Bioinspired Materials for Sensor and Clinical Applications: Two Case Studies
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
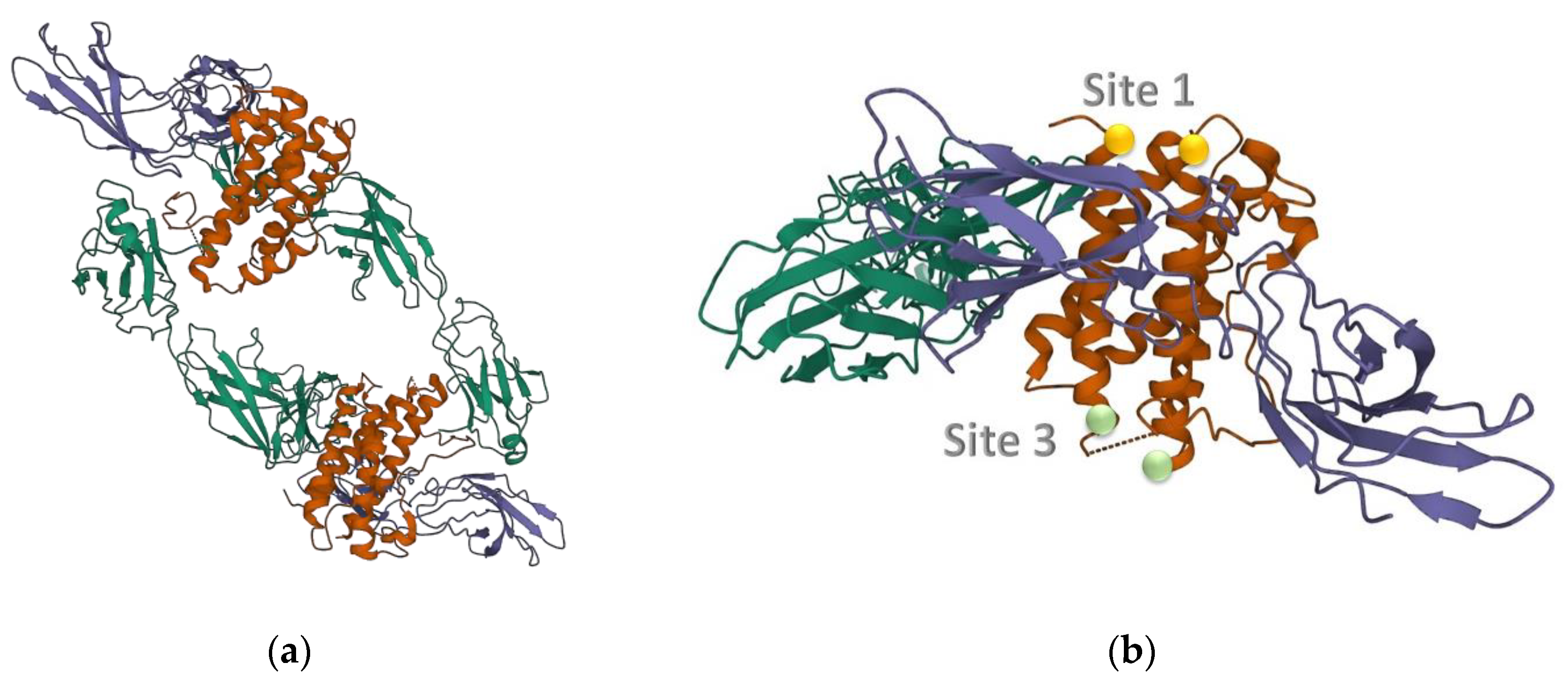
2.1.1. Cytokine IL-6 Receptors
2.1.2. α-Thrombin Receptors
| Receptor/PDB Code | KD ** (nM) | Area (Å) |
|---|---|---|
| TBA-K+/4DII | 34 *** | 540 §/583 ** |
| TBA-Na+/4DIH | 34 *** | 565 §/597 ** |
| mTBA/3QLP | 25 *** | 657 #/703 ** |
| TBAΔT3/4LZ4 | 55 *** | 524 §/548 ** |
| TBAΔT12/4LZ1 | 39 *** | 534 §/545 ** |
| TBA-T4W/6EO6 | 1 *** | 708 #/760 ** |
| TBA-T4K/6EO7 | 0.39 *** | 641 #/662 ** |
| TerΔT3_E/5EW1_E | 55 *** | 501 §§/531 ** |
| NU172-K+/6EVV | 0.29 ** | 591 ##/656 ** |
| NU172-Na+/6GN7 | 35 ** | 588 ##/643 ** |
| RE31/5CMX | 0.56 *** | 552 ##/661 ** |
| HD22/4I7Y | 29 *** | 1079 §§/1294 *** |
| TerΔT3_D/5EW1_D | 29 *** | 1116 §§/1135 *** |
| mToggle-25t/5DO4 | 0.002 *** | 925 #/1344 *** |
| Toggle-25t/3DD2 | 1.9 *** | 755 */1135 *** |
2.2. Methods
3. Results
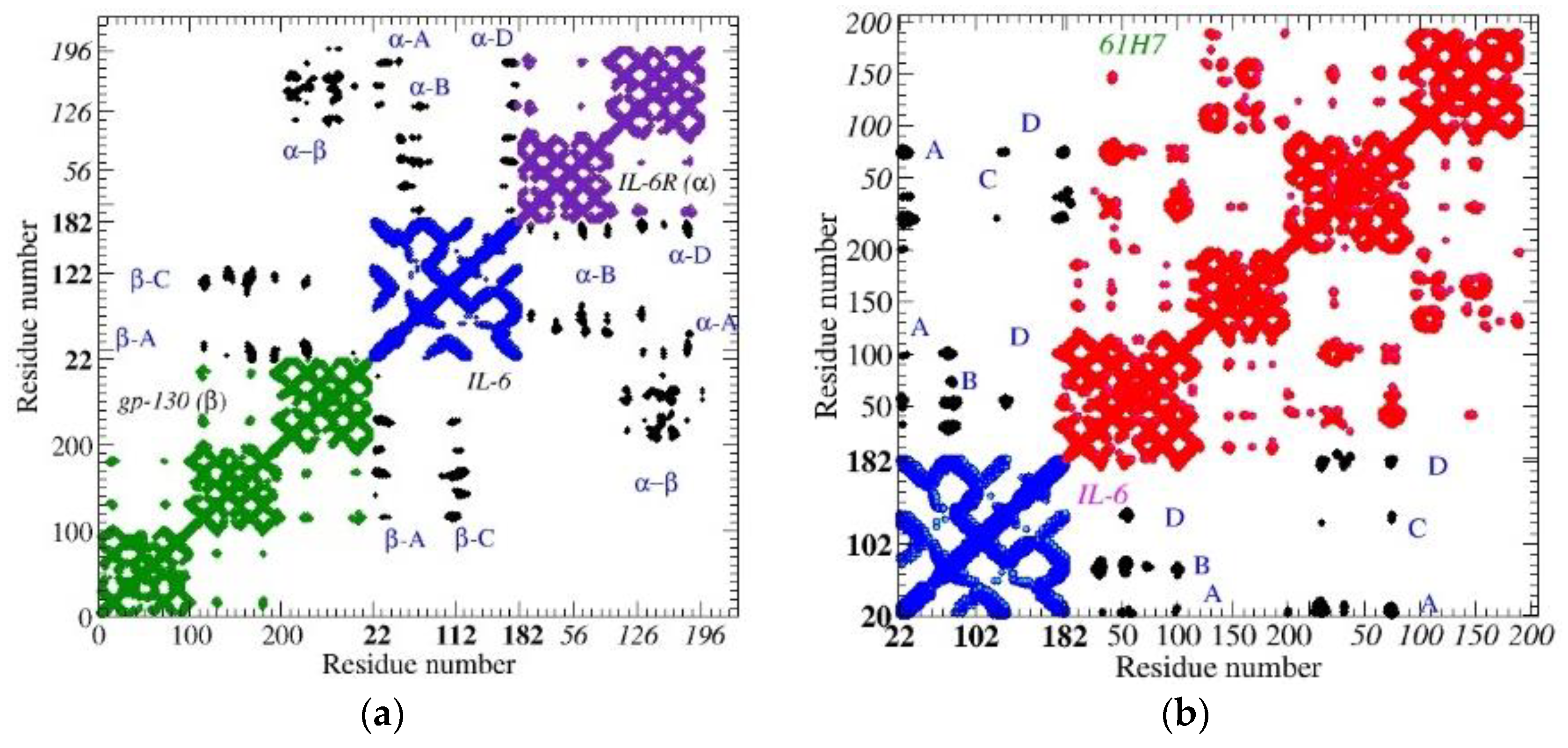
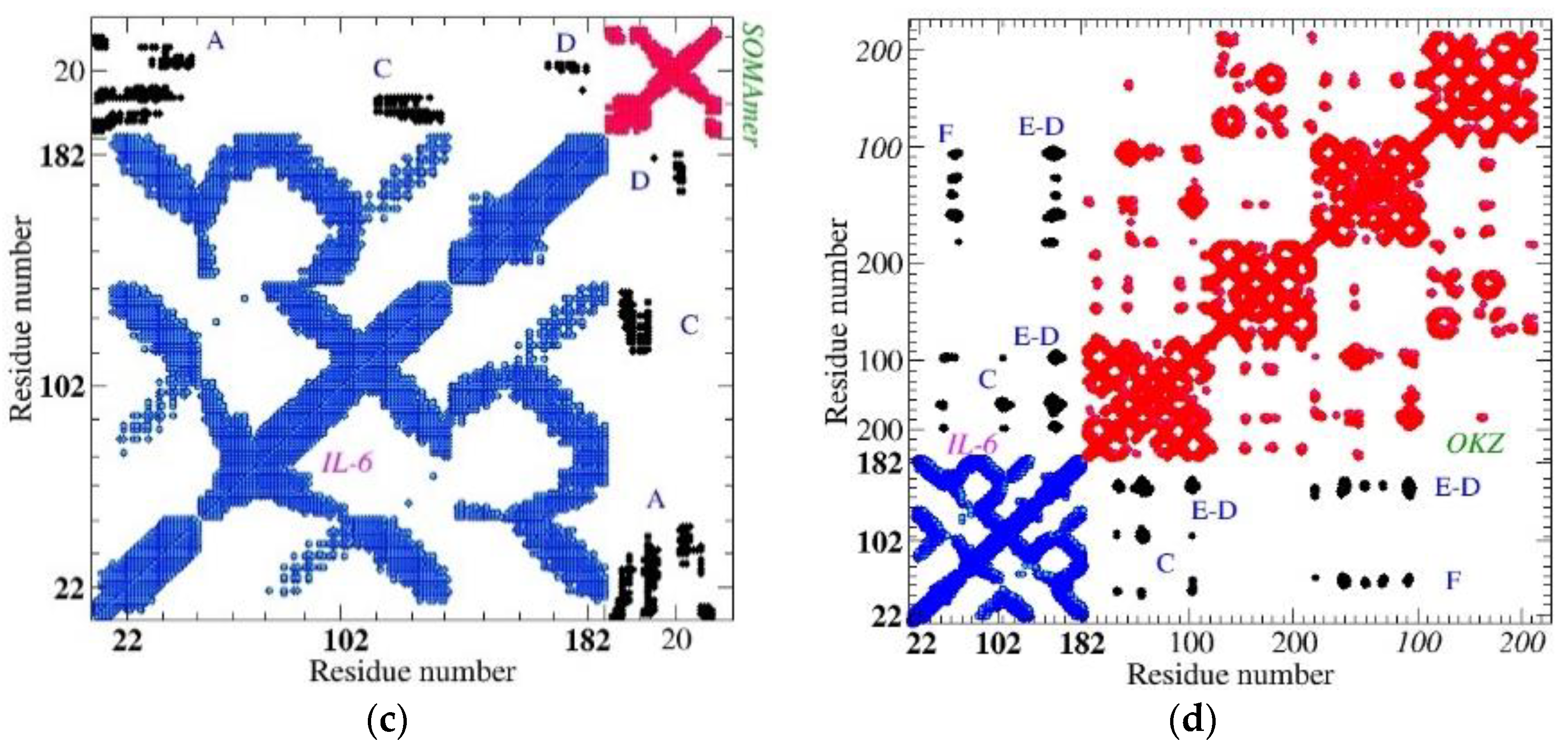
3.1. Topological Investigation
3.2. Electronic Analysis
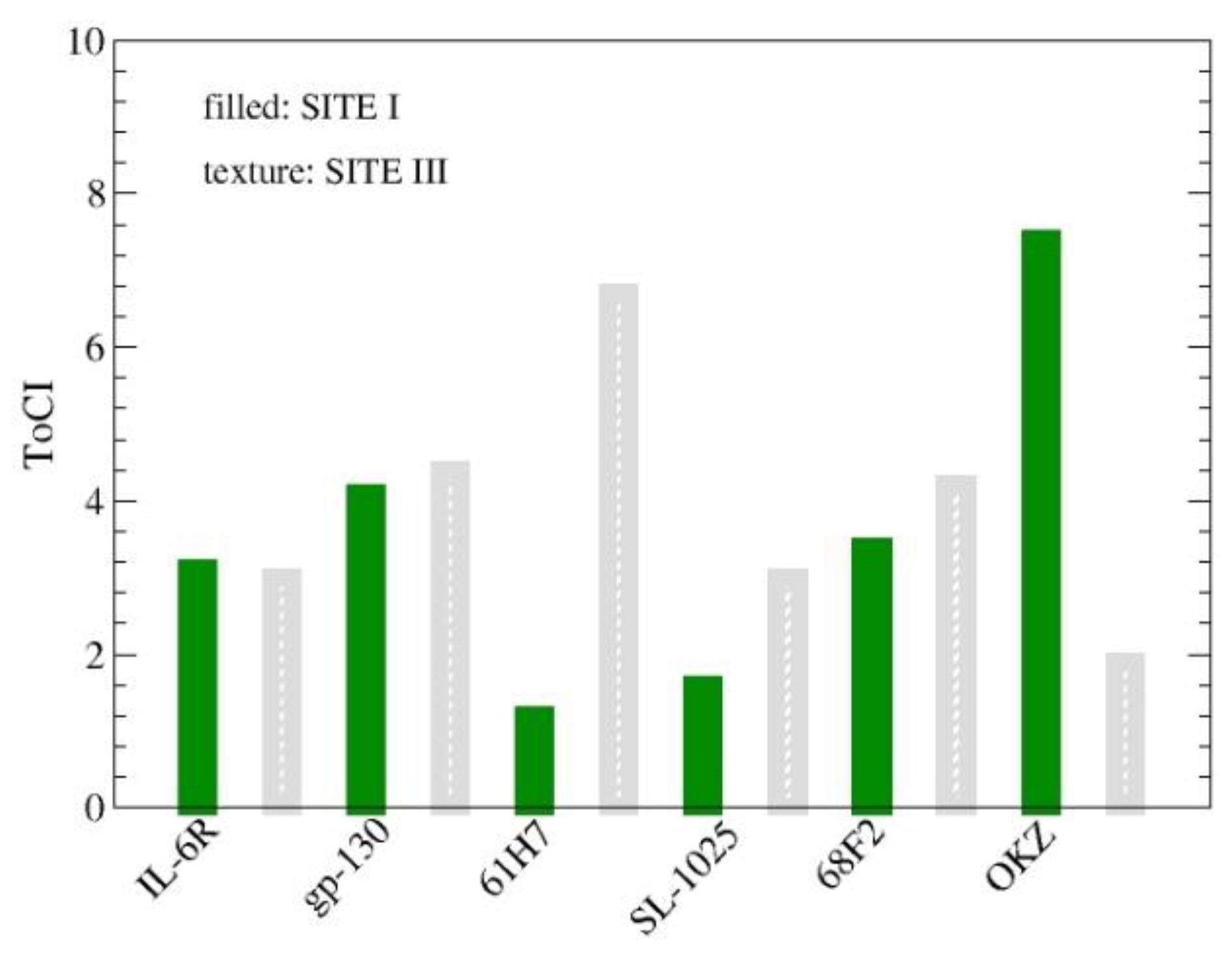
3.2.1. ToCI for Cytokine IL-6 Receptors
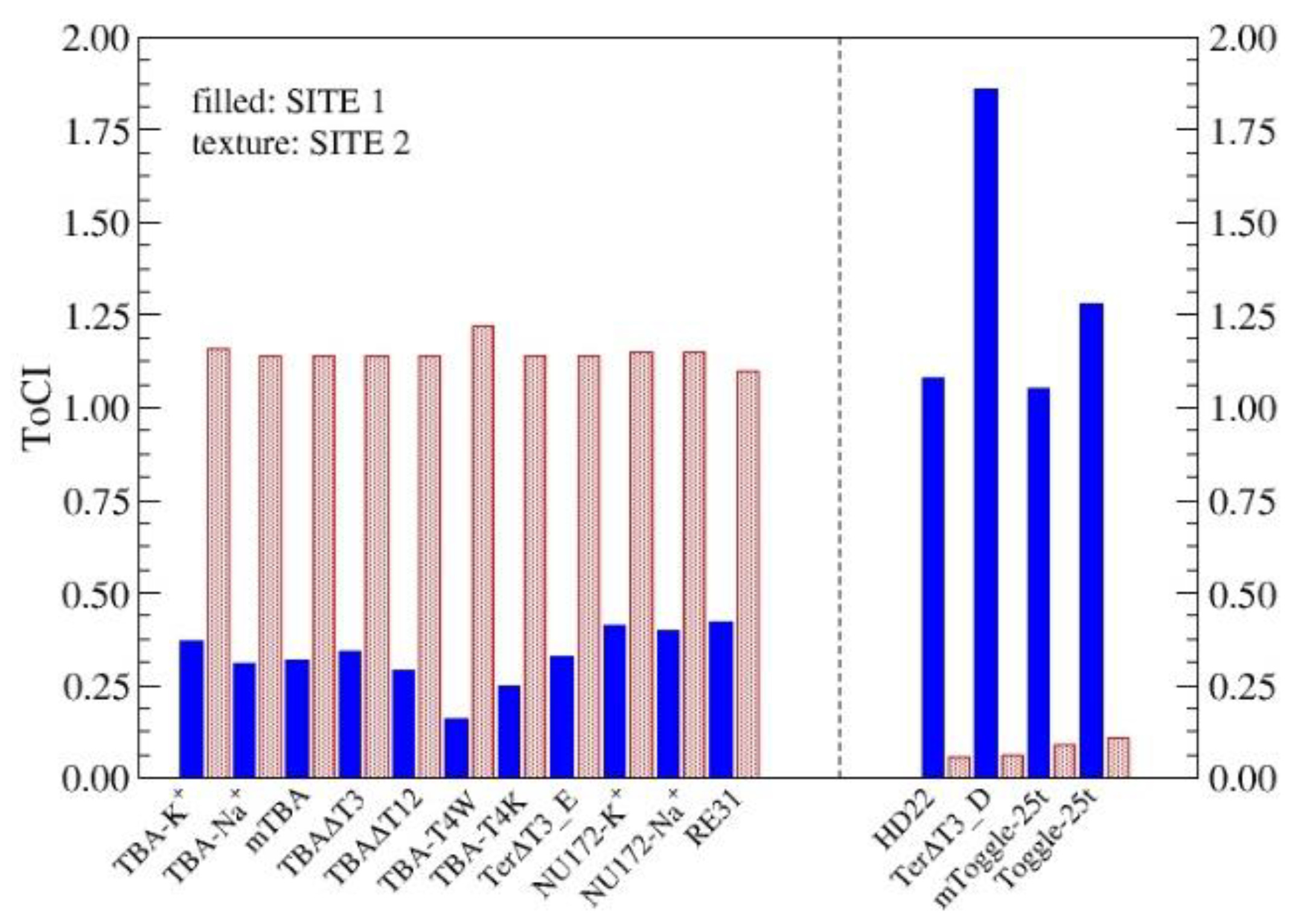
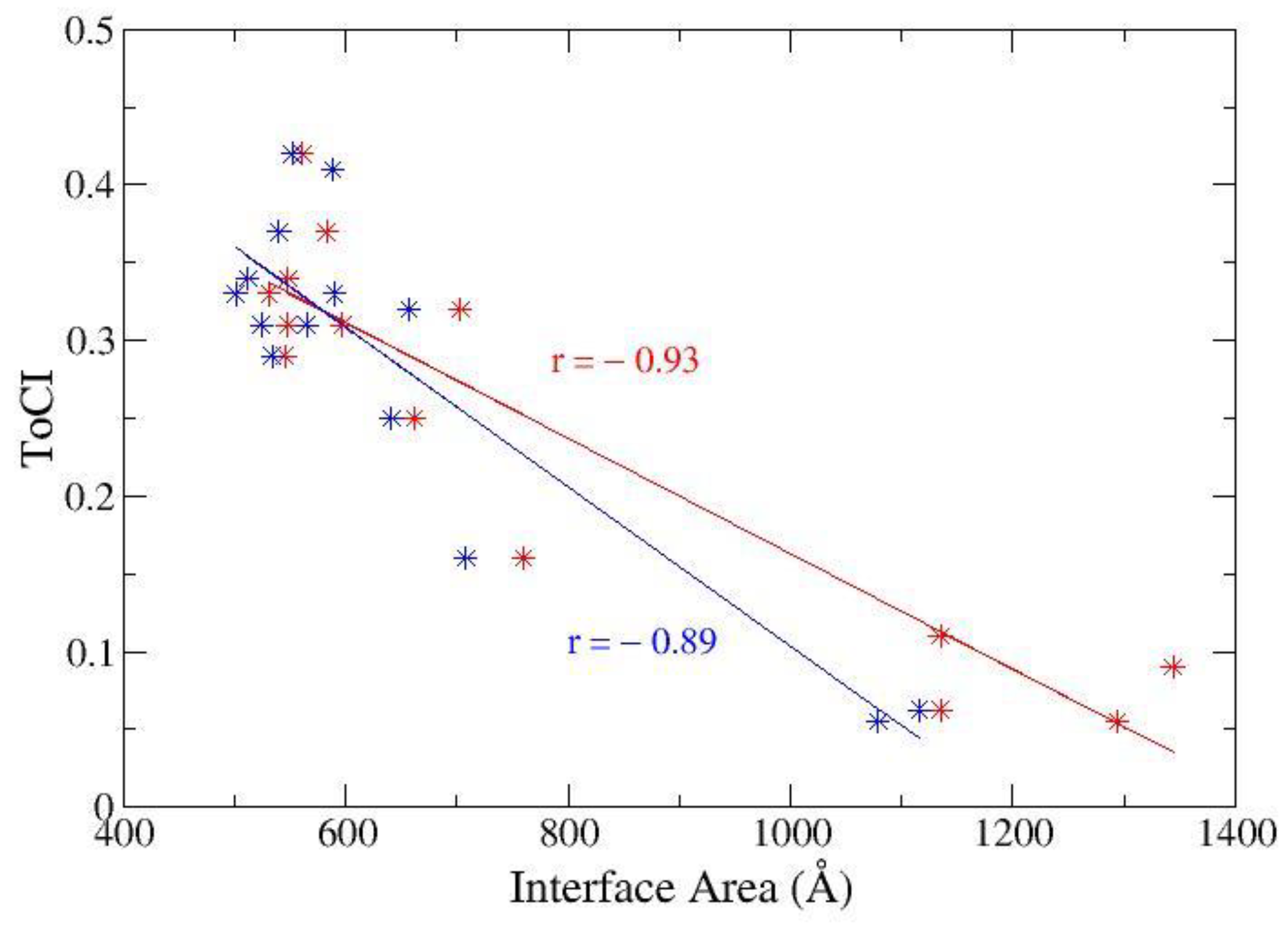
3.2.2. ToCI for α-Thrombin Receptors
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Holzinger, M.; Le Goff, A.; Cosnier, S. Nanomaterials for biosensing applications: A review. Front. Chem. 2014, 2, 63. [Google Scholar] [CrossRef] [PubMed]
- Frokjaer, S.; Otzen, D.E. Protein drug stability: A formulation challenge. Nat. Rev. Drug Discov. 2005, 4, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Chen, X. Aptamer-based targeted therapy. Adv. Drug Deliv. Rev. 2018, 134, 65–78. [Google Scholar] [CrossRef]
- Hawkins, M.J.; Soon-Shiong, P.; Desai, N. Protein nanoparticles as drug carriers in clinical medicine. Drug Deliv. Rev. 2008, 60, 876–885. [Google Scholar] [CrossRef] [PubMed]
- Heath, J.R.; Ratner, M.A. Molecular Electronics; The National Academies Press: Washington, DC, USA, 2003; pp. 43–49. [Google Scholar]
- Maruccio, G.; Cingolani, R.; Rinaldi, R. Projecting the nanoworld: Concepts, results and perspectives of molecular electronics. J. Mater. Chem. 2004, 14, 542–554. [Google Scholar] [CrossRef]
