The Interplay Between Cervicovaginal Microbiota Diversity, Lactobacillus Profiles and Human Papillomavirus in Cervical Cancer: A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Selection
2.2. Statistical Analysis
2.3. Risk of Bias
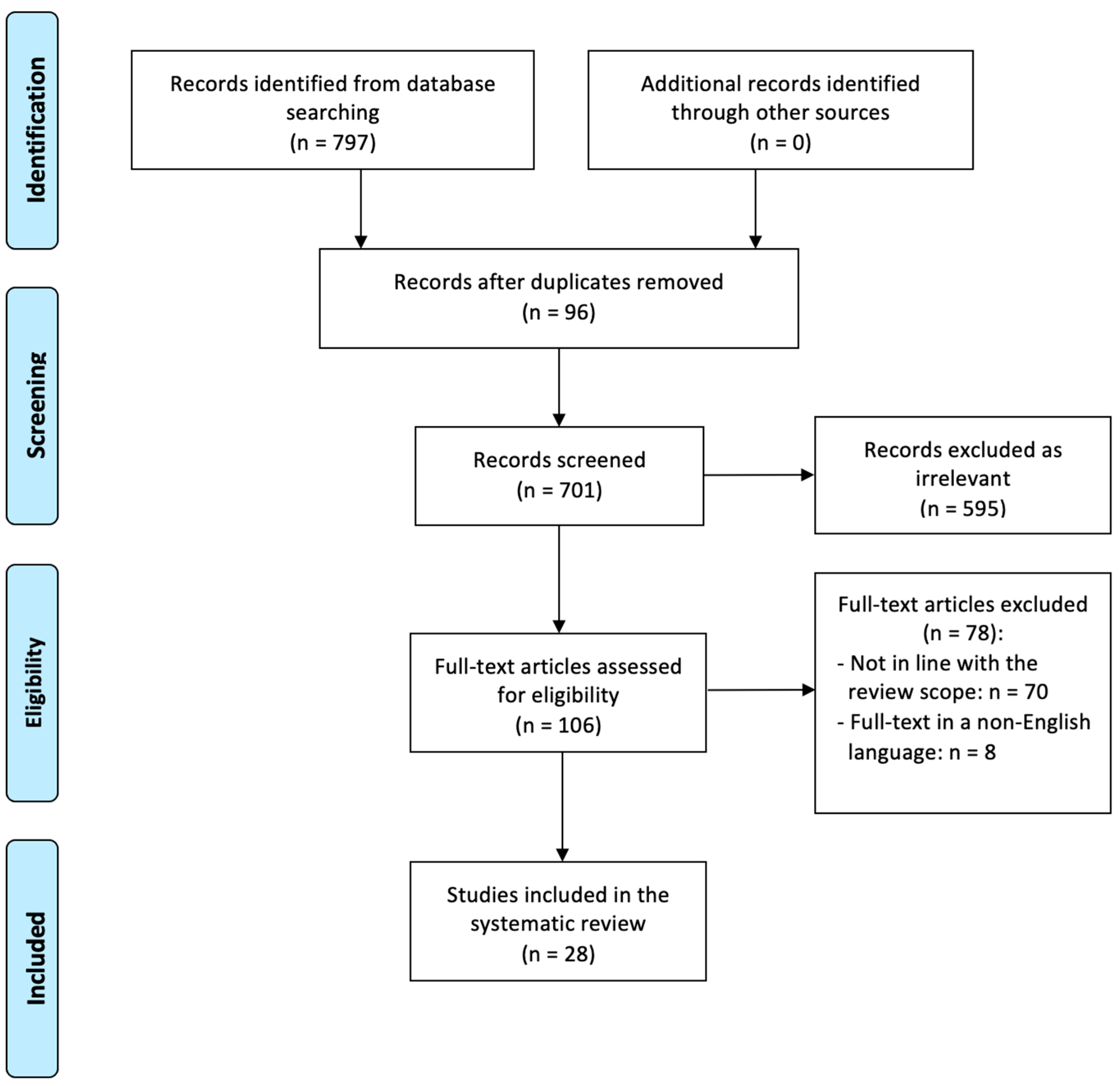
3. Results
4. Discussion
4.1. Cervical Cancer and Microbiota
4.2. Cervical Cancer and Community State Types
4.3. Cervical Cancer and Lactobacillus Profiles
4.4. Cervical Cancer and Microbiota Diversity
4.5. Cervical Cancer and Human Papillomavirus
4.6. Strengths and Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Occhipinti, S.; Incognito, G.G.; Palumbo, M. The influence of the vaginal ecosystem on vaginitis, bacterial vaginosis, and sexually transmitted diseases: An epidemiological study and literature review. Arch. Gynecol. Obstet. 2024; ahead of print. [Google Scholar] [CrossRef]
- Vitali, D.; Wessels, J.M.; Kaushic, C. Role of sex hormones and the vaginal microbiome in susceptibility and mucosal immunity to HIV-1 in the female genital tract. AIDS Res. Ther. 2017, 14, 39. [Google Scholar] [CrossRef] [PubMed]
- Anahtar, M.N.; Gootenberg, D.B.; Mitchell, C.M.; Kwon, D.S. Cervicovaginal Microbiota and Reproductive Health: The Virtue of Simplicity. Cell Host Microbe 2018, 23, 159–168. [Google Scholar] [CrossRef]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.; McCulle, S.L.; Karlebach, S.; Gorle, R.; Russell, J.; Tacket, C.O.; et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4680–4687. [Google Scholar] [CrossRef]
- Onderdonk, A.B.; Delaney, M.L.; Fichorova, R.N. The Human Microbiome during Bacterial Vaginosis. Clin. Microbiol. Rev. 2016, 29, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Ramchander, N.C.; Crosbie, E.J. The vaginal microbiome and gynaecological cancer: Exercise caution when considering causation. BJOG 2018, 125, 316. [Google Scholar] [CrossRef] [PubMed]
- Castellsagué, X. Natural history and epidemiology of HPV infection and cervical cancer. Gynecol. Oncol. 2008, 110 (Suppl. S2), S4–S7. [Google Scholar] [CrossRef]
- Amabebe, E.; Anumba, D.O.C. The Vaginal Microenvironment: The Physiologic Role of Lactobacilli. Front. Med. 2018, 5, 181. [Google Scholar] [CrossRef]
- de Sanjosé, S.; Diaz, M.; Castellsagué, X.; Clifford, G.; Bruni, L.; Muñoz, N.; Bosch, F.X. Worldwide prevalence and genotype distribution of cervical human papillomavirus DNA in women with normal cytology: A meta-analysis. Lancet. Infect. Dis. 2007, 7, 453–459. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ (Clin. Res. Ed.) 2021, 372, n71. [Google Scholar] [CrossRef]
