ECG Enhancement and R-Peak Detection Based on Window Variability
Abstract
1. Introduction
2. Materials and Methods
2.1. Data
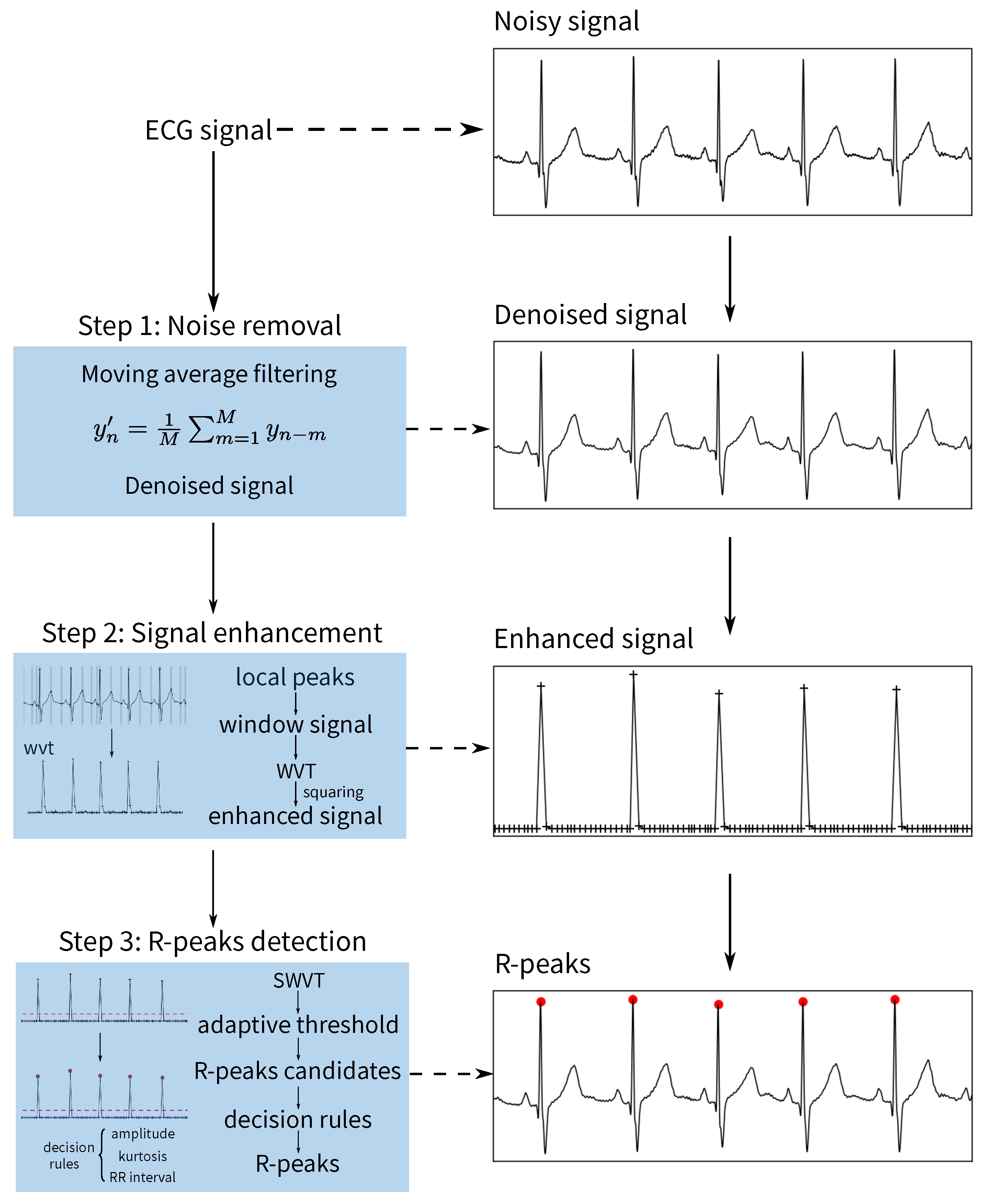
2.2. Overview of the Algorithms
2.2.1. Noise Removal
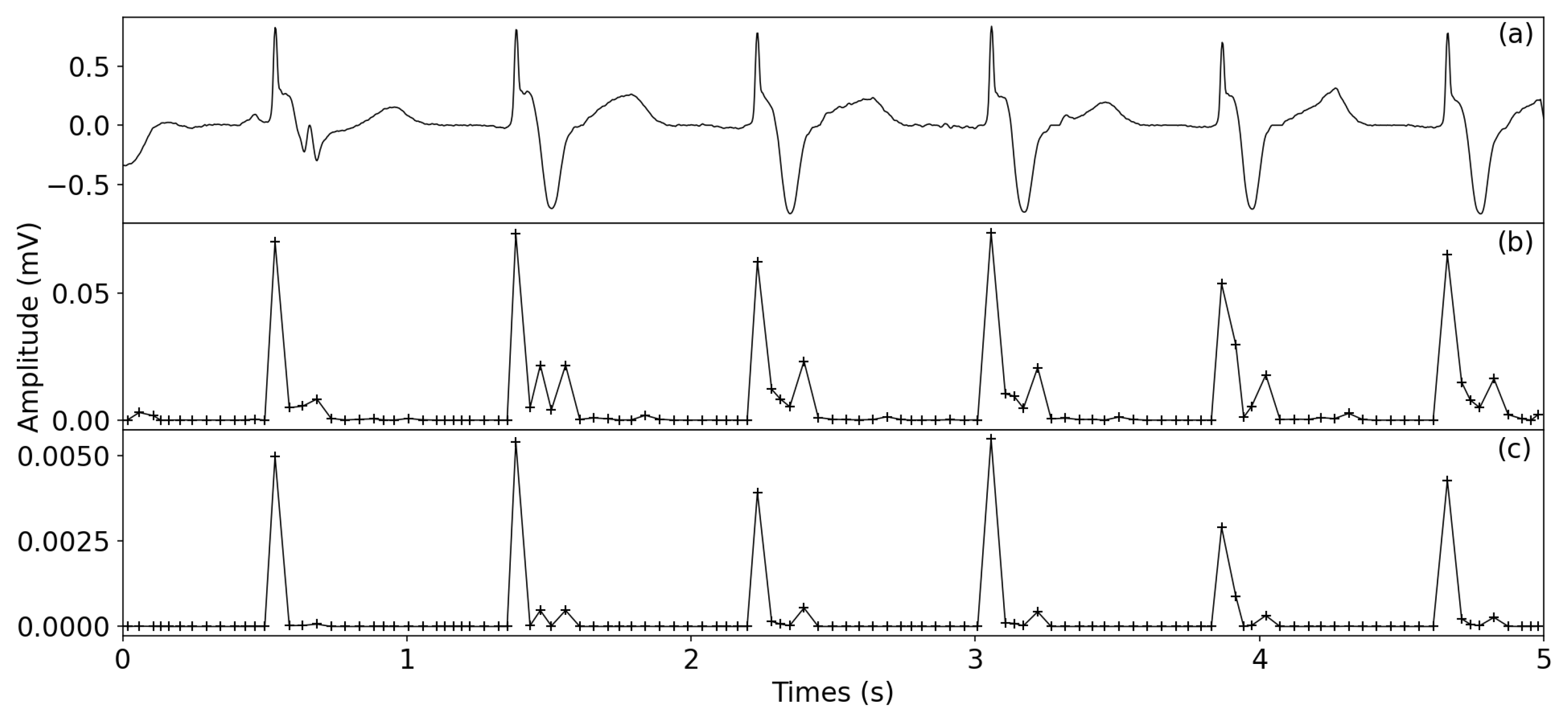
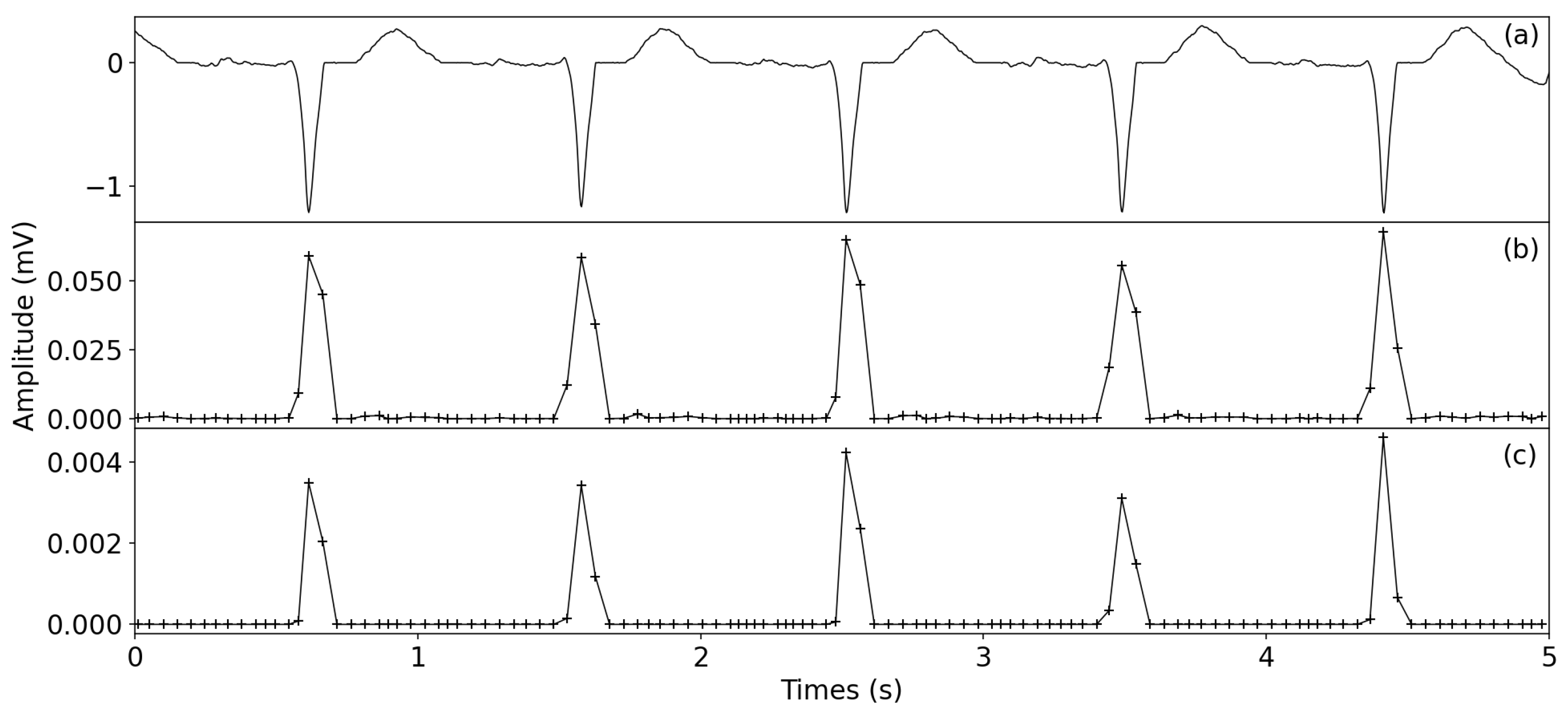
2.2.2. ECG Enhancement
- 1.
- Non-overlapping windowLocal peaks are first detected using the denoised signal. Then, P is applied to generate an non-overlapping window N, , and the ith window is determined by using , where w is the window size. To avoid including multiple waveforms in one window, the minimum width of wave T is used as a reference to set the range of w from 40 to 60 ms. Next, for the non-peak samples between two peaks, they are divided into M non-overlapping windows: ; the window size is . Finally, the and are merged to generate the non-overlapping windows .
- 2.