- Lucas, M.; Riedo, E. Invited review article: Combining scanning probe microscopy with optical spectroscopy for applications in biology and materials science. Rev. Sci. Instrum. 2012, 83, 061101. [Google Scholar] [CrossRef]
- Hawkes, P.W.; Spence, J.C. (Eds.) Springer Handbook of Microscopy; Springer International Publishing: Cham, Switzerland, 2019; pp. 1064–1071. [Google Scholar]
- Koukos, P.I.; Bonvin, A.M.J.J. Integrative modelling of biomolecular complexes. J. Mol. Bio. 2020, 432, 2861–2881. [Google Scholar] [CrossRef]
- Alfinito, E.; Reggiani, L.; Pousset, J. Proteotronics: Electronic devices based on proteins. In Proceedings of the Second National Conference on Sensors, Rome, Italy, 19–21 February 2014; Springer International Publishing: Cham, Switzerland, 2015; pp. 3–7. [Google Scholar]
- Alfinito, E.; Reggiani, L.; Cataldo, R.; De Nunzio, G.; Giotta, L.; Guascito, M.R. Modeling the microscopic electrical properties of thrombin binding aptamer (TBA) for label-free biosensors. Nanotechnology 2017, 28, 065502. [Google Scholar] [CrossRef]
- Cataldo, R.; Ciriaco, F.; Alfinito, E. A validation strategy for in silico generated aptamers. Comput. Biol. Chem. 2018, 77, 123–130. [Google Scholar] [CrossRef]
- Cataldo, R.; Giotta, L.; Guascito, M.R.; Alfinito, E. Assessing the quality of in silico produced biomolecules: The discovery of a new conformer. J. Phys. Chem. B 2019, 123, 1265–1273. [Google Scholar] [CrossRef]
- Cataldo, R.; Leuzzi, M.; Alfinito, E. Modelling and development of electrical aptasensors: A short review. Chemosensors 2018, 6, 20. [Google Scholar] [CrossRef]
- Cataldo, R.; De Nunzio, G.; Millithaler, J.F.; Alfinito, E. Aptamers which Target Proteins: What proteotronics suggests to pharmaceutics. Curr. Pharm. Des. 2020, 26, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Alfinito, E.; Beccaria, M.; Ciccarese, M. Biosensing cytokine IL-6: A comparative analysis of natural and synthetic receptors. Biosensors 2020, 10, 106. [Google Scholar] [CrossRef]
- Alfinito, E.; Cataldo, R.; Reggiani, L. A pH-based bio-rheostat: A proof-of-concept. Appl. Phys. Lett. 2022, 120, 013701. [Google Scholar] [CrossRef]