- Audirac-Chalifour, A.; Torres-Poveda, K.; Bahena-Román, M.; Téllez-Sosa, J.; Martínez-Barnetche, J.; Cortina-Ceballos, B.; López-Estrada, G.; Delgado-Romero, K.; Burguete-García, A.I.; Cantú, D.; et al. Cervical Microbiome and Cytokine Profile at Various Stages of Cervical Cancer: A Pilot Study. PLoS ONE 2016, 11, e0153274. [Google Scholar] [CrossRef]
- Chen, Y.; Qiu, X.; Wang, W.; Li, D.; Wu, A.; Hong, Z.; Di, W.; Qiu, L. Human papillomavirus infection and cervical intraepithelial neoplasia progression are associated with increased vaginal microbiome diversity in a Chinese cohort. BMC Infect. Dis. 2020, 20, 629. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Wu, Y.; Li, M.; An, F.; Yao, L.; Wang, M.; Wang, X.; Yuan, J.; Jiang, K.; Li, W.; et al. Lactobacillus spp. create a protective micro-ecological environment through regulating the core fucosylation of vaginal epithelial cells against cervical cancer. Cell Death Dis. 2021, 12, 1094. [Google Scholar] [CrossRef]
- Han, M.; Wang, N.; Han, W.; Liu, X.; Sun, T.; Xu, J. Specific vaginal and gut microbiome and the anti-tumor effect of butyrate in cervical cancer women. Transl. Oncol. 2024, 44, 101902. [Google Scholar] [CrossRef]
- Ivanov, M.K.; Brenner, E.V.; Hodkevich, A.A.; Dzyubenko, V.V.; Krasilnikov, S.E.; Mansurova, A.S.; Vakhturova, I.E.; Agletdinov, E.F.; Shumeikina, A.O.; Chernyshova, A.L.; et al. Cervicovaginal-Microbiome Analysis by 16S Sequencing and Real-Time PCR in Patients from Novosibirsk (Russia) with Cervical Lesions and Several Years after Cancer Treatment. Diagnostics 2023, 13, 140. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.U.; Jung, D.R.; Lee, Y.H.; Jeon, S.Y.; Han, H.S.; Chong, G.O.; Shin, J.H. Potential Association between Vaginal Microbiota and Cervical Carcinogenesis in Korean Women: A Cohort Study. Microorganisms 2021, 9, 294. [Google Scholar] [CrossRef]
- Kwon, M.; Seo, S.S.; Kim, M.K.; Lee, D.O.; Lim, M.C. Compositional and Functional Differences between Microbiota and Cervical Carcinogenesis as Identified by Shotgun Metagenomic Sequencing. Cancers 2019, 11, 309. [Google Scholar] [CrossRef] [PubMed]
- Łaniewski, P.; Barnes, D.; Goulder, A.; Cui, H.; Roe, D.J.; Chase, D.M.; Herbst-Kralovetz, M.M. Linking cervicovaginal immune signatures, HPV and microbiota composition in cervical carcinogenesis in non-Hispanic and Hispanic women. Sci. Rep. 2018, 8, 7593. [Google Scholar] [CrossRef]
- Li, C.; Zhang, Z.; Yang, Y.; Liao, H. Changes in the cervicovaginal microbiota composition of HPV16-infected patients after clinical treatment. Cancer Med. 2022, 11, 5037–5049. [Google Scholar] [CrossRef]