- Window variance transformTo detect the R-peaks, the window variance transform is used to enhance the QRS complexes; that is, the denoised signal is transformed to the , in whichwhere are the indexes of windows; is the number of samples in jth window; is the amplitude mean of the jth window samples.
- 3.
- SquaringTo further enhance the QRS complexes, the squaring operator is applied in the , that is, the squared window variance transform (SWVT) is described as follows:
2.2.3. R-Peak Detection
- 1.
- Generation of the R-peaks candidate set
- 2.
- Generation of the R-peaks with decision rules
3. Results
3.1. Metrics for Performance Evaluation
3.2. Experimental Data and Environment
3.3. Detailed Accuracy of the MIT-BIH Database
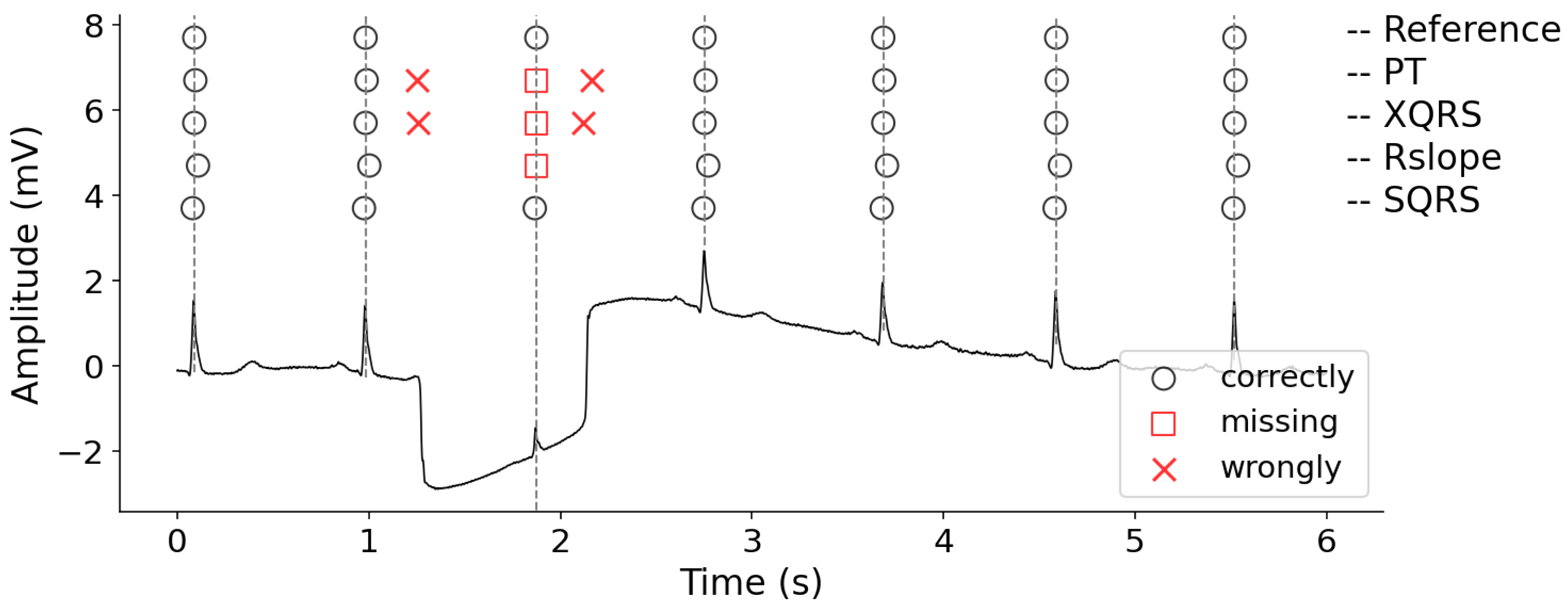
3.4. Accuracy Evaluation and Comparison
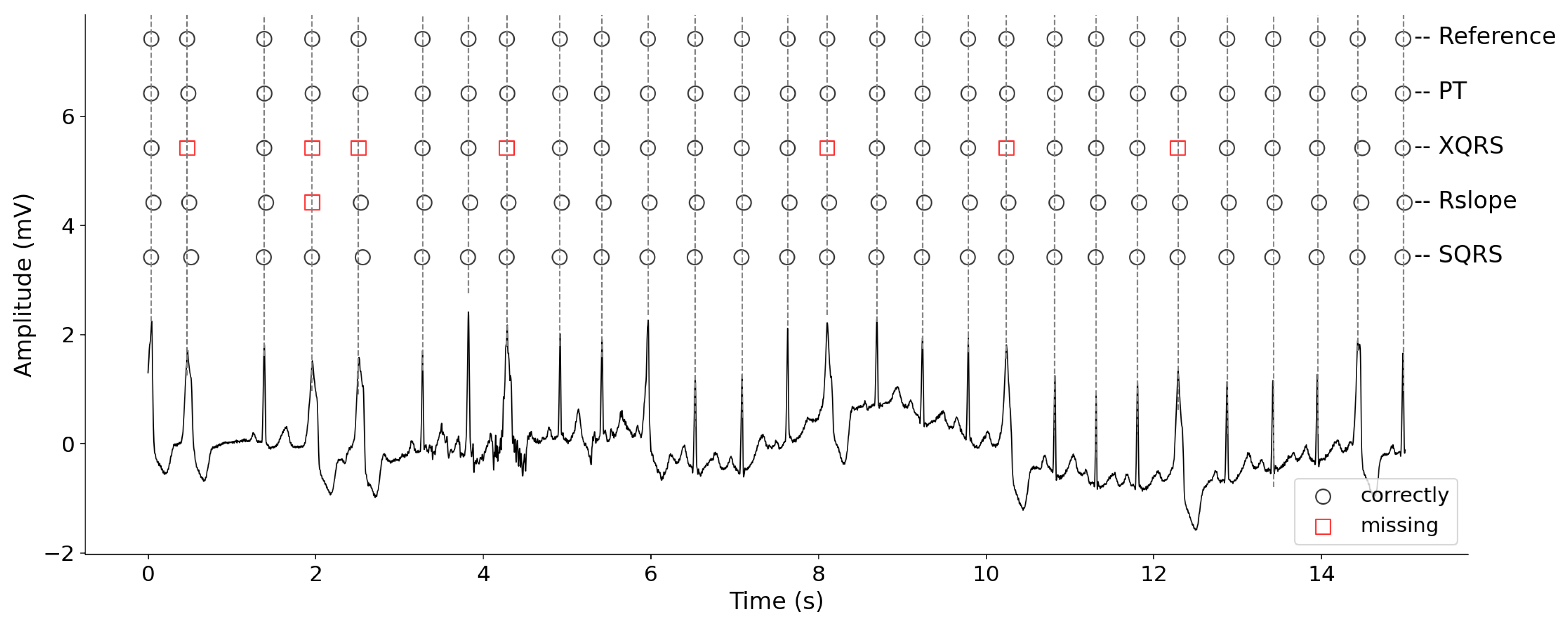
3.5. Visual Display of R-Peak Detection Results
4. Discussion
4.1. ECG Enhancement
4.2. The Effect of Using a Kurtosis Threshold