- Sifniotis, V.; Cruz, E.; Eroglu, B.; Kayser, V. Current advancements in addressing key challenges of therapeutic antibody design, manufacture, and formulation. Antibodies 2019, 8, 36. [Google Scholar] [CrossRef]
- Burkovitz, A.; Ofran, Y. Understanding differences between synthetic and natural antibodies can help improve antibody engineering. In Mabs; Taylor & Francis: Oxford, UK, 2016; Volume 8, pp. 278–287. [Google Scholar]
- Glassy, M.C.; Gupta, R. Technical and ethical limitations in making human monoclonal antibodies (an overview). Human Monoclonal Antibodies: Methods and Protocols. In Human Monoclonal Antibodies; Methods in Molecular Biology; Steinitz, M., Ed.; Humana: Totowa, NJ, USA, 2014; Volume 1060, pp. 9–36. [Google Scholar]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Cui, F.; Zhou, Z.; Zhou, H.S. Molecularly imprinted polymers and surface imprinted polymers based electrochemical biosensor for infectious diseases. Sensors 2020, 20, 996. [Google Scholar] [CrossRef] [PubMed]
- Dabrowski, M.; Lach, P.; Cieplak, M.; Kutner, W. Nanostructured molecularly imprinted polymers for protein chemosensing. Biosens. Bioelectr. 2018, 102, 17–26. [Google Scholar] [CrossRef]
- Erdőssy, J.; Horváth, V.; Yarman, A.; Scheller, F.W.; Gyurcsányi, R.E. Electrosynthesized molecularly imprinted polymers for protein recognition. TrAC 2016, 79, 179–190. [Google Scholar] [CrossRef]
- Gonçalves, L.M. Electropolymerized molecularly imprinted polymers: Perceptions based on recent literature for soon-to-be world-class scientists. Opin. Electrochem. 2021, 25, 100640. [Google Scholar] [CrossRef]
- Crapnell, R.D.; Hudson, A.; Foster, C.W.; Eersels, K.; Grinsven, B.V.; Cleij, T.J.; Banks, C.E.; Peeters, M. Recent advances in electrosynthesized molecularly imprinted polymer sensing platforms for bioanalyte detection. Sensors 2019, 19, 1204. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Guo, T.; Yang, Y.; Zhang, Z. Surface imprinted macroporous film for high performance protein recognition in combination with quartz crystal microbalance. Sens. Actuators B Chem. 2011, 153, 96–102. [Google Scholar] [CrossRef]