- Li, X.; Wu, J.; Wu, Y.; Duan, Z.; Luo, M.; Li, L.; Li, S.; Jia, Y. Imbalance of Vaginal Microbiota and Immunity: Two Main Accomplices of Cervical Cancer in Chinese Women. Int. J. Women’s Health 2023, 15, 987–1002. [Google Scholar] [CrossRef]
- Li, Y.; Cao, L.; Han, X.; Ma, Y.; Liu, Y.; Gao, S.; Zhang, C. Altered vaginal eukaryotic virome is associated with different cervical disease status. Virol. Sin. 2023, 38, 184–197. [Google Scholar] [CrossRef]
- Liu, H.; Liang, H.; Li, D.; Wang, M.; Li, Y. Association of Cervical Dysbacteriosis, HPV Oncogene Expression, and Cervical Lesion Progression. Microbiol. Spectr. 2022, 10, e0015122. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Li, Y.; Liu, Y.; Cao, L.; Han, X.; Gao, S.; Zhang, C. Vaginal Microbiome Dysbiosis is Associated with the Different Cervical Disease Status. J. Microbiol. 2023, 61, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.; MacIntyre, D.A.; Lee, Y.S.; Smith, A.; Marchesi, J.R.; Lehne, B.; Bhatia, R.; Lyons, D.; Paraskevaidis, E.; Li, J.V.; et al. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci. Rep. 2015, 5, 16865. [Google Scholar] [CrossRef] [PubMed]
- Musa, J.; Maiga, M.; Green, S.J.; Magaji, F.A.; Maryam, A.J.; Okolo, M.; Nyam, C.J.; Cosmas, N.T.; Silas, O.A.; Imade, G.E.; et al. Vaginal microbiome community state types and high-risk human papillomaviruses in cervical precancer and cancer in North-central Nigeria. BMC Cancer 2023, 23, 683. [Google Scholar] [CrossRef]
- Ou, J.; Kang, Y.; Medlegeh, F.K.; Zhang, Y.; Yang, W. An analysis of the vaginal microbiota and cervicovaginal metabolomics in cervical lesions and cervical carcinoma. Heliyon 2024, 10, e33383. [Google Scholar] [CrossRef]
- Sekaran, K.; Varghese, R.P.; Gopikrishnan, M.; Alsamman, A.M.; El Allali, A.; Zayed, H.; Doss, C.G.P. Unraveling the Dysbiosis of Vaginal Microbiome to Understand Cervical Cancer Disease Etiology—An Explainable AI Approach. Genes 2023, 14, 936. [Google Scholar] [CrossRef]
- Stoian, I.L.; Botezatu, A.; Fudulu, A.; Ilea, C.G.; Socolov, D.G. Exploring Microbiota Diversity in Cervical Lesion Progression and HPV Infection through 16S rRNA Gene Metagenomic Sequencing. J. Clin. Med. 2023, 12, 4979. [Google Scholar] [CrossRef]
- Teka, B.; Yoshida-Court, K.; Firdawoke, E.; Chanyalew, Z.; Gizaw, M.; Addissie, A.; Mihret, A.; Colbert, L.E.; Napravnik, T.C.; El Alam, M.B.; et al. Cervicovaginal Microbiota Profiles in Precancerous Lesions and Cervical Cancer among Ethiopian Women. Microorganisms 2023, 11, 833. [Google Scholar] [CrossRef]
- Wang, Z.; Xiao, R.; Huang, J.; Qin, X.; Hu, D.; Guo, E.; Liu, C.; Lu, F.; You, L.; Sun, C.; et al. The Diversity of Vaginal Microbiota Predicts Neoadjuvant Chemotherapy Responsiveness in Locally Advanced Cervical Cancer. Microb. Ecol. 2022, 84, 302–313. [Google Scholar] [CrossRef]