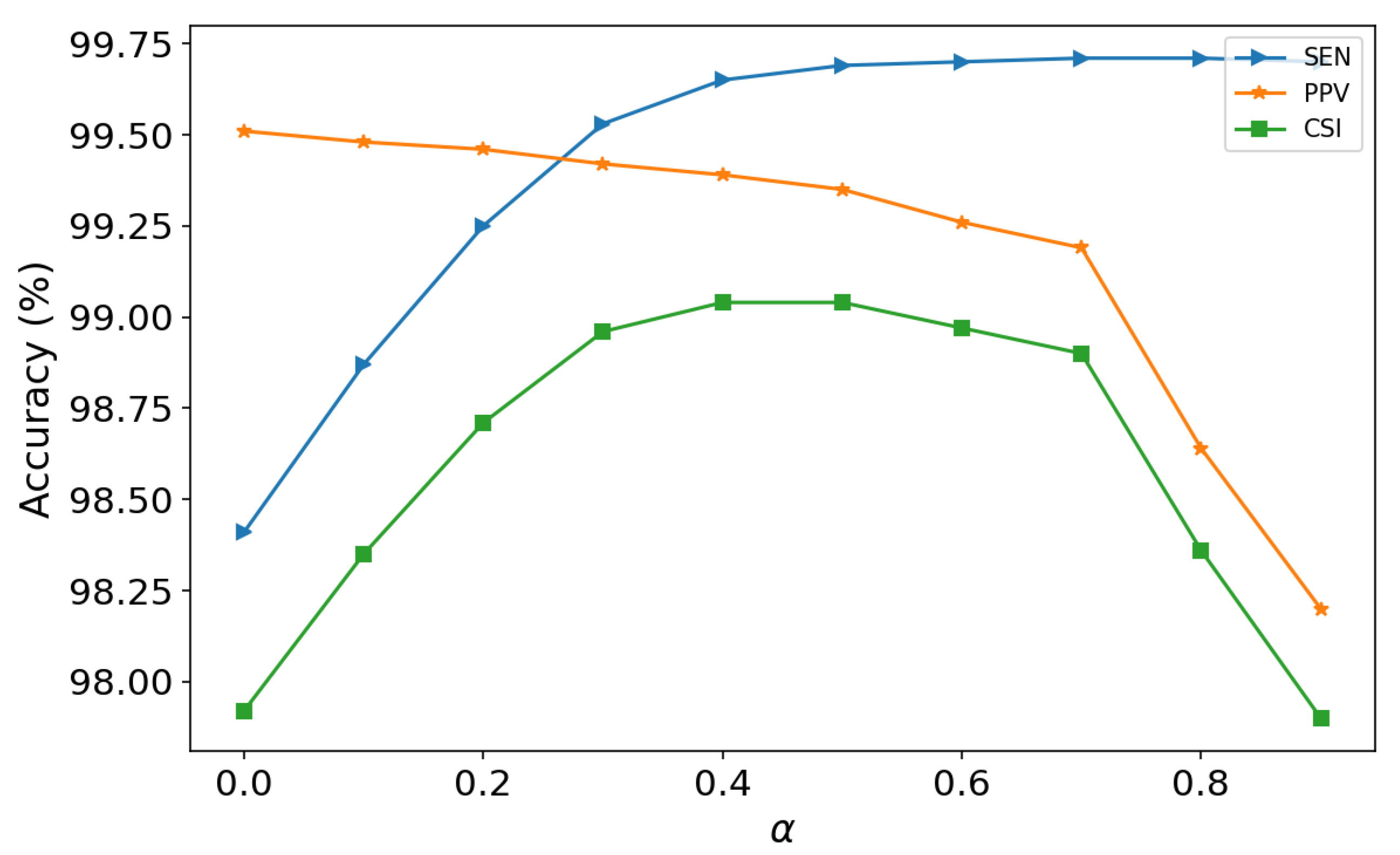
4.3. Selection of the Weight
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Prasad, A.K.; Mishra, V.; Garg, R.K. Electrocardiogram-QRS Point Detection Using Discrete Wavelet Transform. J. Med. Imaging Health Inform. 2012, 2, 206–210. [Google Scholar] [CrossRef]
- Xiang, Y.; Lin, Z.; Meng, J. Automatic QRS Complex Detection Using Two-Level Convolutional Neural Network. Biomed. Eng. 2018, 17, 13. [Google Scholar] [CrossRef]
- Bacharova, L.; Szathmary, V.; Svehlikova, J.; Mateasik, A.; Tysler, M. QRS Complex Waveform Indicators of Ventricular Activation Slowing: Simulation Studies. J. Electrocardiol. 2016, 49, 790–793. [Google Scholar] [CrossRef]
- Wieslander, B.; Nijveldt, R.; Klem, I.; Lokhnygina, Y.; Pura, J.; Wagner, G.S.; Ugander, M.; Atwater, B.D. Evaluation of Selvester QRS Score for Use in Presence of Conduction Abnormalities in a Broad Population. Am. Heart J. 