- Dabrowski, M.; Cieplak, M.; Sharma, P.S.; Borowicz, P.; Noworyta, K.; Lisowski, W.; d’Souza, F.; Kuhn, A.; Kutner, W. Hierarchical templating in deposition of semi-covalently imprinted inverse opal polythiophene film for femtomolar determination of human serum albumin. Biosens. Bioelectr. 2017, 94, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Tchekwagep, P.M.S.; Crapnell, R.D.; Banks, C.E.; Betlem, K.; Rinner, U.; Canfarotta, F.; Lowdon, J.W.; Eersels, K.; van Grinsven, B.; Peeters, M.; et al. A Critical Review on the Use of Molecular Imprinting for Trace Heavy Metal and Micropollutant Detection. Chemosensors 2022, 10, 296. [Google Scholar] [CrossRef]
- Primiceri, E.; Chiriacò, M.S.; Notarangelo, F.M.; Crocamo, A.; Ardissino, D.; Cereda, M.; Bramanti, A.P.; Bianchessi, M.A.; Giannelli, G.; Maruccio, G. Key enabling technologies for point-of-care diagnostics. Sensors 2018, 18, 3607. [Google Scholar] [CrossRef]
- Siciliano, G.; Monteduro, A.G.; Turco, A.; Primiceri, E.; Rizzato, S.; Depalo, N.; Curri, M.L.; Maruccio, G. Polydopamine-Coated Magnetic Iron Oxide Nanoparticles: From Design to Applications. Nanomaterials 2022, 12, 1145. [Google Scholar] [CrossRef]
- Ali, G.K.; Omer, K.M. Molecular imprinted polymer combined with aptamer (MIP-aptamer) as a hybrid dual recognition element for bio (chemical) sensing applications. Rev. Talanta 2022, 236, 122878. [Google Scholar] [CrossRef]
- Csermely, P.; Palotai, R.; Nussinov, R. Induced fit, conformational selection and independent dynamic segments: An extended view of binding events. Nat. Preced. 2010, 35, 539–546. [Google Scholar]
- Levy, Y.; Wolynes, P.G.; Onuchic, J.N. Protein topology determines binding mechanism. PNAS 2004, 101, 511–516. [Google Scholar] [CrossRef]
- Chiriacò, M.S.; Primiceri, E.; De Feo, F.; Montanaro, A.; Monteduro, A.G.; Tinelli, A.; Megha, M.; Carati, D.; Maruccio, G. Simultaneous detection of multiple lower genital tract pathogens by an impedimetric immunochip. Biosens. Bioelectron. 2016, 79, 9–14. [Google Scholar] [CrossRef]
- Buja, I.; Sabella, E.; Monteduro, A.G.; Rizzato, S.; Bellis, L.D.; Elicio, V.; Formica, L.; Luvisi, A.; Maruccio, G. Detection of Ampelovirus and Nepovirus by Lab-on-a-Chip: A Promising Alternative to ELISA Test for Large Scale Health Screening of Grapevine. Biosensors 2022, 12, 147. [Google Scholar] [CrossRef] [PubMed]
- Colombelli, A.; Primiceri, E.; Rizzato, S.; Monteduro, A.G.; Maruccio, G.; Rella, R.; Rella, R.; Manera, M.G. Nanoplasmonic biosensing approach for endotoxin detection in pharmaceutical field. Chemosensors 2021, 9, 10. [Google Scholar] [CrossRef]
