SARS-CoV-2 Detection Methods
Abstract
:1. Introduction
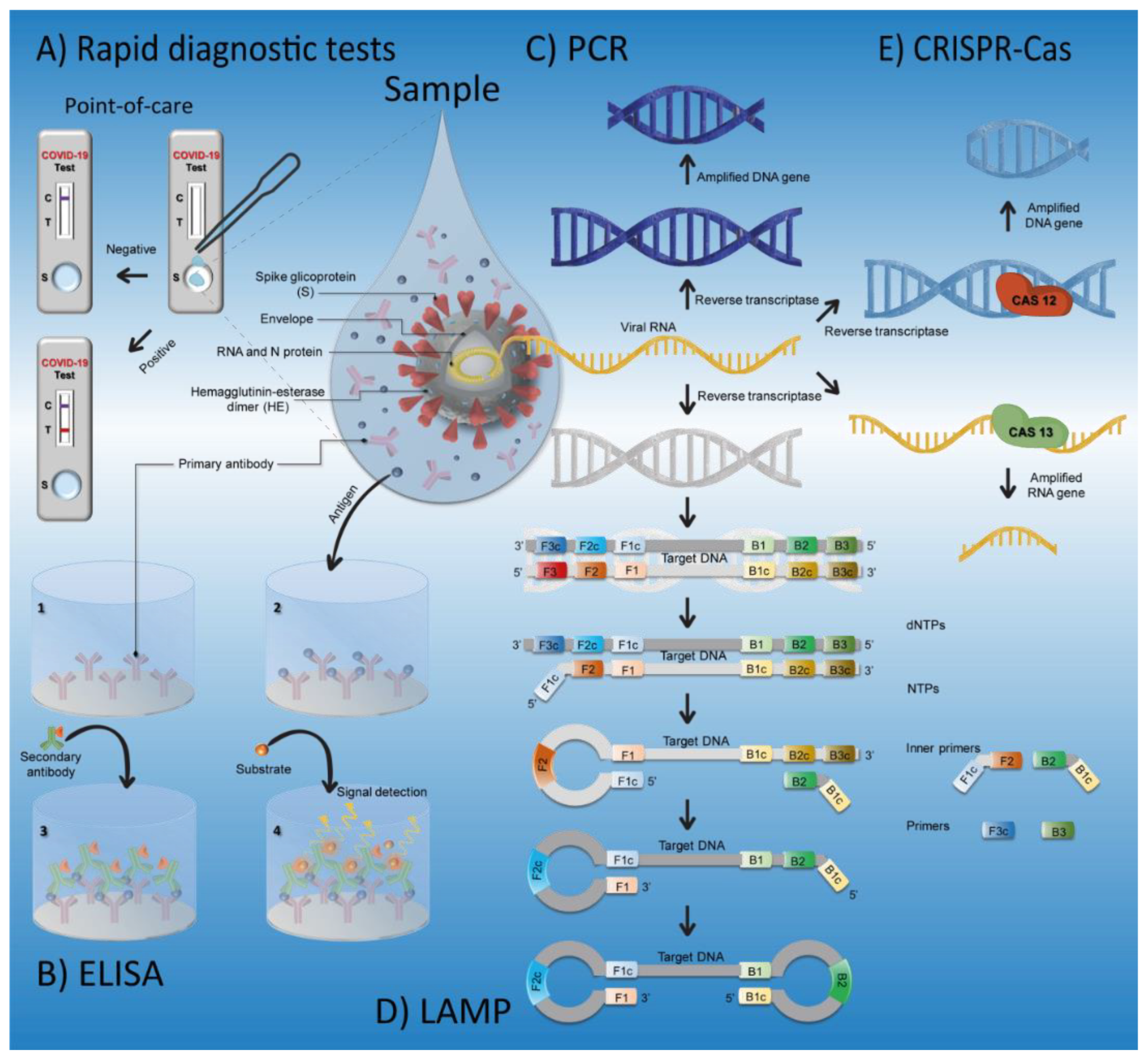
2. SARS-CoV-2 Detection Techniques
2.1. Clinically Useful Methods
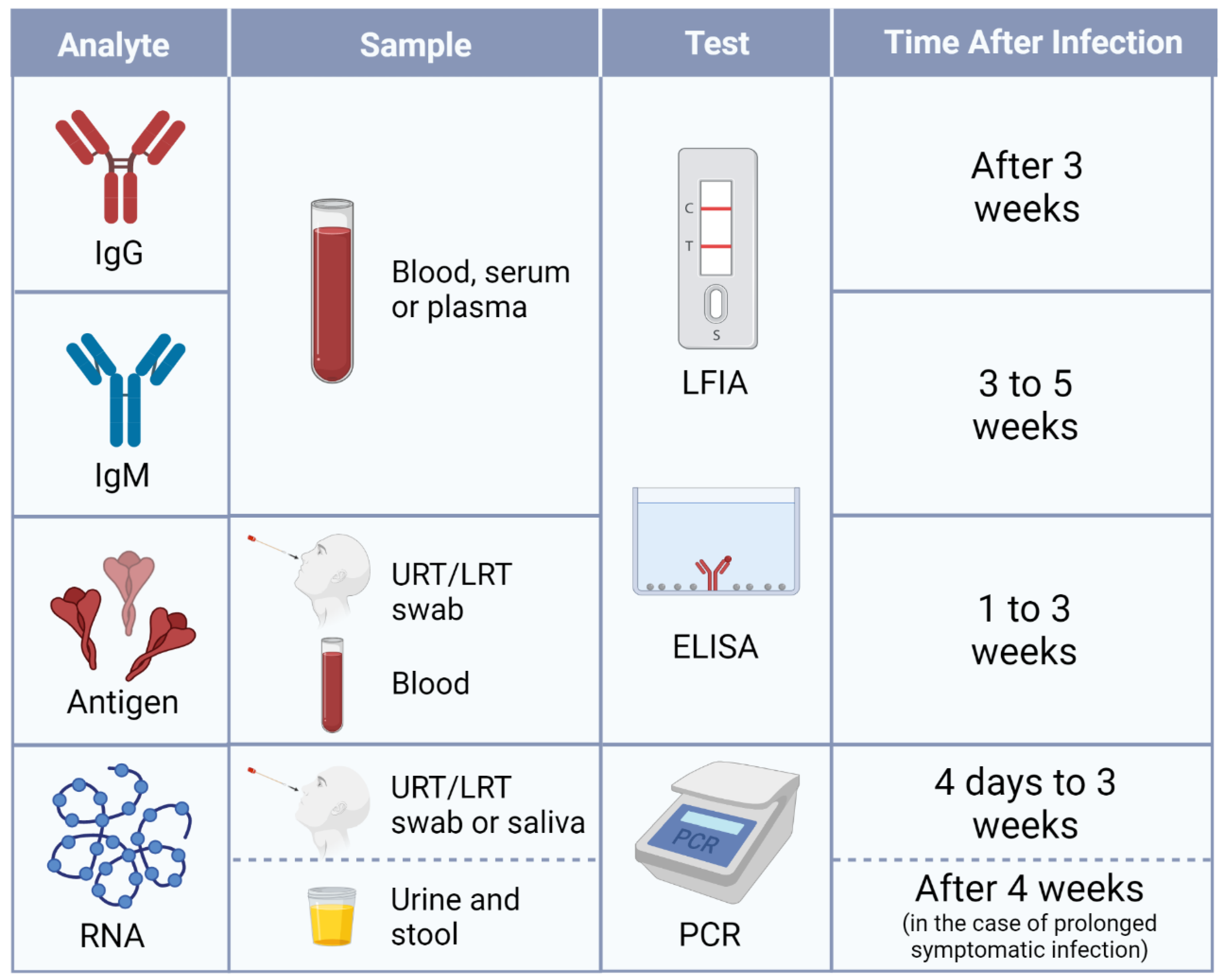
2.1.1. Antibody Tests
2.1.2. Antigen Tests
2.1.3. RT-qPCR
2.2. Potentially Useful Methods
2.2.1. RT-LAMP
2.2.2. CRISPR–Cas
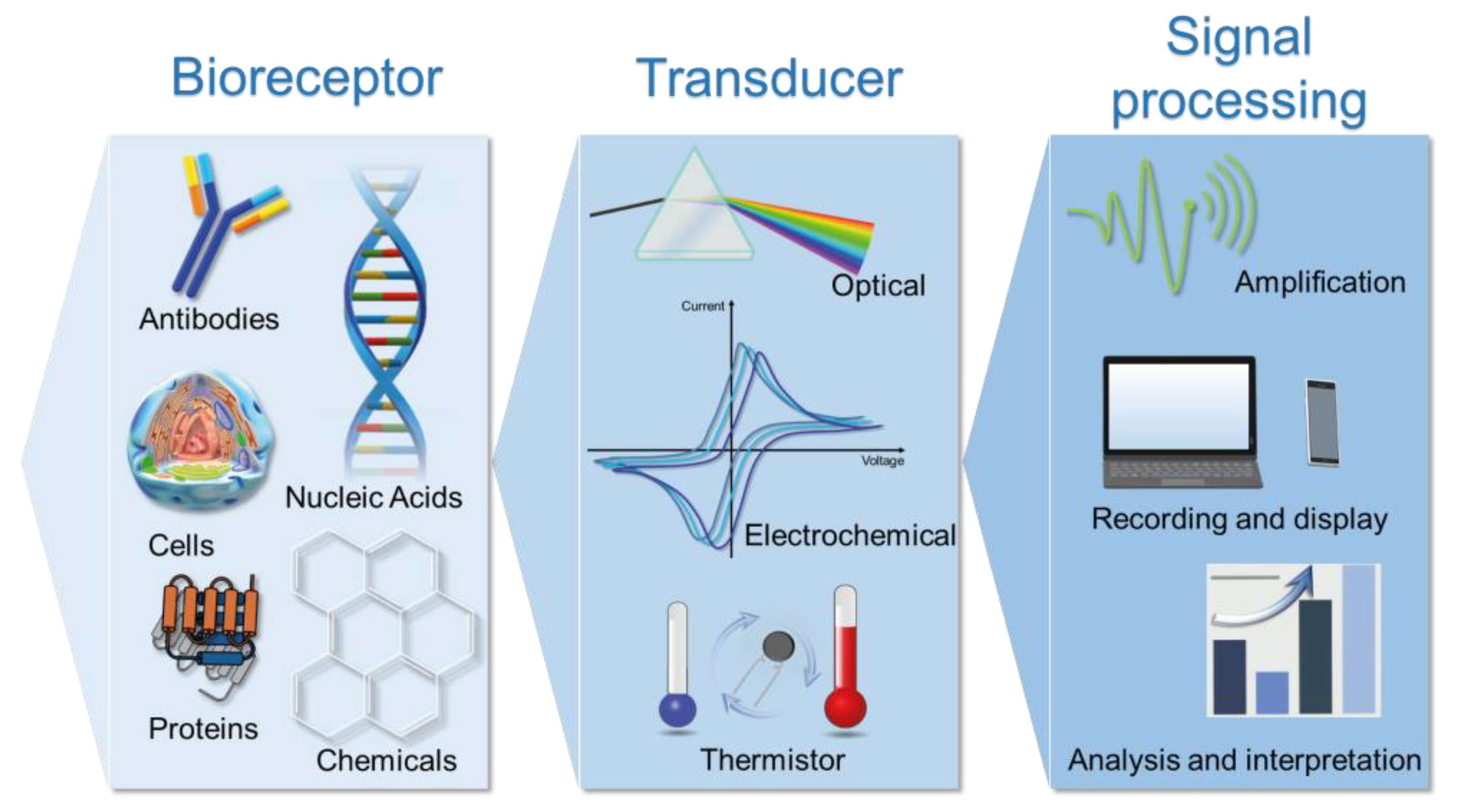
2.2.3. Biosensors
Biosensors Targeting Nucleic Acids
| Type | Bioreceptor | Target | Sample | Sensibility | Time | Ref. |
|---|---|---|---|---|---|---|
| Optical (SPR) | Oligonucleotides | RdRP, E, Orf1a | Synthetic targets | 0.22 pM | - | [47] |
| FET | PMO | RdRP | Throat swab/Serum | 2.29 fM/3.99 fM | 2 min | [48] |
| Electrochemical | Oligonucleotides | RdRP | - | 0.30 pM | 25 min | [49] |
| Electrochemical | Oligonucleotides | N | Nasopharyngeal swab/Saliva | 6.9 copies/μL | 5 min | [50] |
| Colorimetric | cDNA/AuNPs | N | Oropharyngeal swab | 0.18 ng/μL | 10 min | [51] |
| Electrochemiluminescent | Thiol-modified oligonucleotide | ORF1ab | Spiked human serum | 514 aM | - | [52] |
| Electrochemical | Biotinylated probe | ORF1ab | Spiked samples | 807 fM | - | [53] |
| Voltammetric | cDNA-Au@CD bioconjugates | RdRP | Sputum | 0.15 pM | 75 min | [54] |
| Electrochemiluminescent | Y-DNA probe | RdRP | Pharyngeal swab | 59 am | 180 min | [55] |
Biosensors Targeting Viral Proteins
Biosensors Targeting Antibodies
2.2.4. Sequencing
3. Test Selection
4. New Variants
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rezvani Ghomi, E.; Khosravi, F.; Mohseni, M.A.; Nourbakhsh, N.; Haji Mohammad Hoseini, M.; Singh, S.; Hedenqvist, M.S.; Ramakrishna, S. A Collection of the Novel Coronavirus (COVID-19) Detection Assays, Issues, and Challenges. Heliyon 2021, 7, e07247. [Google Scholar] [CrossRef]
- Nguyen, T.; Bang, D.D.; Wolff, A. 2019 Novel Coronavirus Disease (COVID-19): Paving the Road for Rapid Detection and Point-of-Care Diagnostics. Micromachines 2020, 11, 306. [Google Scholar] [CrossRef]
- Zella, D.; Giovanetti, M.; Benedetti, F.; Unali, F.; Spoto, S.; Guarino, M.; Angeletti, S.; Ciccozzi, M. The Variants Question: What Is the Problem? J. Med. Virol. 2021, 93, 6479–6485. [Google Scholar] [CrossRef]