- Wei, B.; Chen, Y.; Lu, T.; Cao, W.; Tang, Z.; Yang, H. Correlation between vaginal microbiota and different progression stages of cervical cancer. Genet. Mol. Biol. 2022, 45, e20200450. [Google Scholar] [CrossRef]
- Wu, S.; Ding, X.; Kong, Y.; Acharya, S.; Wu, H.; Huang, C.; Liang, Y.; Nong, X.; Chen, H. The feature of cervical microbiota associated with the progression of cervical cancer among reproductive females. Gynecol. Oncol. 2021, 163, 348–357. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Feng, Y.; Li, W.; Zhan, F.; Huang, G.; Hu, H.; Xiong, Y.; Tan, B.; Chen, T. Revealing the Disturbed Vaginal Micobiota Caused by Cervical Cancer Using High-Throughput Sequencing Technology. Front. Cell. Infect. Microbiol. 2020, 10, 538336. [Google Scholar] [CrossRef]
- Xu, H.; Liu, L.; Xu, F.; Liu, M.; Song, Y.; Chen, J.; Zhan, H.; Zhang, Y.; Xu, D.; Chen, Y.; et al. Microbiome-metabolome analysis reveals cervical lesion alterations. Acta Biochim. Et Biophys. Sin. 2022, 54, 1552–1560. [Google Scholar] [CrossRef]
- Zeber-Lubecka, N.; Kulecka, M.; Lindner, B.; Krynicki, R.; Paziewska, A.; Nowakowski, A.; Bidzinski, M.; Ostrowski, J. Increased diversity of a cervical microbiome associates with cervical cancer. Front. Oncol. 2022, 12, 1005537. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Liang, Y.; Zhou, J.; Lin, H.; Huang, L.; He, D.; Wen, J.; Wu, B.; Liu, H.; Zhong, Y.; et al. Vaginal microecological changes of different degrees of cervical lesions in Hakka women in Meizhou City. J. Obstet. Gynaecol. 2023, 43, 2186780. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Q.; Zhang, W.; Zhang, Z.; Fu, Y.; Li, Y.; Wang, X.; Li, L.; Meng, Y. Characteristics of the Cervicovaginal Microenvironment in Childbearing-Age Women with Different Degrees of Cervical Lesions and HR-HPV Positivity. Pol. J. Microbiol. 2021, 70, 489–500. [Google Scholar] [CrossRef]
- Zhang, W.; Yin, Y.; Jiang, Y.; Yang, Y.; Wang, W.; Wang, X.; Ge, Y.; Liu, B.; Yao, L. Relationship between vaginal and oral microbiome in patients of human papillomavirus (HPV) infection and cervical cancer. J. Transl. Med. 2024, 22, 396. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, N.; Bosch, F.X.; de Sanjosé, S.; Herrero, R.; Castellsagué, X.; Shah, K.V.; Snijders, P.J.; Meijer, C.J.; International Agency for Research on Cancer Multicenter Cervical Cancer Study Group. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 2003, 348, 518–527. [Google Scholar] [CrossRef]
- Kyrgiou, M.; Mitra, A.; Moscicki, A.B. Does the vaginal microbiota play a role in the development of cervical cancer? Transl. Res. J. Lab. Clin. Med. 2017, 179, 168–182. [Google Scholar] [CrossRef]