2015, 170, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Zidelmal, Z.; Amirou, A.; Adnane, M.; Belouchrani, A. QRS Detection Based on Wavelet Coefficients. Comput. Methods Programs Biomed. 2012, 107, 490–496. [Google Scholar] [CrossRef]
- Hong, Y.; Lian, Y. A Memristor-Based Continuous-Time Digital FIR Filter for Biomedical Signal Processing. IEEE Trans. Circuits Syst. I Regul. Pap. 2015, 62, 1392–1401. [Google Scholar] [CrossRef]
- Ott, G.; Costa, E.A.C.; Almeida, S.J.M.; Fonseca, M.B. IIR Filter Architectures with Truncation Error Feedback for ECG Signal Processing. Circuits Syst. Signal Process. 2019, 38, 329–355. [Google Scholar] [CrossRef]
- Blanco-Velasco, M.; Weng, B.; Barner, K.E. ECG Signal Denoising and Baseline Wander Correction Based on the Empirical Mode Decomposition. Comput. Biol. Med. 2008, 38, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Xiao, Z.; Zhang, N. Removal of Baseline Wander from ECG Signal Based on a Statistical Weighted Moving Average Filter. J. Zhejiang Univ. Sci. C 2011, 12, 397–403. [Google Scholar] [CrossRef]