- Parums, D. Editorial: Revised World Health Organization (WHO) Terminology for Variants of Concern and Variants of Interest of SARS-CoV-2. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2021, 27, e933622. [Google Scholar] [CrossRef]
- Boehm, E.; Kronig, I.; Neher, R.A.; Eckerle, I.; Vetter, P.; Kaiser, L. Novel SARS-CoV-2 Variants: The Pandemics within the Pandemic. Clin. Microbiol. Infect. 2021, 27, 1109–1117. [Google Scholar] [CrossRef]
- WHO Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed on 30 June 2021).
- Ravi, A.; Prabath Singh, V.; Chandran, R.; Venugopal, K.; Haridas, K.; Kavitha, R. COVID-19 Antibody Tests: An Overview. J. Pharm. Bioallied Sci. 2021, 13, 48. [Google Scholar] [CrossRef]
- CDC. Interim Guidelines for COVID-19 Antibody Testing; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2020; pp. 1–8.
- Lv, H.; Wu, N.C.; Tsang, O.T.Y.; Yuan, M.; Perera, R.A.P.M.; Leung, W.S.; So, R.T.Y.; Chan, J.M.C.; Yip, G.K.; Chik, T.S.H.; et al. Cross-Reactive Antibody Response between SARS-CoV-2 and SARS-CoV Infections. bioRxiv 2020, 31, 107725. [Google Scholar] [CrossRef]
- Udugama, B.; Kadhiresan, P.; Kozlowski, H.N.; Malekjahani, A.; Osborne, M.; Li, V.Y.C.; Chen, H.; Mubareka, S.; Gubbay, J.B.; Chan, W.C.W. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 2020, 14, 3822–3835. [Google Scholar] [CrossRef]
- Katsarou, K.; Bardani, E.; Kallemi, P.; Kalantidis, K. Viral Detection: Past, Present, and Future. BioEssays 2019, 41, 1900049. [Google Scholar] [CrossRef]
- Zhao, L.; Sun, L.; Chu, X. Chemiluminescence Immunoassay. TrAC—Trends Anal. Chem. 2009, 28, 404–415. [Google Scholar] [CrossRef]
- Suleman, S.; Shukla, S.K.; Malhotra, N.; Bukkitgar, S.D.; Shetti, N.P.; Pilloton, R.; Narang, J.; Nee Tan, Y.; Aminabhavi, T.M. Point of Care Detection of COVID-19: Advancement in Biosensing and Diagnostic Methods. Chem. Eng. J. 2021, 414, 128759. [Google Scholar] [CrossRef]
