Relationship between Clinical Parameters and Chromosomal Microarray Data in Infants with Developmental Delay
Abstract
1. Introduction
2. Methods
2.1. Infants with DD
2.2. CMA Protocol
2.3. Statistical Analysis
3. Results
3.1. Demographic and Genetic Characterization of the Patients
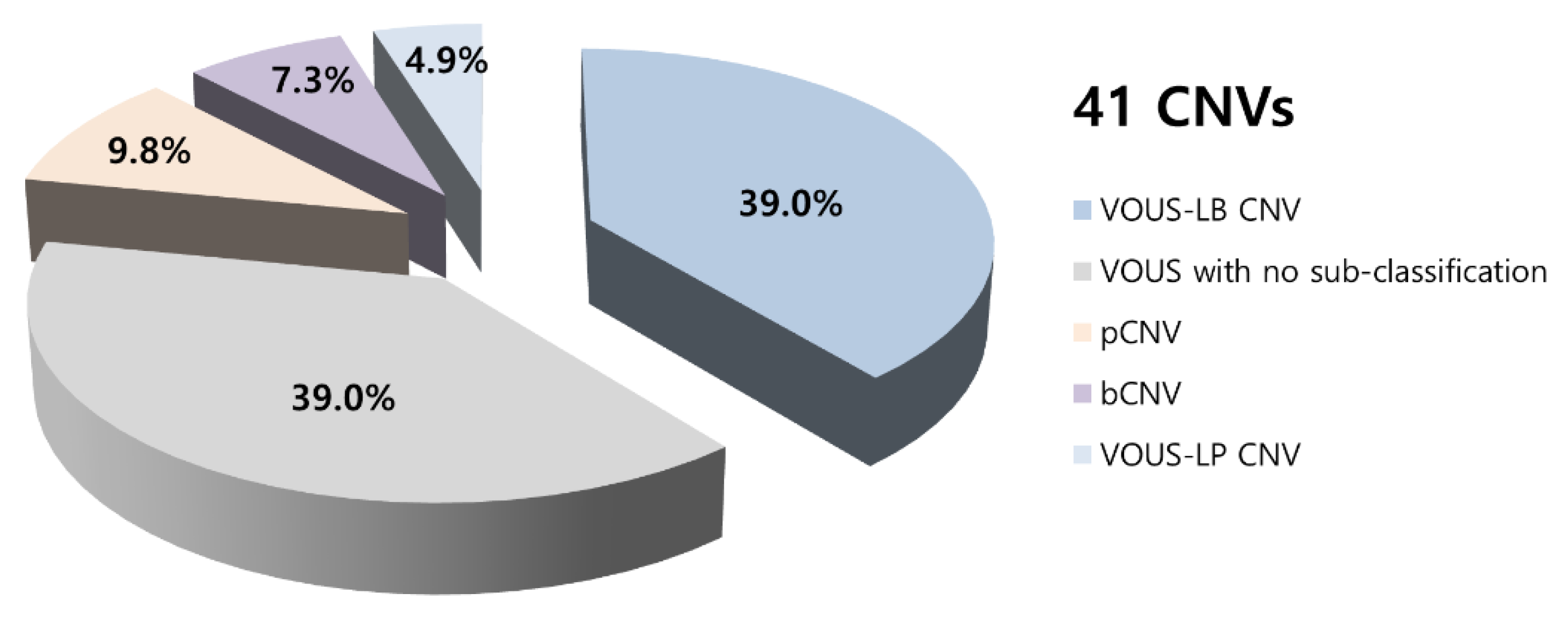
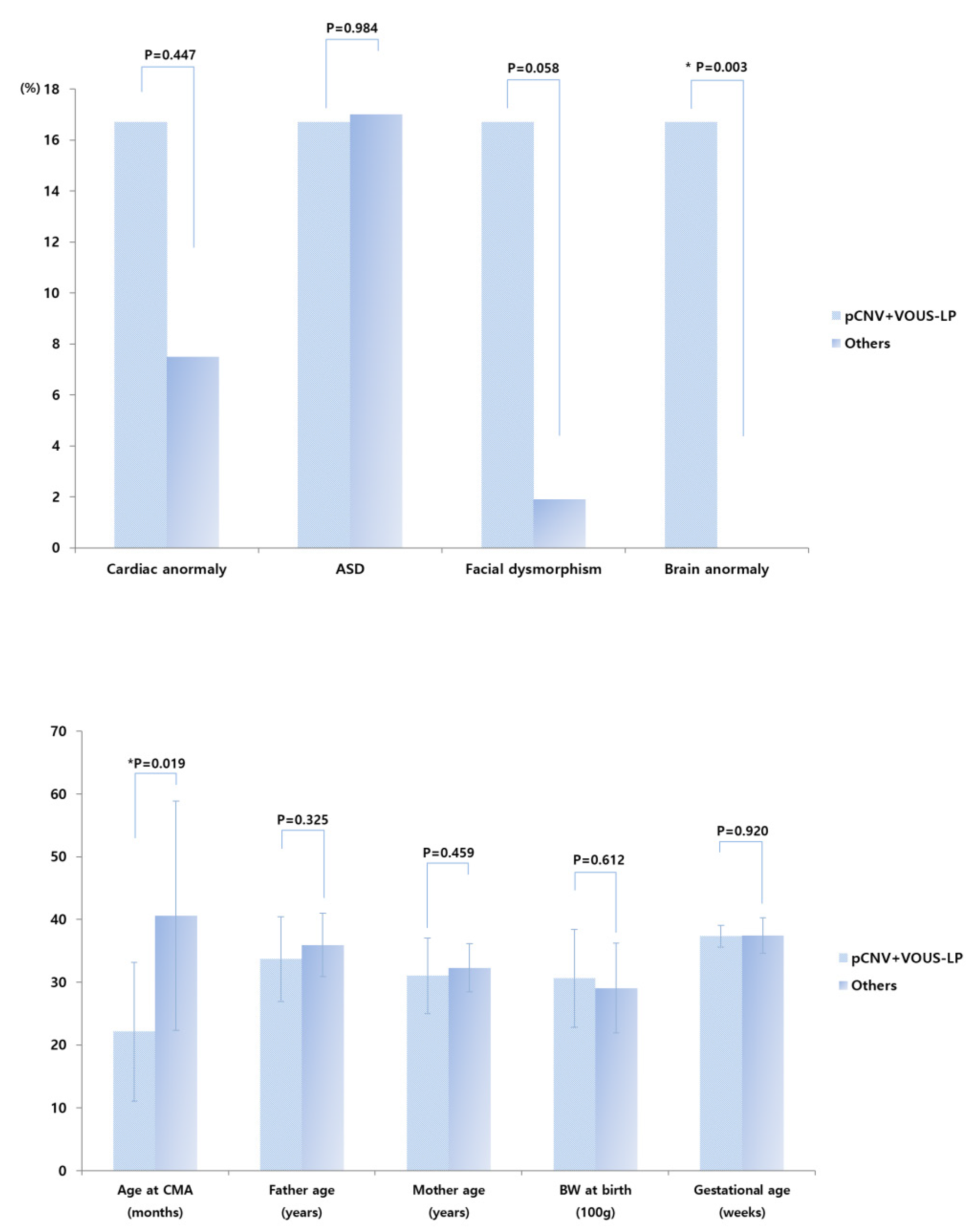
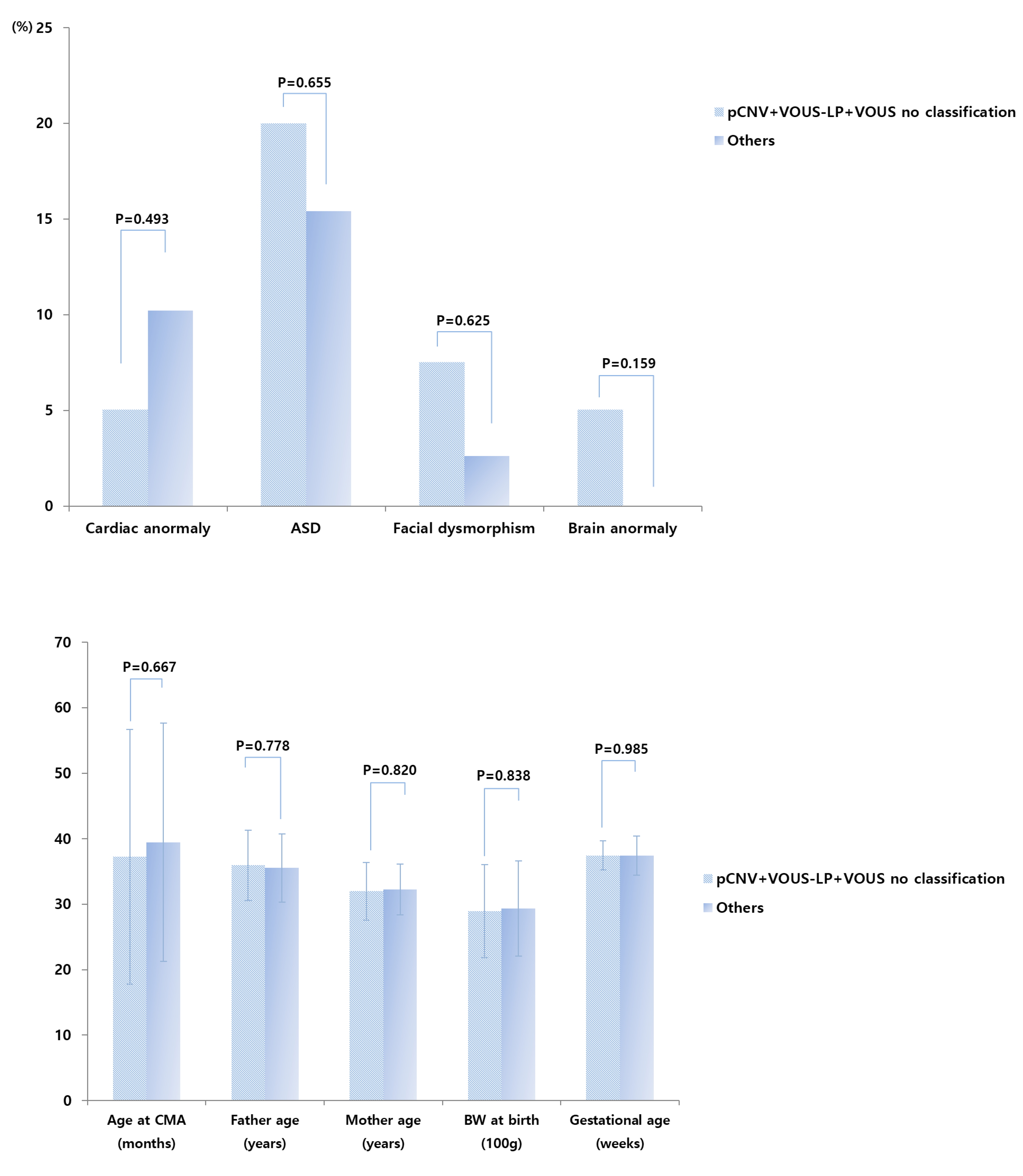
3.2. CMA Results Comparisons
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Petersen, M.C.; Kube, D.A.; Palmer, F.B. Classification of developmental delays. Semin. Pediatr. Neurol. 1998, 5, 2–14. [Google Scholar] [CrossRef]
- Shevell, M.; Ashwal, S.; Donley, D.; Flint, J.; Gingold, M.; Hirtz, D.; Majnemer, A.; Noetzel, M.; Sheth, R.D.; Quality Standards Subcommittee of the American Academy of N; et al. Practice parameter: Evaluation of the child with global developmental delay: Report of the Quality Standards Subcommittee of the American Academy of Neurology and The Practice Committee of the Child Neurology Society. Neurology 2003, 60, 367–380. [Google Scholar] [PubMed]
- Whelen, M.A. Practice parameter: Evaluation of the child with global developmental delay. Neurology 2003, 61, 1315. [Google Scholar] [CrossRef] [PubMed]
- Guralnick, M.J. Early Intervention Approaches to Enhance the Peer-Related Social Competence of Young Children With Developmental Delays: A Historical Perspective. Infants Young Child 2010, 23, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Guralnick, M.J. Why Early Intervention Works: A Systems Perspective. Infants Young Child 2011, 24, 6–28. [Google Scholar] [CrossRef] [PubMed]