- Salih, S.K.; Aljunid, S.A.; Aljunid, S.M.; Maskon, O. Adaptive Filtering Approach for Denoising Electrocardiogram Signal Using Moving Average Filter. J. Med. Imaging Health Inform. 2015, 5, 1065–1069. [Google Scholar] [CrossRef]
- Lynn, P.A. Online Digital Filters for Biological Signals: Some Fast Designs for a Small Computer. Med. Biol. Eng. Comput. 1977, 15, 534–540. [Google Scholar] [CrossRef]
- Xin, Y.; Chen, Y.; Hao, W.T. Ecg Baseline Wander Correction Based on Mean-Median Filter and Empirical Mode Decomposition. Biomed. Mater. Eng. 2014, 24, 365–371. [Google Scholar] [CrossRef]
- Awal, M.A.; Mostafa, S.S.; Ahmad, M.; Rashid, M.A. An Adaptive Level Dependent Wavelet Thresholding for ECG Denoising. Biocybern. Biomed. Eng. 2014, 34, 238–249. [Google Scholar] [CrossRef]
- Lu, G.; Brittain, J.S.; Holland, P.; Yianni, J.; Green, A.L.; Stein, J.F.; Aziz, T.Z.; Wang, S. Removing ECG Noise from Surface EMG Signals Using Adaptive Filtering. Neurosci. Lett. 2009, 462, 14–19. [Google Scholar] [CrossRef] [PubMed]
- An-dong, W.; Lan, L.; Qin, W. An Adaptive Morphologic Filter Applied to ECG De-Noising and Extraction of R Peak at Real-Time. AASRI Procedia 2012, 1, 474–479. [Google Scholar] [CrossRef]
- Wan, X.; Wu, H.; Qiao, F.; Li, F.; Li, Y.; Yan, Y.; Wei, J. Electrocardiogram Baseline Wander Suppression Based on the Combination of Morphological and Wavelet Transformation Based Filtering. Comput. Math. Methods Med. 2019, 2019, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Boudraa, A.O.; Cexus, J.C.; Saidi, Z. EMD-Based Signal Noise Reduction. Signal Process. 2005, 1. Available online: https://www.researchgate.net/publication/228999069_EMD-Based_Signal_Noise_Reduction (accessed on 17 February 2021).