- Boulanger, M.J.; Chow, D.C.; Brevnova, E.E.; Garcia, K.C. Hexameric structure and assembly of the interleukin-6/IL-6 α-receptor/gp130 complex. Science 2003, 300, 2101–2104. [Google Scholar] [CrossRef] [PubMed]
- Shaw, S.; Bourne, T.; Meier, C.; Carrington, B.; Gelinas, R.; Henry, A.; Popplewell, A.; Adams, R.; Baker, T.; Rapecki, S.; et al. Discovery and characterization of olokizumab: A humanized antibody targeting interleukin-6 and neutralizing gp130-signaling. MAbs 2014, 6, 773–781. [Google Scholar] [CrossRef]
- Mihara, M.; Hashizume, M.; Yoshida, H.; Suzuki, M.; Shiina, M. IL-6/IL-6 receptor system and its role in physiological and pathological conditions. Clin. Sci. 2012, 122, 143–159. [Google Scholar] [CrossRef]
- Grivennikov, S.; Karin, M. Autocrine IL-6 signaling: A key event in tumorigenesis? Cancer Cell 2008, 13, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Sansone, P.; Storci, G.; Tavolari, S.; Guarnieri, T.; Giovannini, C.; Taffurelli, M.; Ceccarelli, C.; Santini, D.; Paterini, P.; Marcu, K.B.; et al. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J. Clin. Investig. 2007, 117, 3988–4002. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Klarenbeek, A.; Blanchetot, C.; Schragel, G.; Sadi, A.S.; Ongenae, N.; Hemrika, W.; Wijidenes, H.; Spinelli, S.; Desmyter, A.; Cambillau, C.; et al. Combining somatic mutations present in different in vivo affinity-matured antibodies isolated from immunized Lama glama yields ultra-potent antibody therapeutics. PEDS 2016, 29, 123–133. [Google Scholar] [CrossRef]
- Blanchetot, C.; De Jonge, N.; Desmyter, A.; Ongenae, N.; Hofman, E.; Klarenbeek, A.; Sadi, A.; Hultberg, A.; Spinelli, S.; Loris, R.; et al. Structural mimicry of receptor interaction by antagonistic interleukin-6 (IL-6) antibodies. J. Biol. Chem. 2016, 291, 13846–13854. [Google Scholar] [CrossRef]
- Gelinas, A.D.; Davies, D.R.; Edwards, T.E.; Rohloff, J.C.; Carter, J.D.; Zhang, C.; Gupta, S.; Ishikawa, Y.; Hirota, M.; Jarvis, T.C.; et al. Crystal structure of interleukin-6 in complex with a modified nucleic acid ligand. J. Biol. Chem. 2014, 289, 8720–8734. [Google Scholar] [CrossRef]