- Palfrey, J.S.; Singer, J.D.; Walker, D.K.; Butler, J.A. Early identification of children’s special needs: A study in five metropolitan communities. J. Pediatr. 1987, 111, 651–659. [Google Scholar] [CrossRef]
- Chung, C.Y.; Liu, W.Y.; Chang, C.J.; Chen, C.L.; Tang, S.F.; Wong, A.M. The relationship between parental concerns and final diagnosis in children with developmental delay. J. Child Neurol. 2011, 26, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Moeschler, J.B.; Shevell, M. American Academy of Pediatrics Committee on G: Clinical genetic evaluation of the child with mental retardation or developmental delays. Pediatrics 2006, 117, 2304–2316. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.T.; Adam, M.P.; Aradhya, S.; Biesecker, L.G.; Brothman, A.R.; Carter, N.P.; Church, D.M.; Crolla, J.A.; Eichler, E.E.; Epstein, C.J.; et al. Consensus statement: Chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 2010, 86, 749–764. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Park, C.I.; Lim, J.W.; Lee, G.M.; Cho, E.; Kim, H.J. Phenotypic Analysis of Korean Patients with Abnormal Chromosomal Microarray in Patients with Unexplained Developmental Delay/Intellectual Disability. Yonsei Med. J. 2018, 59, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Farooqi, M.S.; Figueroa, S.; Gotway, G.; Wang, J.; Luu, H.S.; Park, J.Y. Reinterpretation of Chromosomal Microarrays with Detailed Medical History. J. Pediatr. 2020, 222, 180–185.e1. [Google Scholar] [CrossRef] [PubMed]
- Jang, C.H.; Kim, S.W.; Jeon, H.R.; Jung, D.W.; Cho, H.E.; Kim, J.; Lee, J.W. Clinical Usefulness of the Korean Developmental Screening Test (K-DST) for Developmental Delays. Ann. Rehabil. Med. 2019, 43, 490–496. [Google Scholar] [CrossRef] [PubMed]
