomyocyte. Cells 2018, 7, 51. [Google Scholar] [CrossRef]
- Benaissa, H.; Ounoughi, K.; Aujard, I.; Fischer, E.; Goïame, R.; Nguyen, J.; Tebo, A.G.; Li, C.; Le Saux, T.; Bertolin, G.; et al. Engineering of a fluorescent chemogenetic reporter with tunable color for advanced live-cell imaging. Nat. Commun. 2021, 12, 6989. [Google Scholar] [CrossRef]
- Miura, Y.; Senoo, A.; Doura, T.; Kiyonaka, S. Chemogenetics of cell surface receptors: Beyond genetic and pharmacological approaches. RSC Chem. Biol. 2022, 3, 269–287. [Google Scholar] [CrossRef]
- Tanz, S.K.; Castleden, I.; Small, I.D.; Millar, A.H. Fluorescent protein tagging as a tool to define the subcellular distribution of proteins in plants. Front. Plant Sci. 2013, 4, 214. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Gao, C.; Zhao, Q.; Jiang, L. Using Fluorescent Protein Fusions to Study Protein Subcellular Localization and Dynamics in Plant Cells. In High-Resolution Imaging of Cellular Proteins; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2016; pp. 113–123. [Google Scholar]
- Hynes, T.R.; Hughes, T.E.; Berlot, C.H. Cellular Localization of GFP-Tagged α Subunits. In G Protein Signaling; Humana Press: New York, NY, USA, 2003; pp. 233–246. [Google Scholar]
- Chudakov, D.M.; Lukyanov, S.; Lukyanov, K.A. Fluorescent proteins as a toolkit for in vivo imaging. Trends Biotechnol. 2005, 23, 605–613. [Google Scholar] [CrossRef]
- Lippincott-Schwartz, J. Emerging In Vivo Analyses of Cell Function Using Fluorescence Imaging. Annu. Rev. Biochem. 2011, 80, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Borg, R.E.; Rochford, J. Molecular Photoacoustic Contrast Agents: Design Principles & Applications. Photochem. Photobiol. 2018, 94, 1175–1209. [Google Scholar] [CrossRef] [PubMed]
- Perez-Rodriguez, R.; Haitjema, C.; Huang, Q.; Nam, K.H.; Bernardis, S.; Ke, A.; DeLisa, M.P. Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol. Microbiol. 2011, 79, 584–599. [Google Scholar] [CrossRef]
- Dolan, A.E.; Hou, Z.; Xiao, Y.; Gramelspacher, M.J.; Heo, J.; Howden, S.E.; Freddolino, P.L.; Ke, A.; Zhang, Y. Introducing a Spectrum of Long-Range Genomic Deletions in Human Embryonic Stem Cells Using Type I CRISPR-Cas. Mol. Cell 2019, 74, 936–950.e935. [Google Scholar] [CrossRef]
- Hu, C.; Ni, D.; Nam, K.H.; Majumdar, S.; McLean, J.; Stahlberg, H.; Terns, M.P.; Ke, A. Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools. Mol. Cell 2022, 82, 2754–2768.e2755. [Google Scholar] [CrossRef]
- Tan, R.; Krueger, R.K.; Gramelspacher, M.J.; Zhou, X.; Xiao, Y.; Ke, A.; Hou, Z.; Zhang, Y. Cas11 enables genome engineering in human cells with compact CRISPR-Cas3 systems. Mol. Cell 2022, 82, 852–867.e855. [Google Scholar] [CrossRef]
- Shimomura, O.; Johnson, F.H.; Saiga, Y. Extraction, Purification and Properties of Aequorin, a Bioluminescent Protein from the Luminous Hydromedusan, Aequorea. J. Cell. Comp. Physiol. 1962, 59, 223–239. [Google Scholar] [CrossRef]
- Ormö, M.; Cubitt, A.B.; Kallio, K.; Gross, L.A.; Tsien, R.Y.; Remington, S.J. Crystal Structure of the Aequorea victoria Green Fluorescent Protein. Science 1996, 273, 1392–1395. [Google Scholar] [CrossRef]
- Zimmer, M. Green fluorescent protein (GFP): Applications, structure, and related photophysical behavior. Chem. Rev. 2002, 102, 759–781. [Google Scholar] [CrossRef]