- Falzone, L.; Gattuso, G.; Tsatsakis, A.; Spandidos, D.A.; Libra, M. Current and Innovative Methods for the Diagnosis of COVID-19 Infection (Review). Int. J. Mol. Med. 2021, 47, 1–23. [Google Scholar] [CrossRef]
- Serrano, M.M.; Rodríguez, D.N.; Palop, N.T.; Arenas, R.O.; Córdoba, M.M.; Mochón, M.D.O.; Cardona, C.G. Comparison of Commercial Lateral Flow Immunoassays and ELISA for SARS-CoV-2 Antibody Detection. J. Clin. Virol. 2020, 129, 104529. [Google Scholar] [CrossRef]
- Moeller, M.E.; Engsig, F.N.; Bade, M.; Fock, J.; Pah, P.; Soerensen, A.L.; Bang, D.; Donolato, M.; Benfield, T. Rapid Quantitative Point-Of-Care Diagnostic Test for Post COVID-19 Vaccination Antibody Monitoring. Microbiol. Spectr. 2022, 10, e0039622. [Google Scholar] [CrossRef]
- Mardian, Y.; Kosasih, H.; Karyana, M.; Neal, A.; Lau, C.Y. Review of Current COVID-19 Diagnostics and Opportunities for Further Development. Front. Med. 2021, 8, 615099. [Google Scholar] [CrossRef]
- Feng, W.; Newbigging, A.M.; Le, C.; Pang, B.; Peng, H.; Cao, Y.; Wu, J.; Abbas, G.; Song, J.; Wang, D.B.; et al. Molecular Diagnosis of COVID-19: Challenges and Research Needs. Anal. Chem. 2020, 92, 10196–10209. [Google Scholar] [CrossRef]
- Etienne, E.E.; Nunna, B.B.; Talukder, N.; Wang, Y.; Lee, E.S. COVID-19 Biomarkers and Advanced Sensing Technologies for Point-of-Care (POC) Diagnosis. Bioengineering 2021, 8, 98. [Google Scholar] [CrossRef]
- Cerutti, F.; Burdino, E.; Milia, M.G.; Allice, T.; Gregori, G.; Bruzzone, B.; Ghisetti, V. Urgent Need of Rapid Tests for SARS CoV-2 Antigen Detection: Evaluation of the SD-Biosensor Antigen Test for SARS-CoV-2. J. Clin. Virol. 2020, 132, 104654. [Google Scholar] [CrossRef]
- Dinnes, J.; Deeks, J.J.; Adriano, A.; Berhane, S.; Davenport, C.; Dittrich, S.; Emperador, D.; Takwoingi, Y.; Cunningham, J.; Beese, S.; et al. Rapid, Point-of-Care Antigen and Molecular-Based Tests for Diagnosis of SARS-CoV-2 Infection. Cochrane Database Syst. Rev. 2020, 8, CD013705. [Google Scholar] [CrossRef]
- CDC. Interim Guidance for Antigen Testing for SARS-CoV-2. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines.html (accessed on 13 September 2021).