- Yim, C.H.; Kim, G.H.; Eun, B.L. Usefulness of the Korean Developmental Screening Test for infants and children for the evaluation of developmental delay in Korean infants and children: A single-center study. Korean J. Pediatr. 2017, 60, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Watson, C.T.; Marques-Bonet, T.; Sharp, A.J.; Mefford, H.C. The genetics of microdeletion and microduplication syndromes: An update. Annu. Rev. Genom. Hum. Genet. 2014, 15, 215–244. [Google Scholar] [CrossRef] [PubMed]
- Gilissen, C.; Hehir-Kwa, J.Y.; Thung, D.T.; van de Vorst, M.; van Bon, B.W.; Willemsen, M.H.; Kwint, M.; Janssen, I.M.; Hoischen, A.; Schenck, A.; et al. Genome sequencing identifies major causes of severe intellectual disability. Nature 2014, 511, 344–347. [Google Scholar] [CrossRef] [PubMed]
- Gajecka, M. Unrevealed mosaicism in the next-generation sequencing era. Mol. Genet. Genom. 2016, 291, 513–530. [Google Scholar] [CrossRef] [PubMed]
| Parameters | N | Min | Max | Mean | STD |
|---|---|---|---|---|---|
| Age (months) | 59 | 8.3 | 84 | 38.7 | 18.50 |
| 0–12 months | 3 (5.1%) | - | - | - | - |
| 12–24 months | 8 (13.6%) | - | - | - | - |
| 24–36 months | 17 (28.8%) | - | - | - | - |
| 36–48 months | 10 (17.0%) | - | - | - | - |
| 48–60 months | 10 (17.0%) | - | - | - | - |
| 60–72 months | 7 (11.9%) | - | - | - | - |
| 72–84 months | 4 (6.8%) | - | - | - | - |
| Birth weight | 59 | 920 | 4300 | 2921.0 | 716.48 |
| Gestational age | 59 | 28 | 41 | 37.4 | 2.72 |
| Gender | |||||
| Male | 41 (69.5%) | - | - | - | - |
| Female | 18 (30.5%) | - | - | - | - |
| Age of father at birth | 59 | 24 | 52 | 35.7 | 5.23 |
| Age of mother at birth | 59 | 23 | 40 | 32.2 | 4.04 |