- Bae, J.E.; Kim, I.J.; Nam, K.H. Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein ZsYellow by protonation of Glu221. Biochem. Biophys. Res. Commun. 2017, 493, 562–567. [Google Scholar] [CrossRef]
- Tomosugi, W.; Matsuda, T.; Tani, T.; Nemoto, T.; Kotera, I.; Saito, K.; Horikawa, K.; Nagai, T. An ultramarine fluorescent protein with increased photostability and pH insensitivity. Nat. Methods 2009, 6, 351–353. [Google Scholar] [CrossRef]
- Mena, M.A.; Treynor, T.P.; Mayo, S.L.; Daugherty, P.S. Blue fluorescent proteins with enhanced brightness and photostability from a structurally targeted library. Nat. Biotechnol. 2006, 24, 1569–1571. [Google Scholar] [CrossRef]
- Costantini, L.M.; Baloban, M.; Markwardt, M.L.; Rizzo, M.A.; Guo, F.; Verkhusha, V.V.; Snapp, E.L. A palette of fluorescent proteins optimized for diverse cellular environments. Nat. Commun. 2015, 6, 7670. [Google Scholar] [CrossRef]
- Erard, M.; Fredj, A.; Pasquier, H.; Beltolngar, D.-B.; Bousmah, Y.; Derrien, V.; Vincent, P.; Merola, F. Minimum set of mutations needed to optimize cyan fluorescent proteins for live cell imaging. Mol. BioSyst. 2013, 9, 258–267. [Google Scholar] [CrossRef]
- Rizzo, M.A.; Springer, G.H.; Granada, B.; Piston, D.W. An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 2004, 22, 445–449. [Google Scholar] [CrossRef]
- Nguyen, A.W.; Daugherty, P.S. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat. Biotechnol. 2005, 23, 355–360. [Google Scholar] [CrossRef]
- Goedhart, J.; von Stetten, D.; Noirclerc-Savoye, M.; Lelimousin, M.; Joosen, L.; Hink, M.A.; van Weeren, L.; Gadella, T.W.J.; Royant, A. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat. Commun. 2012, 3, 751. [Google Scholar] [CrossRef]
- Zacharias, D.A.; Violin, J.D.; Newton, A.C.; Tsien, R.Y. Partitioning of Lipid-Modified Monomeric GFPs into Membrane Microdomains of Live Cells. Science 2002, 296, 913–916. [Google Scholar] [CrossRef]
- Bajar, B.T.; Wang, E.S.; Lam, A.J.; Kim, B.B.; Jacobs, C.L.; Howe, E.S.; Davidson, M.W.; Lin, M.Z.; Chu, J. Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Sci. Rep. 2016, 6, 20889. [Google Scholar] [CrossRef] [PubMed]
- Shaner, N.C.; Lambert, G.G.; Chammas, A.; Ni, Y.; Cranfill, P.J.; Baird, M.A.; Sell, B.R.; Allen, J.R.; Day, R.N.; Israelsson, M.; et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 2013, 10, 407–409. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Liu, Z.; Suzuki, P.H.; Ahrens, J.F.; Lai, S.; Lu, X.; Guan, S.; St-Pierre, F. Versatile phenotype-activated cell sorting. Sci. Adv. 2020, 6, eabb7438. [Google Scholar] [CrossRef] [PubMed]
- Kremers, G.-J.; Goedhart, J.; van Munster, E.B.; Gadella, T.W.J. Cyan and Yellow Super Fluorescent Proteins with Improved Brightness, Protein Folding, and FRET Förster Radius. Biochemistry 2006, 45, 6570–6580. [Google Scholar] [CrossRef]
- Shaner, N.C.; Campbell, R.E.; Steinbach, P.A.; Giepmans, B.N.G.; Palmer, A.E.; Tsien, R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004, 22, 1567–1572. [Google Scholar] [CrossRef]
- Sakaue-Sawano, A.; Kurokawa, H.; Morimura, T.; Hanyu, A.; Hama, H.; Osawa, H.; Kashiwagi, S.; Fukami, K.; Miyata, T.; Miyoshi, H.; et al. Visualizing Spatiotemporal Dynamics of Multicellular Cell-Cycle Progression. Cell 2008, 132, 487–498. [Google Scholar] [CrossRef]