- Tu, Y.F.; Chien, C.S.; Yarmishyn, A.A.; Lin, Y.Y.; Luo, Y.H.; Lin, Y.T.; Lai, W.Y.; Yang, D.M.; Chou, S.J.; Yang, Y.P.; et al. A Review of Sars-Cov-2 and the Ongoing Clinical Trials. Int. J. Mol. Sci. 2020, 21, 2657. [Google Scholar] [CrossRef]
- Weissleder, R.; Lee, H.; Ko, J.; Pittet, M.J. COVID-19 Diagnostics in Context. Sci. Transl. Med. 2020, 12, 1931. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.; Bleicker, T.; Brünink, S.; Drosten, C.; Landt, O.; Koopmans, M.; Zambon Public Health England, M. Diagnostic Detection of 2019-NCoV by Real-Time RT-RCR. Charité Virol. 2020, 13, 1–13. [Google Scholar]
- Fleige, S.; Pfaffl, M.W. RNA Integrity and the Effect on the Real-Time QRT-PCR Performance. Mol. Aspects Med. 2006, 27, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Afzal, A. Molecular Diagnostic Technologies for COVID-19: Limitations and Challenges. J. Adv. Res. 2020, 26, 149–159. [Google Scholar] [CrossRef]
- Cui, X.; Shi, Y.; Zhao, L.; Gu, S.; Wei, C.; Yang, Y.; Wen, S.; Chen, H.; Ge, J. Application of Real-Time Quantitative PCR to Detect Mink Circovirus in Naturally and Experimentally Infected Minks. Front. Microbiol. 2018, 9, 937. [Google Scholar] [CrossRef]
- Giri, B.; Pandey, S.; Shrestha, R.; Pokharel, K.; Ligler, F.S.; Neupane, B.B. Review of Analytical Performance of COVID-19 Detection Methods. Anal. Bioanal. Chem. 2021, 413, 35–48. [Google Scholar] [CrossRef]
- Kucirka, L.M.; Lauer, S.A.; Laeyendecker, O.; Boon, D.; Lessler, J. Variation in False-Negative Rate of Reverse Transcriptase Polymerase Chain Reaction-Based SARS-CoV-2 Tests by Time Since Exposure. Ann. Intern. Med. 2020, 173, 262–267. [Google Scholar] [CrossRef]
- Binnicker, M.J. Challenges and Controversies to Testing for COVID-19. J. Clin. Microbiol. 2020, 58, e01695-20. [Google Scholar] [CrossRef]
- Safarchi, A.; Fatima, S.; Ayati, Z.; Vafaee, F. An Update on Novel Approaches for Diagnosis and Treatment of SARS-CoV-2 Infection. Cell Biosci. 2021, 11, 164. [Google Scholar] [CrossRef]
- Heijnen, L.; Elsinga, G.; de Graaf, M.; Molenkamp, R.; Koopmans, M.P.G.; Medema, G. Droplet Digital RT-PCR to Detect SARS-CoV-2 Signature Mutations of Variants of Concern in Wastewater. Sci. Total Environ. 2021, 799, 149456. [Google Scholar] [CrossRef]
- Fassy, J.; Lacoux, C.; Leroy, S.; Noussair, L.; Hubac, S.; Degoutte, A.; Vassaux, G.; Leclercq, V.; Rouquié, D.; Marquette, C.H.; et al. Versatile and Flexible Microfluidic QPCR Test for High-Throughput SARS-CoV-2 and Cellular Response Detection in Nasopharyngeal Swab Samples. PLoS ONE 2021, 16, e0243333. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Gjorgjieva, T.; Attieh, Z.; Dieng, M.M.; Arnoux, M.; Khair, M.; Moussa, Y.; Al Jallaf, F.; Rahiman, N.; Jackson, C.A.; et al. Microfluidic Nano-Scale QPCR Enables Ultra-Sensitive and Quantitative Detection of SARS-CoV-2. Processes 2020, 8, 1425. [Google Scholar] [CrossRef]
- Thompson, D.; Lei, Y. Mini Review: Recent Progress in RT-LAMP Enabled COVID-19 Detection. Sens. Actuators Rep. 2020, 2, 100017. [Google Scholar] [CrossRef] [PubMed]
- Amaral, C.; Antunes, W.; Moe, E.; Duarte, A.G.; Lima, L.M.P.; Santos, C.; Gomes, I.L.; Afonso, G.S.; Vieira, R.; Teles, H.S.S.; et al. A Molecular Test Based on RT-LAMP for Rapid, Sensitive and Inexpensive Colorimetric Detection of SARS-CoV-2 in Clinical Samples. Sci. Rep. 2021, 11, 16430. [Google Scholar] [CrossRef]
- Lamb, L.E.; Bartolone, S.N.; Ward, E.; Chancellor, M.B. Rapid Detection of Novel Coronavirus/Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by Reverse Transcription-Loop-Mediated Isothermal Amplification. PLoS ONE 2020, 15, e0234682. [Google Scholar] [CrossRef]
- Kline, E.C.; Panpradist, N.; Hull, I.T.; Wang, Q.; Oreskovic, A.K.; Han, P.D.; Starita, L.M.; Lutz, B.R. Multiplex Target-Redundant RT-LAMP for Robust Detection of SARS-CoV-2 Using Fluorescent Universal Displacement Probes. medRxiv 2021. [Google Scholar] [CrossRef]
- Ali, Z.; Aman, R.; Mahas, A.; Rao, G.S.; Tehseen, M.; Marsic, T.; Salunke, R.; Subudhi, A.K.; Hala, S.M.; Hamdan, S.M.; et al. ISCAN: An RT-LAMP-Coupled CRISPR-Cas12 Module for Rapid, Sensitive Detection of SARS-CoV-2. Virus Res. 2020, 288, 198129. [Google Scholar] [CrossRef]
- Wang, R.; Qian, C.; Pang, Y.; Li, M.; Yang, Y.; Ma, H.; Zhao, M.; Qian, F.; Yu, H.; Liu, Z.; et al. OpvCRISPR: One-Pot Visual RT-LAMP-CRISPR Platform for SARS-Cov-2 Detection. Biosens. Bioelectron. 2021, 172, 112766. [Google Scholar] [CrossRef]
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T.; et al. Clinical Validation of a Cas13-Based Assay for the Detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1140–1149. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR–Cas12-Based Detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef]