| Delivery methods | |||||
| Vaginal delivery | 23 (39%) | - | - | - | - |
| Caesarean section | 36 (61%) | - | - | - | - |
| Mode of conception | |||||
| Natural pregnancy | 50 (84.7%) | ||||
| Intra-uterine insemination | 2 (3.4%) | ||||
| In vitro fertilization | 7 (11.9) | ||||
| Comorbidity | |||||
| Cerebral palsy | 1 (1.7%) | - | - | - | - |
| Autism Spectrum Disorder | 10 (16.9%) | - | - | - | - |
| Cardiac anormaly | 5 (8.5%) | - | - | - | - |
| Dysmorphism | 2 (3.4%) | - | - | - | - |
| Brain anormaly | 1 (1.7%) | - | - | - | - |
| Presence of any anormaly | 5 (18.7%) | - | - | - | - |
| No | Sex | Age (m) | Result of CMA | OMIM Data Base | OMIM Gene | Clinical Features | Age of Father | Age of Mother | Birth Weight | Delivery | GA |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 28.6 | Yq11.221q11.222 duplication 1.5 Mb | Likely benign CNV | XKRY, CDY2A, HSFY1 | DD | 33 | 29 | 3350 | C-sec | 38 |
| 2 | F | 14.3 | 2q13 duplication 861 Kb | VOUS | RGPD6, MALL, NPHP1 | DD | 49 | 24 | 2600 | C-sec | 38 |
| 3 | F | 77.3 | 8p23.2 duplication 2.4 Mb | VOUS | CSMD1 | DD, AuSD | 34 | 33 | 3790 | NVD | 40 |
| 4 | M | 42 | Yq11.223q11.23 duplication 3.1 Mb | Likely benign CNV | Multiple OMIM gene | DD, AuSD | 47 | 36 | 3010 | C-sec | 39 |
| 5 | M | 15.1 | 10q11.22 duplication 1.3 Mb | Likely benign CNV | SYT15, GPRIN2, NPY4R | DD, Polydactyly | 34 | 36 | 3900 | C-sec | 40 |
| 6 | M | 20.6 | Yq11.223q11.23 duplication 1.6 Mb | Likely benign CNV | Multiple OMIM gene | DD, Umblicus fistula to intestine | 37 | 34 | 2500 | C-sec | 39 |
| 7 | M | 70.7 | 10p15.3 deletion 1.4 Mb | VOUS | DIP2C, IDI2, IDI2-AS1, ADARB2 | DD | 38 | 33 | 2800 | NVD | 36 |