- Merzlyak, E.M.; Goedhart, J.; Shcherbo, D.; Bulina, M.E.; Shcheglov, A.S.; Fradkov, A.F.; Gaintzeva, A.; Lukyanov, K.A.; Lukyanov, S.; Gadella, T.W.J.; et al. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat. Methods 2007, 4, 555–557. [Google Scholar] [CrossRef]
- Shaner, N.C.; Lin, M.Z.; McKeown, M.R.; Steinbach, P.A.; Hazelwood, K.L.; Davidson, M.W.; Tsien, R.Y. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods 2008, 5, 545–551. [Google Scholar] [CrossRef]
- Bindels, D.S.; Haarbosch, L.; van Weeren, L.; Postma, M.; Wiese, K.E.; Mastop, M.; Aumonier, S.; Gotthard, G.; Royant, A.; Hink, M.A.; et al. mScarlet: A bright monomeric red fluorescent protein for cellular imaging. Nat. Methods 2016, 14, 53–56. [Google Scholar] [CrossRef]
- Nishizawa, K.; Kita, Y.; Kitayama, M.; Ishimoto, M. A red fluorescent protein, DsRed2, as a visual reporter for transient expression and stable transformation in soybean. Plant Cell Rep. 2006, 25, 1355–1361. [Google Scholar] [CrossRef]
- Wang, L.; Jackson, W.C.; Steinbach, P.A.; Tsien, R.Y. Evolution of new nonantibody proteins via iterative somatic hypermutation. Proc. Natl. Acad. Sci. USA 2004, 101, 16745–16749. [Google Scholar] [CrossRef]
- Lin, M.Z.; McKeown, M.R.; Ng, H.-L.; Aguilera, T.A.; Shaner, N.C.; Campbell, R.E.; Adams, S.R.; Gross, L.A.; Ma, W.; Alber, T.; et al. Autofluorescent Proteins with Excitation in the Optical Window for Intravital Imaging in Mammals. Chem. Biol. 2009, 16, 1169–1179. [Google Scholar] [CrossRef]
- Morozova, K.S.; Piatkevich, K.D.; Gould, T.J.; Zhang, J.; Bewersdorf, J.; Verkhusha, V.V. Far-Red Fluorescent Protein Excitable with Red Lasers for Flow Cytometry and Superresolution STED Nanoscopy. Biophys. J. 2010, 99, L13–L15. [Google Scholar] [CrossRef]
- Lambert, T.J. FPbase: A community-editable fluorescent protein database. Nat. Methods 2019, 16, 277–278. [Google Scholar] [CrossRef]
- Kogure, T.; Karasawa, S.; Araki, T.; Saito, K.; Kinjo, M.; Miyawaki, A. A fluorescent variant of a protein from the stony coral Montipora facilitates dual-color single-laser fluorescence cross-correlation spectroscopy. Nat. Biotechnol. 2006, 24, 577–581. [Google Scholar] [CrossRef]
- Piatkevich, K.D.; Hulit, J.; Subach, O.M.; Wu, B.; Abdulla, A.; Segall, J.E.; Verkhusha, V.V. Monomeric red fluorescent proteins with a large Stokes shift. Proc. Natl. Acad. Sci. USA 2010, 107, 5369–5374. [Google Scholar] [CrossRef]
- Zapata-Hommer, O.; Griesbeck, O. Efficiently folding and circularly permuted variants of the Sapphire mutant of GFP. BMC Biotechnol. 2003, 3, 5. [Google Scholar] [CrossRef]
- Gunewardene, M.S.; Subach, F.V.; Gould, T.J.; Penoncello, G.P.; Gudheti, M.V.; Verkhusha, V.V.; Hess, S.T. Superresolution Imaging of Multiple Fluorescent Proteins with Highly Overlapping Emission Spectra in Living Cells. Biophys. J. 2011, 101, 1522–1528. [Google Scholar] [CrossRef]
- Subach, F.V.; Patterson, G.H.; Manley, S.; Gillette, J.M.; Lippincott-Schwartz, J.; Verkhusha, V.V. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat. Methods 2009, 6, 153–159. [Google Scholar] [CrossRef]