- Schiffman, M.; Wentzensen, N.; Wacholder, S.; Kinney, W.; Gage, J.C.; Castle, P.E. Human papillomavirus testing in the prevention of cervical cancer. J. Natl. Cancer Inst. 2011, 103, 368–383. [Google Scholar] [CrossRef]
- Shulzhenko, N.; Lyng, H.; Sanson, G.F.; Morgun, A. Menage a trois: An evolutionary interplay between human papillomavirus, a tumor, and a woman. Trends Microbiol. 2014, 22, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Norenhag, J.; Du, J.; Olovsson, M.; Verstraelen, H.; Engstrand, L.; Brusselaers, N. The vaginal microbiota, human papillomavirus and cervical dysplasia: A systematic review and network meta-analysis. BJOG Int. J. Obstet. Gynaecol. 2020, 127, 171–180. [Google Scholar] [CrossRef]
- Lee, J.E.; Lee, S.; Lee, H.; Song, Y.M.; Lee, K.; Han, M.J.; Sung, J.; Ko, G. Association of the vaginal microbiota with human papillomavirus infection in a Korean twin cohort. PLoS ONE 2013, 8, e63514. [Google Scholar] [CrossRef]
- Guijon, F.; Paraskevas, M.; Rand, F.; Heywood, E.; Brunham, R.; McNicol, P. Vaginal microbial flora as a cofactor in the pathogenesis of uterine cervical intraepithelial neoplasia. Int. J. Gynaecol. Obstet. 1992, 37, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Capozzi, V.A.; Incognito, G.G.; Scarpelli, E.; Palumbo, M.; Randazzo, C.L.; Pino, A.; La Verde, M.; Ronsini, C.; Riemma, G.; Gaiano, M.; et al. Exploring the Relationship between Ovarian Cancer and Genital Microbiota: A Systematic Review and Meta-Analysis. J. Pers. Med. 2024, 14, 351. [Google Scholar] [CrossRef]
- Mhatre, M.; McAndrew, T.; Carpenter, C.; Burk, R.D.; Einstein, M.H.; Herold, B.C. Cervical intraepithelial neoplasia is associated with genital tract mucosal inflammation. Sex. Transm. Dis. 2012, 39, 591–597. [Google Scholar] [CrossRef] [PubMed]
- van de Wijgert, J.H.H.M.; Gill, A.C.; Chikandiwa, A.; Verwijs, M.C.; Kelly, H.A.; Omar, T.; Delany-Moretlwe, S.; Segondy, M.; Francis, S.; Darby, A.C.; et al. Human papillomavirus infection and cervical dysplasia in HIV-positive women: Potential role of the vaginal microbiota. AIDS 2020, 34, 115–125. [Google Scholar] [CrossRef]
- Gardella, B.; Pasquali, M.F.; La Verde, M.; Cianci, S.; Torella, M.; Dominoni, M. The Complex Interplay between Vaginal Microbiota, HPV Infection, and Immunological Microenvironment in Cervical Intraepithelial Neoplasia: A Literature Review. Int. J. Mol. Sci. 2022, 23, 7174. [Google Scholar] [CrossRef]
- Brotman, R.M.; Shardell, M.D.; Gajer, P.; Tracy, J.K.; Zenilman, J.M.; Ravel, J.; Gravitt, P.E. Interplay between the temporal dynamics of the vaginal microbiota and human papillomavirus detection. J. Infect. Dis. 2014, 210, 1723–1733. [Google Scholar] [CrossRef]