- Misra, R.; Acharya, S.; Sushmitha, N. Nanobiosensor-Based Diagnostic Tools in Viral Infections: Special Emphasis on COVID-19. Rev. Med. Virol. 2021, 32, e2267. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, N.; Jamaluddin, N.D.; Tan, L.L.; Mohd Yusof, N.Y. A Review on the Development of Gold and Silver Nanoparticles-Based Biosensor as a Detection Strategy of Emerging and Pathogenic RNA Virus. Sensors 2021, 21, 5114. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Dong, S. Nucleic Acid Biosensors: Recent Advances and Perspectives. Anal. Chem. 2017, 89, 189–215. [Google Scholar] [CrossRef] [PubMed]
- Qiu, G.; Gai, Z.; Tao, Y.; Schmitt, J.; Kullak-Ublick, G.A.; Wang, J. Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano 2020, 14, 5268–5277. [Google Scholar] [CrossRef]
- Li, J.; Wu, D.; Yu, Y.; Li, T.; Li, K.; Xiao, M.M.; Li, Y.; Zhang, Z.Y.; Zhang, G.J. Rapid and Unamplified Identification of COVID-19 with Morpholino-Modified Graphene Field-Effect Transistor Nanosensor. Biosens. Bioelectron. 2021, 183, 113206. [Google Scholar] [CrossRef]
- Farzin, L.; Sadjadi, S.; Sheini, A.; Mohagheghpour, E. A Nanoscale Genosensor for Early Detection of COVID-19 by Voltammetric Determination of RNA-Dependent RNA Polymerase (RdRP) Sequence of SARS-CoV-2 Virus. Microchim. Acta 2021, 188, 121. [Google Scholar] [CrossRef]
- Alafeef, M.; Dighe, K.; Moitra, P.; Pan, D. Rapid, Ultrasensitive, and Quantitative Detection of SARS-CoV-2 Using Antisense Oligonucleotides Directed Electrochemical Biosensor Chip. ACS Nano 2020, 14, 17028–17045. [Google Scholar] [CrossRef]
- Moitra, P.; Alafeef, M.; Alafeef, M.; Alafeef, M.; Dighe, K.; Frieman, M.B.; Pan, D.; Pan, D.; Pan, D. Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano 2020, 14, 7617–7627. [Google Scholar] [CrossRef]
- Gutiérrez-Gálvez, L.; del Caño, R.; Menéndez-Luque, I.; García-Nieto, D.; Rodríguez-Peña, M.; Luna, M.; Pineda, T.; Pariente, F.; García-Mendiola, T.; Lorenzo, E. Electrochemiluminescent Nanostructured DNA Biosensor for SARS-CoV-2 Detection. Talanta 2022, 240, 123203. [Google Scholar] [CrossRef]
- Cajigas, S.; Alzate, D.; Fernández, M.; Muskus, C.; Orozco, J. Electrochemical Genosensor for the Specific Detection of SARS-CoV-2. Talanta 2022, 245, 123482. [Google Scholar] [CrossRef]
- Ali Farzin, M.; Abdoos, H.; Saber, R. Graphite Nanocrystals Coated Paper-Based Electrode for Detection of SARS-Cov-2 Gene Using DNA-Functionalized Au@carbon Dot Core-Shell Nanoparticles. Microchem. J. 2022, 179, 107585. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Fan, Z.; Huang, Y.; Ding, Y.; Xie, M.; Wang, M. Hybridization Chain Reaction Circuit-Based Electrochemiluminescent Biosensor for SARS-Cov-2 RdRp Gene Assay. Talanta 2022, 240, 123207. [Google Scholar] [CrossRef] [PubMed]
- Cieplak, M.; Kutner, W. Artificial Biosensors: How Can Molecular Imprinting Mimic Biorecognition? Trends Biotechnol. 2016, 34, 922–941. [Google Scholar] [CrossRef]
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.H.; Choi, M.; Ku, K.B.; Lee, C.S.; Jun, S.; Park, D.; Kim, H.G.; et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142. [Google Scholar] [CrossRef]
- de Lima, L.F.; Ferreira, A.L.; Torres, M.D.T.; de Araujo, W.R.; de la Fuente-Nunez, C. Minute-Scale Detection of SARS-CoV-2 Using a Low-Cost Biosensor Composed of Pencil Graphite Electrodes. Proc. Natl. Acad. Sci. USA 2021, 118, e2106724118. [Google Scholar] [CrossRef] [PubMed]
- Mavrikou, S.; Moschopoulou, G.; Tsekouras, V.; Kintzios, S. Development of a Portable, Ultra-Rapid and Ultra-Sensitive Cell-Based Biosensor for the Direct Detection of the SARS-COV-2 S1 Spike Protein Antigen. Sensors 2020, 20, 3121. [Google Scholar] [CrossRef]
- McClements, J.; Bar, L.; Singla, P.; Canfarotta, F.; Thomson, A.; Czulak, J.; Johnson, R.E.; Crapnell, R.D.; Banks, C.E.; Payne, B.; et al. Molecularly Imprinted Polymer Nanoparticles Enable Rapid, Reliable, and Robust Point-of-Care Thermal Detection of SARS-CoV-2. ACS Sensors 2022, 7, 1122–1131. [Google Scholar] [CrossRef]
- Perdomo, S.A.; Ortega, V.; Jaramillo-Botero, A.; Mancilla, N.; Mosquera-Delacruz, J.H.; Valencia, D.P.; Quimbaya, M.; Contreras, J.D.; Velez, G.E.; Loaiza, O.A.; et al. SenSARS: A Low-Cost Portable Electrochemical System for Ultra-Sensitive, near Real-Time, Diagnostics of SARS-CoV-2 Infections. IEEE Trans. Instrum. Meas. 2021, 70, 4007710. [Google Scholar] [CrossRef]
- Wu, Q.; Wu, W.; Chen, F.; Ren, P. Highly Sensitive and Selective Surface Plasmon Resonance Biosensor for the Detection of SARS-CoV-2 Spike S1 Protein. Analyst 2022, Advance Article. [Google Scholar] [CrossRef]