- Kabir, M.A.; Shahnaz, C. Denoising of ECG Signals Based on Noise Reduction Algorithms in EMD and Wavelet Domains. Biomed. Signal Process. Control 2012, 7, 481–489. [Google Scholar] [CrossRef]
- Hashim, F.; Adnan, J.; Daud, N.G.N.; Mokhtar, A.; Rashidi, A.; Rizman, Z. Electrocardiogram Noise Cancellation Using Wavelet Transform. J. Fundam. Appl. Sci. 2017, 9, 131. [Google Scholar] [CrossRef][Green Version]
- Arrais Junior, E.; de Medeiros Valentim, R.A.; Bezerra Brandao, G. Real Time QRS Detection Based on Redundant Discrete Wavelet Transform. IEEE Lat. Am. Trans. 2016, 14, 1662–1668. [Google Scholar] [CrossRef]
- Chen, H.; Chen, S. A Moving Average Based Filtering System with Its Application to Real-Time QRS Detection. In Computers in Cardiology; IEEE: Piscataway, NJ, USA, 2003; pp. 585–588. [Google Scholar]
- Burguera, A. Fast QRS Detection and ECG Compression Based on Signal Structural Analysis. IEEE J. Biomed. Health Inform. 2018, 23, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.S.; Lingyat, N.S. SVM-Based Algorithm for Recognition of QRS Complexes in Electrocardiogram. Irbm 2008, 29, 310–317. [Google Scholar] [CrossRef]
- Pan, J.; Tompkins, W.J. A Real-Time QRS Detection Algorithm. IEEE Trans. Biomed. Eng. 1985, BME-32, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zheng, C.; Tai, C. Detection of ECG Characteristic Points Using Wavelet Transforms. IEEE Trans. Biomed. Eng. 1995, 42, 21–28. [Google Scholar] [PubMed]
- Pal, S.; Mitra, M. Empirical Mode Decomposition Based ECG Enhancement and QRS Detection. Comput. Biol. Med. 2012, 42, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Moody, G.; Mark, R. The Impact of the MIT-BIH Arrhythmia Database. IEEE Eng. Med. Biol. Mag. 2001, 20, 45–50. [Google Scholar] [CrossRef]
- Romagnoli, S.; Sbrollini, A.; Burattini, L.; Marcantoni, I.; Morettini, M.; Burattini, L. Digital Cardiotocography: What Is the Optimal Sampling Frequency? Biomed. Signal Process. Control 2019, 51, 210–215. [Google Scholar] [CrossRef]
- Moody, G.B. WFDB Applications Guide. Available online: https://www.physionet.org/physiotools/wag/ (accessed on 8 March 2019).
- Gieraltowski, J.; Ciuchcinski, K.; Grzegorczyk, I.; Kosna, K.; Solinski, M.; Podziemski, P. RS Slope Detection Algorithm for Extraction of Heart Rate from Noisy, Multimodal Recordings. Physiol. Meas. 2015, 36, 1743–1761. [Google Scholar] [CrossRef] [PubMed]
- Yeh, Y.C.; Wang, W.J. QRS Complexes Detection for ECG Signal: The Difference Operation Method. Comput. Methods Programs Biomed. 2008, 9, 245–254. [Google Scholar] [CrossRef]
- Laguna, P.; Mark, R.; Goldberg, A.; Moody, G. A Database for Evaluation of Algorithms for Measurement of QT and Other Waveform Intervals in the ECG. In Computers in Cardiology; IEEE: Piscataway, NJ, USA, 1997; pp. 673–676. [Google Scholar]


| Record | TP | FP | FN | SEN (%) | PPV (%) | CSI (%) |
|---|---|---|---|---|---|---|
| 100 | 2273 | 1 | 0 | 100 | 99.96 | 99.96 |
| 101 | 1863 | 3 | 2 | 99.89 | 99.84 | 99.73 |
| 102 | 2174 | 13 | 13 | 99.41 | 99.41 | 98.81 |
| 103 | 2084 | 0 | 0 | 100 | 100 | 100 |
| 107 | 2128 | 5 | 9 | 99.58 | 99.77 | 99.34 |
| 112 | 2539 | 0 | 0 | 100 | 100 | 100 |
| 113 | 1795 | 0 | 0 | 100 | 100 | 100 |