| 8 | M | 54.3 | X121.2 deletion 523 Kb | Likely benign CNV | DACH2 | DD | 45 | 37 | 3150 | NVD | 40 |
| 9 | F | 24.6 | 2p16.3 deletion 142 Kb | Likely pathogenic CNV | NRXN1 | DD, Occipital meningocele, RDS | 41 | 36 | 2100 | C-sec | 34 |
| 10 | M | 34.2 | 7p21.1 duplication 484 Kb Xq21.33 duplication 611 Kb | Likely benign CNV VOUS | AHR DIAPH2, RPA4 | DD | 30 | 28 | 2500 | C-sec | 38 |
| 11 | M | 51.5 | 2q13 deletion 861 Kb Yq11.223q11.23 duplication 4.0 Mb | VOUS Likely benign CNV | RGPD6, MALL, NPHP1 Multiple OMIM gene | DD | 33 | 32 | 3200 | C-sec | 38 |
| 12 | M | 46 | 1q43q44 duplication 5.8 Mb | VOUS | Multiple OMIM gene | DD, Hydronephrosis, Placenta previa | 34 | 34 | 2920 | NVD | 38 |
| 13 | M | 35.6 | 3q29 duplication 1.7 Mb 15q13.3 duplication 440 Kb | Likely pathogenic CNV VOUS | Multiple OMIM gene CHRNA7 | DD, AuSD | 28 | 24 | 4180 | NVD | 37 |
| 14 | F | 42.9 | Xp22.33 duplication 413 Kb | Likely benign CNV | GTPBP6, PPP2R3B | DD, AuSD | 33 | 31 | 3990 | C-sec | 41 |
| 15 | M | 32.2 | 2q13 duplication 861 Kb | VOUS | RGPD6, MALL, NPHP1 | DD, RDS | 41 | 37 | 920 | C-sec | 30 |
| 16 | M | 14.1 | 7q11.21 deletion 443 Kb | Likely benign CNV | ZNF92 | DD, PDA, RDS | 32 | 29 | 1380 | C-sec | 28 |
| 17 | M | 14.1 | 10q11.22 duplication 1.1 Mb | Likely benign CNV | NPY4R | CP, VSD, RDS, Neonatal jaundice | 33 | 33 | 1120 | NVD | 30 |
| 18 | M | 72.1 | 9q31.1 duplication 497 Kb Xp22.33 duplication 436 Kb | VOUS VOUS | SMC2 SHOX | DD, Neonatal jaundice | 38 | 35 | 2500 | NVD | 38 |
| 19 | M | 41.3 | 10q11.22 deletion 1.1 Mb | Likely benign CNV | NPY4R | DD, AuSD | 36 | 36 | 3660 | C-sec | 41 |