- Ahmadivand, A.; Gerislioglu, B.; Ramezani, Z.; Kaushik, A.; Manickam, P.; Ghoreishi, S.A. Functionalized Terahertz Plasmonic Metasensors: Femtomolar-Level Detection of SARS-CoV-2 Spike Proteins. Biosens. Bioelectron. 2021, 177, 112971. [Google Scholar] [CrossRef]
- Novodchuk, I.; Kayaharman, M.; Prassas, I.; Soosaipillai, A.; Karimi, R.; Goldthorpe, I.A.; Abdel-Rahman, E.; Sanderson, J.; Diamandis, E.P.; Bajcsy, M.; et al. Electronic Field Effect Detection of SARS-CoV-2 N-Protein before the Onset of Symptoms. Biosens. Bioelectron. 2022, 210, 114331. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, P.G.; Guy, C.S.; Hasan, M.; Ahmad, A.; Richards, S.J.; Baker, A.N.; Thakkar, N.V.; Walker, M.; Pandey, S.; Anderson, N.R.; et al. Plasmonic Detection of SARS-CoV-2 Spike Protein with Polymer-Stabilized Glycosylated Gold Nanorods. ACS Macro Lett. 2022, 11, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Divagar, M.; Gayathri, R.; Rasool, R.; Shamlee, J.K.; Bhatia, H.; Satija, J.; Sai, V.V.R. Plasmonic Fiberoptic Absorbance Biosensor (P-FAB) for Rapid Detection of SARS-CoV-2 Nucleocapsid Protein. IEEE Sens. J. 2021, 21, 22758–22766. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-C.; Chiang, Y.-H.; Chiang, H.-Y. A Label-Free Electrochemical Impedimetric Immunosensor with Biotinylated-Antibody for SARS-CoV-2 Nucleoprotein Detection in Saliva. Biosensors 2022, 12, 265. [Google Scholar] [CrossRef]
- Kizek, R.; Krejcova, L.; Michalek, P.; Merlos Rodrigo, M.; Heger, Z.; Krizkova, S.; Vaculovicova, M.; Hynek, D.; Adam, V. Nanoscale Virus Biosensors: State of the Art. Nanobiosensors Dis. Diagn. 2015, 4, 47. [Google Scholar] [CrossRef]
- Djaileb, A.; Charron, B.; Jodaylami, M.H.; Thibault, V.; Coutu, J.; Stevenson, K.; Forest, S.; Live, L.S.; Boudreau, D.; Pelletier, J.N.; et al. A Rapid and Quantitative Serum Test for SARS-CoV-2 Antibodies with Portable Surface Plasmon Resonance Sensing. ChemRxiv 2020, preprint. [Google Scholar] [CrossRef]
- Liv, L.; Kayabay, H. An Electrochemical Biosensing Platform for the SARS-CoV-2 Spike Antibody Detection Based on the Functionalised SARS-CoV-2 Spike Antigen Modified Electrode. ChemistrySelect 2022, 7, e202200256. [Google Scholar] [CrossRef]
- Sadique, M.A.; Yadav, S.; Ranjan, P.; Khan, R.; Khan, F.; Kumar, A.; Biswas, D. Highly Sensitive Electrochemical Immunosensor Platforms for Dual Detection of SARS-CoV-2 Antigen and Antibody Based on Gold Nanoparticle Functionalized Graphene Oxide Nanocomposites. ACS Appl. Bio Mater. 2022, 5, 2421–2430. [Google Scholar] [CrossRef]
- Liv, L. Electrochemical Immunosensor Platform Based on Gold-Clusters, Cysteamine and Glutaraldehyde Modified Electrode for Diagnosing COVID-19. Microchem. J. 2021, 168, 106445. [Google Scholar] [CrossRef]
- Liustrovaite, V.; Drobysh, M.; Rucinskiene, A.; Baradoke, A.; Ramanaviciene, A.; Plikusiene, I.; Samukaite-Bubniene, U.; Viter, R.; Chen, C.-F.; Ramanavicius, A. Towards an Electrochemical Immunosensor for the Detection of Antibodies against SARS-CoV-2 Spike Protein. J. Electrochem. Soc. 2022, 169, 37523. [Google Scholar] [CrossRef]
- Zheng, Y.; Song, K.; Cai, K.; Liu, L.; Tang, D.; Long, W.; Zhai, B.; Chen, J.; Tao, Y.; Zhao, Y.; et al. B-Cell-Epitope-Based Fluorescent Quantum Dot Biosensors for SARS-CoV-2 Enable Highly Sensitive COVID-19 Antibody Detection. Viruses 2022, 14, 1031. [Google Scholar] [CrossRef] [PubMed]
- Ameku, W.A.; Provance, D.W.; Morel, C.M.; De-Simone, S.G. Rapid Detection of Anti-SARS-CoV-2 Antibodies with a Screen-Printed Electrode Modified with a Spike Glycoprotein Epitope. Biosensors 2022, 12, 272. [Google Scholar] [CrossRef]
- Jiang, M.; Dong, T.; Han, C.; Liu, L.; Zhang, T.; Kang, Q.; Wang, P.; Zhou, F. Regenerable and High-Throughput Surface Plasmon Resonance Assay for Rapid Screening of Anti-SARS-CoV-2 Antibody in Serum Samples. Anal. Chim. Acta 2022, 1208, 339830. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.H.; Leirs, K.; Maes, W.; Imbrechts, M.; Callewaert, N.; Lagrou, K.; Geukens, N.; Lammertyn, J.; Spasic, D. Innovative FO-SPR Label-Free Strategy for Detecting Anti-RBD Antibodies in COVID-19 Patient Serum and Whole Blood. ACS Sensors 2022, 7, 477–487. [Google Scholar] [CrossRef]
- Ali, M.A.; Hu, C.; Zhang, F.; Jahan, S.; Yuan, B.; Saleh, M.S.; Gao, S.J.; Panat, R. N Protein-Based Ultrasensitive SARS-CoV-2 Antibody Detection in Seconds via 3D Nanoprinted, Microarchitected Array Electrodes. J. Med. Virol. 2022, 94, 2067–2078. [Google Scholar] [CrossRef]
- GISAID. GISAID Submission Tracker Global. 2021. Available online: https://www.gisaid.org (accessed on 1 June 2022).