- Gelinas, A.D.; Davies, D.R.; Janjic, N. Embracing proteins: Structural themes in aptamer–protein complexes. Curr. Opin. Struct. Biol. 2016, 36, 122–132. [Google Scholar] [CrossRef]
- Pagano, B.; Martino, L.; Randazzo, A.; Giancola, C. Stability and binding properties of a modified thrombin binding aptamer. Biophys. J. 2008, 94, 562–569. [Google Scholar] [CrossRef]
- Ferroni, P.; Martini, F.; Portarena, I.; Grenga, I.; Riondino, S.; La Farina, F.; Laudisi, A.; Roselli, M.; Guadagni, F. An activated protein C-dependent thrombin generation assay predicts chemotherapy-associated venous thromboembolism in cancer patients. Thromb. Haemos. 2011, 105, 931–932. [Google Scholar]
- Saadeh, F.A.; Langhe, R.; Galvin, D.M.; Toole, S.O.; O’Donnell, D.M.; Gleeson, N.; Norris, L.A. Procoagulant activity in gynaecological cancer patients; the effect of surgery and chemotherapy. Thromb. Res. 2016, 139, 135–141. [Google Scholar] [CrossRef]
- Reddel, C.J.; Tan, C.W.; Chen, V.M. Thrombin generation and cancer: Contributors and consequences. Cancers 2019, 11, 100. [Google Scholar] [CrossRef]
- Lee, C.J.; Ansell, J.E. Direct thrombin inhibitors. Br. J. Clin. Pharmacol. 2011, 72, 581–592. [Google Scholar] [CrossRef]
- Pica, A.; Russo Krauss, I.; Parente, V.; Tateishi-Karimata, H.; Nagatoishi, S.; Tsumoto, K.; Sugimoto, N.; Sica, F. Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers. Nucleic Acids Res. 2017, 45, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Schultze, P.; Macaya, R.F.; Feigon, J. Three-dimensional solution structure of the thrombin-binding DNA aptamer d (GGTTGGTGTGGTTGG). J. Mol. Biol. 1994, 235, 1532–1547. [Google Scholar] [CrossRef] [PubMed]
- Macaya, R.F.; Schultze, P.; Smith, F.W.; Roe, J.A.; Feigon, J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. PNAS 1993, 90, 3745–3749. [Google Scholar] [CrossRef]
- Pica, A.; Russo Krauss, I.; Merlino, A.; Nagatoishi, S.; Sugimoto, N.; Sica, F. Dissecting the contribution of thrombin exosite I in the recognition of thrombin binding aptamer. FEBS J. 2013, 280, 6581–6588. [Google Scholar] [CrossRef] [PubMed]
- Russo Krauss, I.; Merlino, A.; Randazzo, A.; Mazzarella, L.; Sica, F. Crystallization and preliminary X-ray analysis of the complex of human α-thrombin with a modified thrombin-binding aptamer. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 961–963. [Google Scholar] [CrossRef] [PubMed]