| 20 | M | 10.8 | 7q11.21 deletion 456 Kb Yq11.221q11.222 duplication 1.5 Mb | Likely benign CNV Likely benign CNV | ZNF92 XKRY, CDY2A, HSFY1 | DD, RDS | 29 | 28 | 2610 | C-sec | 37 |
| 21 | M | 51.1 | 2p24.3 duplication 814 Kb 11q25 duplication 477 Kb | Likely benign CNV Likely benign CNV | B3GAT1 | DD, ID | 36 | 31 | 3200 | NVD | 38 |
| 22 | M | 31.3 | 2q23.3 deletion 467 Kb Duplication, overall area of X chromosome | Benign CNV Pathogenic CNV(Klinefelter syndrome) | Multiple OMIM gene | DD | 32 | 32 | 2500 | C-sec | 38 |
| 23 | M | 31 | 1p32.3 duplication 461 Kb | Likely benign CNV | ACOT11, TTC4, PARS2, DHCR24 | DD | 31 | 29 | 2700 | NVD | 38 |
| 24 | F | 22.9 | 3p26.3p26.1 duplication 2.2 Mb Loss mosaicism, overall area of X chromosome | VOUS Pathogenic CNV(Turner syndrome) | CNTN4, IL5RA, TRNT1 Multiple OMIM gene | DD, Neonatal jaundice | 38 | 35 | 2700 | C-sec | 38 |
| 25 | M | 30.3 | 3q26.31 duplication 706 Kb 17p13.3 duplication 210 Kb | VOUS VOUS | NLGN1, NAALADL2 ABR, BHLHA9, TUSC5, YWHAE | DD, AuSD | 36 | 27 | 2650 | NVD | 38 |
| 26 | M | 37.3 | 17p13.3 duplication 306 Kb | VOUS | ABR, BHLHA9, TUSC5 | DD | 34 | 33 | 2600 | C-sec | 38 |
| 27 | M | 29 | 2p25.3 duplication 133 Kb 2p25.3 duplication 191 Kb | VOUS VOUS | SNTG2 PXDN, MYT1L | DD | 40 | 36 | 2660 | C-sec | 37 |
| 28 | F | 29 | 2p25.3 duplication 139 Kb 2p25.3 duplication 191 Kb | VOUS VOUS | SNTG2 PXDN, MYT1L | DD | 40 | 36 | 3400 | C-sec | 37 |
| 29 | F | 10 | 2q37.1q37.3 deletion 8.8 Mb | Pathogenic CNV | Multiple OMIM gene | DD | 39 | 36 | 3700 | NVD | 39 |
| 30 | M | 51.9 | 2q13 duplication 861 Kb 5p15.33 duplication 592 Kb Yq11.222 duplication 428 Kb | VOUS Likely benign CNV Likely benign CNV | RGPD6, MALL, NPHP1 IRX1 HSFY1 | DD, AuSD | 35 | 33 | 3260 | C-sec | 41 |