- Subach, F.V.; Patterson, G.H.; Renz, M.; Lippincott-Schwartz, J.; Verkhusha, V.V. Bright Monomeric Photoactivatable Red Fluorescent Protein for Two-Color Super-Resolution sptPALM of Live Cells. J. Am. Chem. Soc. 2010, 132, 6481–6491. [Google Scholar] [CrossRef]
- Ando, R.; Mizuno, H.; Miyawaki, A. Regulated Fast Nucleocytoplasmic Shuttling Observed by Reversible Protein Highlighting. Science 2004, 306, 1370–1373. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Zhang, M.; Ji, W.; Chen, J.; Zhang, Y.; Liu, B.; Lu, J.; Zhang, J.; Xu, P.; Xu, T. A unique series of reversibly switchable fluorescent proteins with beneficial properties for various applications. Proc. Natl. Acad. Sci. USA 2012, 109, 4455–4460. [Google Scholar] [CrossRef] [PubMed]
- Pletnev, S.; Subach, F.V.; Dauter, Z.; Wlodawer, A.; Verkhusha, V.V. A Structural Basis for Reversible Photoswitching of Absorbance Spectra in Red Fluorescent Protein rsTagRFP. J. Mol. Biol. 2012, 417, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Ando, R.; Hama, H.; Yamamoto-Hino, M.; Mizuno, H.; Miyawaki, A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc. Natl. Acad. Sci. USA 2002, 99, 12651–12656. [Google Scholar] [CrossRef]
- Zhang, M.; Chang, H.; Zhang, Y.; Yu, J.; Wu, L.; Ji, W.; Chen, J.; Liu, B.; Lu, J.; Liu, Y.; et al. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat. Methods 2012, 9, 727–729. [Google Scholar] [CrossRef]
- Labas, Y.A.; Gurskaya, N.G.; Yanushevich, Y.G.; Fradkov, A.F.; Lukyanov, K.A.; Lukyanov, S.A.; Matz, M.V. Diversity and evolution of the green fluorescent protein family. Proc. Natl. Acad. Sci. USA 2002, 99, 4256–4261. [Google Scholar] [CrossRef]
- Miyawaki, A.; Llopis, J.; Heim, R.; McCaffery, J.M.; Adams, J.A.; Ikura, M.; Tsien, R.Y. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 1997, 388, 882–887. [Google Scholar] [CrossRef]
- Kim, I.J.; Xu, Y.; Nam, K.H. Spectroscopic and Structural Analysis of Cu2+-Induced Fluorescence Quenching of ZsYellow. Biosensors 2020, 10, 29. [Google Scholar] [CrossRef]
- Kim, I.J.; Xu, Y.; Nam, K.H. Metal-Induced Fluorescence Quenching of Photoconvertible Fluorescent Protein DendFP. Molecules 2022, 27, 2922. [Google Scholar] [CrossRef]
- Isarankura-Na-Ayudhya, C.; Tantimongcolwat, T.; Galla, H.J.; Prachayasittikul, V. Fluorescent protein-based optical biosensor for copper ion quantitation. Biol. Trace Elem. Res. 2010, 134, 352–363. [Google Scholar] [CrossRef]
- Richmond, T.A.; Takahashi, T.T.; Shimkhada, R.; Bernsdorf, J. Engineered metal binding sites on green fluorescence protein. Biochem. Biophys. Res. Commun. 2000, 268, 462–465. [Google Scholar] [CrossRef]
- Mizuno, T.; Murao, K.; Tanabe, Y.; Oda, M.; Tanaka, T. Metal-ion-dependent GFP emission in vivo by combining a circularly permutated green fluorescent protein with an engineered metal-ion-binding coiled-coil. J. Am. Chem. Soc. 2007, 129, 11378–11383. [Google Scholar] [CrossRef]
- Eli, P.; Chakrabartty, A. Variants of DsRed fluorescent protein: Development of a copper sensor. Protein Sci. 2006, 15, 2442–2447. [Google Scholar] [CrossRef]
- Kim, I.J.; Kim, S.; Park, J.; Eom, I.; Kim, S.; Kim, J.H.; Ha, S.C.; Kim, Y.G.; Hwang, K.Y.; Nam, K.H. Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development. FEBS Lett. 2016, 590, 2982–2990. [Google Scholar] [CrossRef]