| 114 | 1875 | 118 | 4 | 99.79 | 94.08 | 93.87 |
| 115 | 1953 | 0 | 0 | 100 | 100 | 100 |
| 116 | 2385 | 1 | 27 | 98.88 | 99.96 | 98.84 |
| 117 | 1535 | 0 | 0 | 100 | 100 | 100 |
| 118 | 2278 | 0 | 0 | 100 | 100 | 100 |
| 119 | 1987 | 1 | 0 | 100 | 99.95 | 99.95 |
| 121 | 1859 | 4 | 4 | 99.79 | 99.79 | 99.57 |
| 122 | 2475 | 1 | 1 | 99.96 | 99.96 | 99.92 |
| 123 | 1518 | 0 | 0 | 100 | 100 | 100 |
| 124 | 1617 | 2 | 2 | 99.88 | 99.88 | 99.75 |
| 200 | 2598 | 1 | 3 | 99.88 | 99.96 | 99.85 |
| 202 | 2114 | 18 | 22 | 98.97 | 99.16 | 98.13 |
| 205 | 2639 | 0 | 17 | 99.36 | 100 | 99.36 |
| 207 | 1842 | 133 | 18 | 99.03 | 93.27 | 92.30 |
| 208 | 2893 | 5 | 62 | 97.90 | 99.83 | 97.73 |
| 209 | 3005 | 0 | 0 | 100 | 100 | 100 |
| 210 | 2607 | 2 | 43 | 98.38 | 99.92 | 98.30 |
| 212 | 2748 | 0 | 0 | 100 | 100 | 100 |
| 213 | 3243 | 5 | 8 | 99.75 | 99.85 | 99.60 |
| 214 | 2247 | 37 | 15 | 99.34 | 98.38 | 97.72 |
| 215 | 3357 | 2 | 6 | 99.82 | 99.94 | 99.76 |
| 217 | 2190 | 22 | 18 | 99.18 | 99.01 | 98.19 |
| 219 | 2148 | 1 | 6 | 99.72 | 99.95 | 99.67 |
| 220 | 2048 | 0 | 0 | 100 | 100 | 100 |
| 221 | 2419 | 4 | 8 | 99.67 | 99.83 | 99.51 |
| 222 | 2463 | 0 | 20 | 99.19 | 100.00 | 99.19 |
| 228 | 2034 | 29 | 19 | 99.07 | 98.59 | 97.67 |
| 230 | 2256 | 0 | 0 | 100 | 100 | 100 |
| 231 | 1571 | 0 | 0 | 100 | 100 | 100 |
| 232 | 1779 | 69 | 1 | 99.94 | 96.27 | 96.21 |
| 233 | 3072 | 5 | 7 | 99.77 | 99.84 | 99.61 |
| 234 | 2753 | 0 | 0 | 100 | 100 | 100 |
| Total | 88,364 | 482 | 335 | 99.65 | 99.39 | 99.03 |
| Method | SEN | PPV | CSI |
|---|---|---|---|
| PT | 99.04 | 99.38 | 98.49 |
| RSlope | 99.23 | 98.58 | 97.83 |
| XQRS | 99.19 | 99.21 | 98.41 |
| SQRS | 99.65 | 99.39 | 99.04 |
| Method | Total | FP | FN | |
|---|---|---|---|---|
| PT | 88,699 | 512 (0.58%) | 754 (0.85%) | 1.43% |
| RSlope | 88,699 | 1198 (1.35%) | 721 (0.81%) | 2.16% |
| XQRS | 88,699 | 715 (0.81%) | 807 (0.91%) | 1.72% |
| SQRS | 88,699 | 482 (0.54%) | 335 (0.38%) | 0.92% |
| Method | Preprocessing | Enhancement |
|---|---|---|
| PT | bandpass | Derivative Squaring Moving-window integration |
| XQRS | bandpass | Moving-window integration |
| RSlope | detrending lowpass | Separate heart beat detection Quality assessment |
| SQRS | moving-average | window variance transform |
| Using Kurtosis | TP | FP | FN | SEN | PPV | CSI |
|---|---|---|---|---|---|---|
| No | 87,190 | 374 | 1509 | 98.44 | 99.52 | 97.96 |
| Yes | 88,364 | 482 | 335 | 99.65 | 99.39 | 99.04 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, L.; Xie, X.; Wang, Y. ECG Enhancement and R-Peak Detection Based on Window Variability. Healthcare 2021, 9, 227. https://doi.org/10.3390/healthcare9020227
Wu L, Xie X, Wang Y. ECG Enhancement and R-Peak Detection Based on Window Variability. Healthcare. 2021; 9(2):227. https://doi.org/10.3390/healthcare9020227
Chicago/Turabian StyleWu, Lu, Xiaoyun Xie, and Yinglong Wang. 2021. "ECG Enhancement and R-Peak Detection Based on Window Variability" Healthcare 9, no. 2: 227. https://doi.org/10.3390/healthcare9020227
APA StyleWu, L., Xie, X., & Wang, Y. (2021). ECG Enhancement and R-Peak Detection Based on Window Variability. Healthcare, 9(2), 227. https://doi.org/10.3390/healthcare9020227