| 31 | M | 8.3 | LOH, overall area of autosome 22q11.21 duplication 3.3 Mb Yq11.222 duplication 823 Kb | VOUS Pathogenic CNV VOUS | Multiple OMIM gene USP18, DGCR6, PRODH, DGCR2, DGCR14 | DD, ASD, Megalencephaly, Soft claft palate | 24 | 23 | 3200 | NVD | 38 |
| 32 | F | 49.6 | 10q11.22 duplication 1.2 Mb | Benign CNV | SYT15, GPRIN2, NPY4R | DD | 38 | 31 | 3700 | NVD | 40 |
| 33 | F | 36.2 | 2q13 duplication 861 Kb | VOUS | RGPD6, MALL, NPHP1 | DD | 35 | 33 | 3700 | NVD | 38 |
| 34 | F | 71.1 | 10q11.22 duplication 1.1 Mb | Benign CNV | NPY4R | DD | 33 | 32 | 3080 | C-sec | 38 |
| 35 | M | 26.8 | 2q11.2 duplication 431 Kb | Benign CNV | - | DD | 39 | 39 | 2900 | C-sec | 36 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Z.; Lee, B.J.; Park, S.; Park, D. Relationship between Clinical Parameters and Chromosomal Microarray Data in Infants with Developmental Delay. Healthcare 2020, 8, 305. https://doi.org/10.3390/healthcare8030305
Lee Z, Lee BJ, Park S, Park D. Relationship between Clinical Parameters and Chromosomal Microarray Data in Infants with Developmental Delay. Healthcare. 2020; 8(3):305. https://doi.org/10.3390/healthcare8030305
Chicago/Turabian StyleLee, Zeeihn, Byung Joo Lee, Sungwon Park, and Donghwi Park. 2020. "Relationship between Clinical Parameters and Chromosomal Microarray Data in Infants with Developmental Delay" Healthcare 8, no. 3: 305. https://doi.org/10.3390/healthcare8030305
APA StyleLee, Z., Lee, B. J., Park, S., & Park, D. (2020). Relationship between Clinical Parameters and Chromosomal Microarray Data in Infants with Developmental Delay. Healthcare, 8(3), 305. https://doi.org/10.3390/healthcare8030305