- Bae, J.E.; Kim, I.J.; Nam, K.H. Spectroscopic Analysis of the Cu2+-Induced Fluorescence Quenching of Fluorescent Proteins AmCyan and mOrange2. Mol. Biotechnol. 2018, 60, 485–491. [Google Scholar] [CrossRef]
- Kim, I.J.; Xu, Y.; Nam, K.H. Spectroscopic Analysis of Fe Ion-Induced Fluorescence Quenching of the Green Fluorescent Protein ZsGreen. J. Fluoresc. 2021, 31, 307–314. [Google Scholar] [CrossRef]
- Matz, M.V.; Fradkov, A.F.; Labas, Y.A.; Savitsky, A.P.; Zaraisky, A.G.; Markelov, M.L.; Lukyanov, S.A. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 1999, 17, 969–973. [Google Scholar] [CrossRef]
- Sumner, J.P.; Westerberg, N.M.; Stoddard, A.K.; Hurst, T.K.; Cramer, M.; Thompson, R.B.; Fierke, C.A.; Kopelman, R. DsRed as a highly sensitive, selective, and reversible fluorescence-based biosensor for both Cu+ and Cu2+ ions. Biosens. Bioelectron. 2006, 21, 1302–1308. [Google Scholar] [CrossRef]
- Rahimi, Y.; Goulding, A.; Shrestha, S.; Mirpuri, S.; Deo, S.K. Mechanism of copper induced fluorescence quenching of red fluorescent protein, DsRed. Biochem. Biophys. Res. Commun. 2008, 370, 57–61. [Google Scholar] [CrossRef]
- Yarbrough, D.; Wachter, R.M.; Kallio, K.; Matz, M.V.; Remington, S.J. Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-Å resolution. Proc. Natl. Acad. Sci. USA 2001, 98, 462–467. [Google Scholar] [CrossRef]
- Taraska, J.W.; Puljung, M.C.; Olivier, N.B.; Flynn, G.E.; Zagotta, W.N. Mapping the structure and conformational movements of proteins with transition metal ion FRET. Nat. Methods 2009, 6, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.H.; Kwon, O.Y.; Sugiyama, K.; Lee, W.H.; Kim, Y.K.; Song, H.K.; Kim, E.E.; Park, S.Y.; Jeon, H.; Hwang, K.Y. Structural characterization of the photoswitchable fluorescent protein Dronpa-C62S. Biochem. Biophys. Res. Commun. 2007, 354, 962–967. [Google Scholar] [CrossRef] [PubMed]
- Guryev, O.; Jaimes, M.C.; Edinger, M.G.; Matvienko, M.; Abrams, B.; Dubrovsky, T. Use of a new violet-excitable AmCyan variant as a label in cell analysis. Cytom. Part A 2012, 81A, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Remington, S.J.; Wachter, R.M.; Yarbrough, D.K.; Branchaud, B.; Anderson, D.C.; Kallio, K.; Lukyanov, K.A. zFP538, a yellow-fluorescent protein from Zoanthus, contains a novel three-ring chromophore. Biochemistry 2005, 44, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Wenck, A.; Pugieux, C.; Turner, M.; Dunn, M.; Stacy, C.; Tiozzo, A.; Dunder, E.; van Grinsven, E.; Khan, R.; Sigareva, M.; et al. Reef-coral proteins as visual, non-destructive reporters for plant transformation. Plant Cell Rep. 2003, 22, 244–251. [Google Scholar] [CrossRef]
- Pletneva, N.V.; Pletnev, S.; Pakhomov, A.A.; Chertkova, R.V.; Martynov, V.I.; Muslinkina, L.; Dauter, Z.; Pletnev, V.Z. Crystal structure of the fluorescent protein from Dendronephthya sp. in both green and photoconverted red forms. Acta Crystallogr. D Struct. Biol. 2016, 72, 922–932. [Google Scholar] [CrossRef]
- Pakhomov, A.A.; Chertkova, R.V.; Martynov, V.I. Ph-Sensor Properties of a Fluorescent Protein from Dendronephthya sp. Bioorg. Khim. 2015, 41, 669–674. [Google Scholar] [CrossRef]