- Dolot, R.; Lam, C.H.; Sierant, M.; Zhao, Q.; Liu, F.W.; Nawrot, B.; Egli, M.; Yang, X. Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity. Nucleic Acids Res. 2018, 46, 4819–4830. [Google Scholar] [CrossRef] [PubMed]
- Troisi, R.; Napolitano, V.; Spiridonova, V.; Russo Krauss, I.; Sica, F. Several structural motifs cooperate in determining the highly effective anti-thrombin activity of NU172 aptamer. Nucleic Acids Res. 2018, 46, 12177–12185. [Google Scholar] [CrossRef] [PubMed]
- Russo Krauss, I.; Spiridonova, V.; Pica, A.; Napolitano, V.; Sica, F. Different duplex/quadruplex junctions determine the properties of anti-thrombin aptamers with mixed folding. Nucleic Acids Res. 2016, 44, 983–991. [Google Scholar] [CrossRef] [PubMed]
- White, R.; Rusconi, C.; Scardino, E.; Wolberg, A.; Lawson, J.; Hoffman, M.; Sullenger, B. Generation of species cross-reactive aptamers using “toggle” SELEX. Mol. Ther. 2001, 4, 567–573. [Google Scholar] [CrossRef]
- Russo Krauss, I.; Pica, A.; Merlino, A.; Mazzarella, L.; Sica, F. Duplex–quadruplex motifs in a peculiar structural organization cooperatively contribute to thrombin binding of a DNA aptamer. Acta Crystallogr. D 2013, 69, 2403–2411. [Google Scholar] [CrossRef] [PubMed]
- Zavyalova, E.G.; Legatova, V.A.; Alieva, R.S.; Zalevsky, A.O.; Tashlitsky, V.N.; Arutyunyan, A.M.; Kopylov, A.M. Putative mechanisms underlying high inhibitory activities of bimodular DNA aptamers to thrombin. Biomolecules 2019, 9, 41. [Google Scholar] [CrossRef]
- Novoseltseva, A.; Zavyalova, E.; Golovin, A.; Kopylov, A. An insight into aptamer–protein complexes. Aptamers 2018, 2, 55–63. [Google Scholar]
- Troisi, R.; Balasco, N.; Autiero, I.; Vitagliano, L.; Sica, F. Exosite Binding in Thrombin: A Global Structural/Dynamic Overview of Complexes with Aptamers and Other Ligands. Int. J. Mol. Sci. 2021, 22, 10803. [Google Scholar] [CrossRef]
- Bender, B.J.; Gahbauer, S.; Luttens, A.; Lyu, J.; Webb, C.M.; Stein, R.M.; Fink, E.A.; Balius, T.E.; Carlsson, J.; Irwin, J.J.; et al. A practical guide to large-scale docking. Nat. Protoc. 2021, 16, 4799–4832. [Google Scholar] [CrossRef] [PubMed]