- Maxmen, A. One Million Coronavirus Sequences: Popular Genome Site Hits Mega Milestone. Nature 2021, 593, 21. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Zhang, D.; Yang, P.; Poon, L.L.M.; Wang, Q. Viral Load of SARS-CoV-2 in Clinical Samples. Lancet Infect. Dis. 2020, 20, 411–412. [Google Scholar] [CrossRef]
- Mesoraca, A.; Margiotti, K.; Viola, A.; Cima, A.; Sparacino, D.; Giorlandino, C. Evaluation of SARS-CoV-2 Viral RNA in Fecal Samples. Virol. J. 2020, 17, 86. [Google Scholar] [CrossRef]
- Bisoffi, Z.; Pomari, E.; Deiana, M.; Piubelli, C.; Ronzoni, N.; Beltrame, A.; Bertoli, G.; Riccardi, N.; Perandin, F.; Formenti, F.; et al. Sensitivity, Specificity and Predictive Values of Molecular and Serological Tests for COVID-19. A Longitudinal Study in Emergency Room. Diagnostics 2020, 10, 669. [Google Scholar] [CrossRef]
- CDC. Interim Guidelines for Clinical Specimens for COVID-19; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2020; pp. 1–5.
- FDA. Policy for Evaluating Impact of Viral Mutations on COVID-19 Tests; FDA: Silver Spring, MD, USA, 2021; pp. 1–13.
- Brown, K.A.; Gubbay, J.; Hopkins, J.; Patel, S.; Buchan, S.A.; Daneman, N.; Goneau, L.W. S-Gene Target Failure as a Marker of Variant B.1.1.7 among SARS-CoV-2 Isolates in the Greater Toronto Area, December 2020 to March 2021. J. Am. Med. Assoc. 2021, 325, 2115–2116. [Google Scholar] [CrossRef]
- Metzger, C.M.J.A.; Lienhard, R.; Seth-Smith, H.M.B.; Roloff, T.; Wegner, F.; Sieber, J.; Bel, M.; Greub, G.; Egli, A. PCR Performance in the SARS-CoV-2 Omicron Variant of Concern? Swiss Med. Wkly. 2021, 151, w30120. [Google Scholar] [CrossRef]
- Gand, M.; Vanneste, K.; Thomas, I.; Van Gucht, S.; Capron, A.; Herman, P.; Roosens, N.H.C.; De Keersmaecker, S.C.J. Deepening of in Silico Evaluation of Sars-Cov-2 Detection Rt-Qpcr Assays in the Context of New Variants. Genes 2021, 12, 565. [Google Scholar] [CrossRef]
- Artesi, M.; Bontems, S.; Göbbels, P.; Franckh, M.; Maes, P.; Boreux, R.; Meex, C.; Melin, P.; Hayette, M.P.; Bours, V.; et al. A Recurrent Mutation at Position 26340 of SARS-CoV-2 Is Associated with Failure of the E Gene Quantitative Reverse Transcription-PCR Utilized in a Commercial Dual-Target Diagnostic Assay. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef]
- Vogels, C.B.F.; Brito, A.F.; Wyllie, A.L.; Fauver, J.R.; Ott, I.M.; Kalinich, C.C.; Petrone, M.E.; Casanovas-Massana, A.; Muenker, M.C.; Moore, A.J.; et al. Analytical Sensitivity and Efficiency Comparisons of SARS-CoV-2 RT–QPCR Primer–Probe Sets. Nat. Microbiol. 2020, 5, 1299–1305. [Google Scholar] [CrossRef]
- Khan, K.A.; Cheung, P. Presence of Mismatches between Diagnostic PCR Assays and Coronavirus SARS-CoV-2 Genome. R. Soc. Open Sci. 2020, 7, 200636. [Google Scholar] [CrossRef]
- Ayyadevara, S.; Thaden, J.; Reis, R. Discrimination of Primer 3’-Nucleotide Mismatch by Taq DNA Polymerase during Polymerase Chain Reaction. Anal. Biochem. 2000, 284, 11–18. [Google Scholar] [CrossRef]
- Zimmerman, P.A.; King, C.L.; Ghannoum, M.; Bonomo, R.A.; Procop, G.W. Molecular Diagnosis of SARS-CoV-2: Assessing and Interpreting Nucleic Acid and Antigen Tests. Pathog. Immun. 2021, 6, 135–156. [Google Scholar] [CrossRef]
- Chan, J.F.W.; Yip, C.C.Y.; To, K.K.W.; Tang, T.H.C.; Wong, S.C.Y.; Leung, K.H.; Fung, A.Y.F.; Ng, A.C.K.; Zou, Z.; Tsoi, H.W.; et al. Improved Molecular Diagnosis of COVID-19 by the Novel, Highly Sensitive and Specific COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR Assay Validated in Vitro and with Clinical Specimens. J. Clin. Microbiol. 2020, 58, e00310-20. [Google Scholar] [CrossRef]
- CDC. Real-Time RT-PCR Diagnostic Panel for Emergency Use Only; CDC-006-00019, Revision: 03; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2020.