- Brown, D.R.; Qin, K.; Herms, J.W.; Madlung, A.; Manson, J.; Strome, R.; Fraser, P.E.; Kruck, T.; von Bohlen, A.; Schulz-Schaeffer, W.; et al. The cellular prion protein binds copper in vivo. Nature 1997, 390, 684–687. [Google Scholar] [CrossRef]
- Atwood, C.S.; Moir, R.D.; Huang, X.; Scarpa, R.C.; Bacarra, N.M.E.; Romano, D.M.; Hartshorn, M.A.; Tanzi, R.E.; Bush, A.I. Dramatic Aggregation of Alzheimer Aβ by Cu(II) Is Induced by Conditions Representing Physiological Acidosis. J. Biol. Chem. 1998, 273, 12817–12826. [Google Scholar] [CrossRef]
- Cherny, R.A.; Atwood, C.S.; Xilinas, M.E.; Gray, D.N.; Jones, W.D.; McLean, C.A.; Barnham, K.J.; Volitakis, I.; Fraser, F.W.; Kim, Y.-S.; et al. Treatment with a Copper-Zinc Chelator Markedly and Rapidly Inhibits β-Amyloid Accumulation in Alzheimer’s Disease Transgenic Mice. Neuron 2001, 30, 665–676. [Google Scholar] [CrossRef]
- Qin, K.; Yang, Y.; Mastrangelo, P.; Westaway, D. Mapping Cu(II) Binding Sites in Prion Proteins by Diethyl Pyrocarbonate Modification and Matrix-assisted Laser Desorption Ionization-Time of Flight (MALDI-TOF) Mass Spectrometric Footprinting. J. Biol. Chem. 2002, 277, 1981–1990. [Google Scholar] [CrossRef]
- Huang, A.; Li, W.; Shi, S.; Yao, T. Quantitative Fluorescence Quenching on Antibody-conjugated Graphene Oxide as a Platform for Protein Sensing. Sci. Rep. 2017, 7, 40772. [Google Scholar] [CrossRef]
- Zhao, H.; Zastrow, M.L. Transition Metals Induce Quenching of Monomeric Near-Infrared Fluorescent Proteins. Biochemistry 2022, 61, 494–504. [Google Scholar] [CrossRef]


| Application | Biological Research | Reference |
|---|---|---|
| Förster or fluorescence resonance energy transfer (FRET) | Protein-protein interactions or conformational changes within proteins | [30,31,32] |
| Biosensors | Monitoring of small biomolecules or other physiological intracellular processes | [33,34,35,36,37] |
| Optogenetics | Measuring or controlling molecular signals, cells, or groups of cells | [38,39,40] |
| Chemogenetics | Monitoring cellular receptors that affect signal pathways within a cell | [41,42] |
| Subcellular localization | Monitoring the location of the target molecule in cells | [43,44,45] |
| In vivo imaging | Imaging plasmids or protein-protein interactions in organs | [46,47,48] |
| Genome editing | Monitoring genome editing | [49,50,51,52] |
| Color | Protein | λEx (nm) | λEm (nm) | QY | Oligomerization | Reference |
|---|---|---|---|---|---|---|
| Blue/UV | Sirius | 355 | 424 | 0.24 | Monomer | [57] |
| Azurite | 383 | 447 | 0.55 | Weak dimer | [58] | |
| moxBFP | 385 | 448 | 0.56 | Monomer | [59] | |
| Cyan | Aquamarine | 430 | 474 | 0.89 | Monomer | [60] |
| Cerulean | 433 | 475 | 0.62 | Weak dimer | [61] | |
| CyPet | 435 | 477 | 0.51 | Weak dimer | [62] | |
| mTurquoise2 | 434 | 474 | 0.93 | Monomer | [63] | |
| Green | mEGFP | 488 | 507 | 0.60 | Monomer | [64] |
| mClover3 | 506 | 518 | 0.78 | Monomer | [65] | |
| mNeonGreen | 506 | 517 | 0.80 | Monomer | [66] | |
| Yellow | mGold | 515 | 530 | 0.64 | Monomer | [67] |
| mCitrine | 516 | 529 | 0.74 | Monomer | [64] | |
| mVenus | 515 | 527 | 0.64 | Monomer | [68] | |
| Orange | mOrange | 548 | 562 | 0.69 | Monomer | [69] |
| mKO2 | 551 | 565 | 0.62 | Monomer | [70] | |
| TurboRFP | 553 | 574 | 0.67 | Dimer | [71] | |