- Nasonov, E.; Fatenejad, S.; Feist, E.; Ivanova, M.; Korneva, E.; Krechikova, D.G.; Maslyanskiy, A.L.; Samsonov, M.; Stoilov, R.; Zonova, E.V.; et al. Olokizumab, a monoclonal antibody against interleukin 6, in combination with methotrexate in patients with rheumatoid arthritis inadequately controlled by methotrexate: Efficacy and safety results of a randomised controlled phase III study. ARD 2022, 81, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Russo Krauss, I.; Merlino, A.; Randazzo, A.; Novellino, E.; Mazzarella, L.; Sica, F. High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity. Nucleic Acids Res. 2012, 40, 8119–8128. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.V.; Allabush, F.; Bunka, D.; Tolley, A.; Mendes, P.M.; Tucker, J.H.; Turner, N.W. Hybrid aptamer-molecularly imprinted polymer (AptaMIP) nanoparticles selective for the antibiotic moxifloxacin. Polym. Chem. 2021, 12, 4394–4405. [Google Scholar] [CrossRef]


| Receptor | KD (pM) | Area * (Å) |
|---|---|---|
| OKZ | 10 1 | 840 2 |
| 61H7 | <10 2 | 940 2 |
| 68F2 | 10–20 2 | 1160 2 |
| SL1025 | 200 3 | >1200 3 |
| Receptor | Helix |
|---|---|
| IL-6R | A, B, D |
| gp-130 | A, C |
| Fab 68F2 | A, D |
| Fab 61HZ | A, D, B, C |
| Antibody-OKZ | C, D, E, F |
| SOMAmer-SL1025 | A, C, D |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alfinito, E.; Ciccarese, M.; Maruccio, G.; Monteduro, A.G.; Rizzato, S. Bioinspired Materials for Sensor and Clinical Applications: Two Case Studies. Chemosensors 2023, 11, 195. https://doi.org/10.3390/chemosensors11030195
Alfinito E, Ciccarese M, Maruccio G, Monteduro AG, Rizzato S. Bioinspired Materials for Sensor and Clinical Applications: Two Case Studies. Chemosensors. 2023; 11(3):195. https://doi.org/10.3390/chemosensors11030195
Chicago/Turabian StyleAlfinito, Eleonora, Mariangela Ciccarese, Giuseppe Maruccio, Anna Grazia Monteduro, and Silvia Rizzato. 2023. "Bioinspired Materials for Sensor and Clinical Applications: Two Case Studies" Chemosensors 11, no. 3: 195. https://doi.org/10.3390/chemosensors11030195
APA StyleAlfinito, E., Ciccarese, M., Maruccio, G., Monteduro, A. G., & Rizzato, S. (2023). Bioinspired Materials for Sensor and Clinical Applications: Two Case Studies. Chemosensors, 11(3), 195. https://doi.org/10.3390/chemosensors11030195