- Hasan, M.R.; Sundararaju, S.; Manickam, C.; Mirza, F.; Al-Hail, H.; Lorenz, S.; Tang, P. A Novel Point Mutation in the N Gene of SARS-CoV-2 May Affect the Detection of the Virus by Reverse Transcription-Quantitative PCR. J. Clin. Microbiol. 2021, 59, e03278-20. [Google Scholar] [CrossRef]
- Bourassa, L.; Perchetti, G.A.; Phung, Q.; Lin, M.J.; Mills, M.G.; Roychoudhury, P.; Harmon, K.G.; Reed, J.C.; Greninger, A.L. A SARS-CoV-2 Nucleocapsid Variant That Affects Antigen Test Performance. J. Clin. Virol. 2021, 141, 104900. [Google Scholar] [CrossRef]
| Type | Bioreceptor | Target | Sample | Sensibility | Time | Ref. |
|---|---|---|---|---|---|---|
| Field-Transistor Effect (FET) | Antibodies | S protein | Nasopharyngeal swab | 242 copies/mL | - | [57] |
| Electrochemical | ACE2 | S protein | Nasopharyngeal swab/Saliva | 229 fg/mL | 6 min | [58] |
| Electrochemical | Antibodies | S protein | Engineered Vero cells | 1 fg/mL | 3 min | [59] |
| Thermal | MIP | S protein | Nasal/Throat Swab | <10 fg/mL | 15 min | [60] |
| Electrochemical impedance | Antibodies | S protein | Nasopharyngeal swab/Saliva | 1fg/mL | 10 min | [61] |
| Surface Plasmon Resonance (SPR) | AgNP/Antibody nanoconjugate | S protein | Serum | 12 fg/mL | - | [62] |
| Plasmonic biosensor | AuNP/Antibody nanoconjugate | S protein | Solution containing S proteins | 4.2 fmol | 80 min | [63] |
| FET | Antibodies | N protein | Solution containing N proteins | 10 ag/mL | 4 min | [64] |
| Plasmonic | NeuNAc | S protein | Nasal swab | - | - | [65] |
| Plasmonic Fiberoptic Absorbance | Anti-SARS-CoV-2 N-protein IgG1 | N protein | Solution containing N proteins | 2.5 ng/mL | 10 min | [66] |
| Electrochemical impedance | Biotinylated antibody | N protein | PBS-diluted saliva | 6 pg/mL | - | [67] |
| Type | Bioreceptor | Target | Sample | Sensibility | Time | Ref. |
|---|---|---|---|---|---|---|
| SPR | N protein | Antibodies | Serum | 30 nM | 15 min | [69] |
| Electrochemical | S protein | Antibodies | Antibodies in synthetic media | 9.3 ag/mL | 20 min | [70] |
| Electrochemical | GO-Au-antigen nanocomposite | Antibodies | Nasopharyngeal swab/Serum | 1 fg/mL | - | [71] |
| Square Wave Voltammetry (SWV) | S protein + gold clusters | S protein antibody | Oropharyngeal swab/saliva | 0.03 fg/mL | 30 min | [72] |
| Electrochemical Impedance | Recombinant S protein | Antibodies | Serum | 1.99 nM | - | [73] |
| Cyclic Voltammetry | Recombinant S protein | Antibodies | Serum | 2.53 nM | - | [73] |
| Fluorescent | B-cell epitopes | Antibodies | Serum | 100 pM | 5 min | [74] |
| SWV | S protein epitope | IgG | Serum | - | 22 min | [75] |
| SPR | Histidine-tagged S protein | Antibodies | Serum | 0.057 μg/mL | - | [76] |
| SPR | RBD | Antibodies | Serum/Whole Blood | - | 30 min | [77] |
| Electrochemical | N protein | Antibody | Solution with antibodies | 13 fM | <1 min | [78] |
| Test | Advantages | Disadvantages |
|---|---|---|
| RT-PCR | High specificity and sensitivity, detects multiple targets | Time consuming, high cost, needs a laboratory setting and technicians |
| ELISA | High sensitivity, faster and cheaper than RT-PCR | Only detects 1 target, risk of cross-reactivity, needs a laboratory setting and technicians |
| LFIA | Very fast, portable, and cheap | Low specificity and sensitivity |
| CRISPR-Based | Fast, high sensitivity and specificity | Only detects 1 target, needs a laboratory setting and technicians |
| RT-LAMP | Fast, high specificity and sensitivity, no bulky equipment | Difficulty in primer design |
| Biosensors | Fast, portable, cheap, high specificity and sensitivity | Needs optimization |
| Sequencing | Most complete, detects all mutations | Time consuming and very high cost, needs a laboratory setting and technicians |
| Mutation | Gene | Assay | VOC/VOI | Ref. |
|---|---|---|---|---|
| C16289T | ORF1ab | Chan-ORF1ab | - | [94] |
| A22812C | S | COVID-19-RdRp/Hel | Theta | [88] |
| G22813T | S | COVID-19-RdRp/Hel | Iota | [88] |
| ΔH69/V70 | S | ThermoFisher TaqPath COVID-19 | Alpha, Omicron | [86,87] |
| C26340T | E | Cobas SARS-CoV-2 | - | [89] |
| T28688C | N | US-CDC-N-3 | - | [95] |
| C28887T | N | China CDC | Beta, Eta, Mu | [88] |
| C28977T | N | China CDC | Alpha | [88] |
| C29200A | N | Cepheid Xpert Xpress SARS-CoV-2 | - | [96] |
| C29311T | N | US-CDC-N-1 | Lambda | [95] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lino, A.; Cardoso, M.A.; Gonçalves, H.M.R.; Martins-Lopes, P. SARS-CoV-2 Detection Methods. Chemosensors 2022, 10, 221. https://doi.org/10.3390/chemosensors10060221
Lino A, Cardoso MA, Gonçalves HMR, Martins-Lopes P. SARS-CoV-2 Detection Methods. Chemosensors. 2022; 10(6):221. https://doi.org/10.3390/chemosensors10060221
Chicago/Turabian StyleLino, Alexandra, Marita A. Cardoso, Helena M. R. Gonçalves, and Paula Martins-Lopes. 2022. "SARS-CoV-2 Detection Methods" Chemosensors 10, no. 6: 221. https://doi.org/10.3390/chemosensors10060221
APA StyleLino, A., Cardoso, M. A., Gonçalves, H. M. R., & Martins-Lopes, P. (2022). SARS-CoV-2 Detection Methods. Chemosensors, 10(6), 221. https://doi.org/10.3390/chemosensors10060221