| tdTomato | 554 | 581 | 0.69 | Tandem dimer | [69] | |
| Red | mApple | 568 | 592 | 0.49 | Monomer | [72] |
| mScarlet | 569 | 594 | 0.70 | Monomer | [73] | |
| mCherry | 587 | 610 | 0.22 | Monomer | [69] | |
| DsRed2 | 561 | 587 | 0.55 | Tetramer | [74] | |
| Far-Red | mPlum | 590 | 649 | 0.10 | Monomer | [75] |
| mRaspberry | 598 | 625 | 0.15 | Monomer | [75] | |
| mNeptune | 600 | 650 | 0.20 | Monomer | [76] | |
| TagRFP657 | 611 | 657 | 0.10 | Monomer | [77] |
| Property | Protein | λEx (nm) | λEm (nm) | QY | Brightness | Oligomerization | Ref. | |
|---|---|---|---|---|---|---|---|---|
| Large Stokes Shift | tKeima | 440 | 616 | 0.22 | 3.19 | Tetramer | [79] | |
| LSSmKate2 | 460 | 605 | 0.17 | 4.42 | Monomer | [80] | ||
| T-sapphire | 399 | 511 | 0.60 | 26.4 | Weak dimer | [81] | ||
| Photoactivatable | PAmKate | 586 | 628 | 0.18 | 4.5 | Monomer | [82] | |
| PAmCherry1 | 564 | 595 | 0.46 | 8.28 | Monomer | [83] | ||
| PATagRFP | 562 | 595 | 0.38 | 25.08 | Monomer | [84] | ||
| Photoswitchable | Dronpa | 503 | 518 | 0.85 | 80.75 | Monomer | [85] | |
| mGeos-M | 503 | 514 | 0.85 | 43.86 | Monomer | [86] | ||
| rsTagRFP | 567 | 585 | 0.11 | 4.05 | Monomer | [87] | ||
| Photoconvertible | Kaede | Green | 508 | 518 | 0.88 | 86.94 | Tetramer | [88] |
| Red | 572 | 580 | 0.33 | 19.93 | ||||
| mEos3.2 | Green | 507 | 516 | 0.84 | 53.26 | Monomer | [89] | |
| Red | 572 | 580 | 0.55 | 17.71 | ||||
| Dendra | Green | 492 | 508 | 0.65 | 58.5 | Tetramer | [90] | |
| Red | 557 | 575 | 0.68 | 23.8 | ||||
| FP | λex | λem | Chromophore Sequence | Quenchable Metal Ion | Kd (μM) | Reference |
|---|---|---|---|---|---|---|
| DsRed | 558 | 583 | QYG | Cu2+ | 14.80 | [97] |
| mBFP1 | 374 | 447 | THG | Cu2+ | 24 | [35] |
| Iq-mEmerald | 450 | 509 | TYG | Cu2+ | 0.2 | [9] |
| Dronpa 1 | 448 | 518 | CYG | Cu2+ | N/A | [98] |
| AmCyan | 474 | 486 | MYG | Cu2+ | 56.10 | [99] |
| mOrange2 | 549 | 565 | TYG | Cu2+ | 21.46 | [99] |
| ZsYellow 1 | 529 | 539 | KYG | Cu2+ | N/A | [92] |
| ZsGreen | 496 | 506 | NYG | Fe2+ | 11.5 | [100] |
| Fe3+ | 16.3 | |||||
| Cu2+ | 68.2 | |||||
| DendFP (green) | 492 | 508 | HYG | Fe2+ | 24.59 | [93] |
| Fe3+ | 41.66 | |||||
| Cu2+ | 137.18 |
| FP | PDB Code | Resolution (Å) | Metal Ion | Interaction Residues | Reference |
|---|---|---|---|---|---|
| DsRed 1 | Cu2+ | Cys117 | [97] | ||
| mBFP1 2 | 1KYR | 1.50 | Cu2+ | Chromophore | [35] |
| 1KYS | 1.44 | Zn2+ | Chromophore | ||
| Iq-mEmerald 2 | 4KW8 | 2.46 | Ni2+ | H202-H204 | [9] |
| 4KW9 | 1.80 | Zn2+ | H202-H204 | ||
| Dronpa 2 | 5HZS | 2.17 | Co2+ | H202-H212 | [98] |
| 5HZT | 2.84 | Cu2+ | H194-H202 | ||
| 5HZU | 1.89 | Ni2+ | H202-H212 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nam, K.H. Fluorescent Protein-Based Metal Biosensors. Chemosensors 2023, 11, 216. https://doi.org/10.3390/chemosensors11040216
Nam KH. Fluorescent Protein-Based Metal Biosensors. Chemosensors. 2023; 11(4):216. https://doi.org/10.3390/chemosensors11040216
Chicago/Turabian StyleNam, Ki Hyun. 2023. "Fluorescent Protein-Based Metal Biosensors" Chemosensors 11, no. 4: 216. https://doi.org/10.3390/chemosensors11040216
APA StyleNam, K. H. (2023). Fluorescent Protein-Based Metal Biosensors. Chemosensors, 11(4), 216. https://doi.org/10.3390/chemosensors11040216