- Verhoeven, V.; Renard, N.; Makar, A.; Van Royen, P.; Bogers, J.P.; Lardon, F.; Peeters, M.; Baay, M. Probiotics enhance the clearance of human papillomavirus-related cervical lesions: A prospective controlled pilot study. Eur. J. Cancer Prev. 2013, 22, 46–51. [Google Scholar] [CrossRef]
- Mitra, A.; MacIntyre, D.A.; Ntritsos, G.; Smith, A.; Tsilidis, K.K.; Marchesi, J.R.; Bennett, P.R.; Moscicki, A.B.; Kyrgiou, M. The vaginal microbiota associates with the regression of untreated cervical intraepithelial neoplasia 2 lesions. Nat. Commun. 2020, 11, 1999. [Google Scholar] [CrossRef] [PubMed]
- Tango, C.N.; Seo, S.-S.; Kwon, M.; Lee, D.-O.; Chang, H.K.; Kim, M.K. Taxonomic and functional differences in cervical microbiome associated with cervical cancer development. Sci. Rep. 2020, 10, 9720. [Google Scholar] [CrossRef] [PubMed]
- Piyathilake, C.J.; Ollberding, N.J.; Kumar, R.; Macaluso, M.; Alvarez, R.D.; Morrow, C.D. Cervical Microbiota Associated with Higher Grade Cervical Intraepithelial Neoplasia in Women Infected with High-Risk Human Papillomaviruses. Cancer Prev. Res. 2016, 9, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Koliopoulos, G.; Nyaga, V.N.; Santesso, N.; Bryant, A.; Martin-Hirsch, P.P.; Mustafa, R.A.; Schünemann, H.; Paraskevaidis, E.; Arbyn, M. Cytology versus HPV testing for cervical cancer screening in the general population. Cochrane Database Syst. Rev. 2017, 8, CD008587. [Google Scholar] [CrossRef]
- Son, Y.M.; Kim, J. The Microbiome-Immune Axis Therapeutic Effects in Cancer Treatments. J. Microbiol. Biotechnol. 2022, 32, 1086–1097. [Google Scholar] [CrossRef]
| Author, Year | Country | Cases (n) | Controls (n) | HPV Genotypes | Cases+ (n, %) | Controls+ (n, %) | Sample Type | Microbial Analysis | CSTs Cases (n, %) | CSTs Controls (n, %) | Lactobacillus Profiles Cases (n, %) | Lactobacillus Profiles Controls (n, %) |
α-Diversity (Index) |
β-Diversity (Index) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Audirac-Chalifour, 2016 [11] | Mexico | 8 ICC+ | 10 NILM-, 10 NILM+ | NR | 8 (100) | 10 (50) | cervical (swab, biopsy) | V3-V4 16S rRNA | IV: 2 (25), VI: 1 (12.5), VII: 2 (25), VIII: 3 (37.5) | NILM-: I: 4 (57), II: 1 (14), V: 1 (14), VI: 1 (14); NILM+: I: 2 (20), II: 4 (40), III: 3 (30), V: 1 (10) | NR | NR | ↔ cases 3.08 ± 1.28 vs. NILM- 2.00 ± 0.63: p = 0.498, ↔ cases 3.08 ± 1.28 vs. NILM+ 2.49 ± 0.70: p = 1 (Shannon index), ↑ cases 4.14 ± 1.49 vs. NILM- 1.55 ± 0.99: p = 0.036, ↔ cases 4.14 ± 1.49) vs. NILM+ 2.49 ± 1.61): p = 0.318 (PD whole tree) | p < 0.00001 (cases vs. NILM-) (weighted Unifrac) |
| Chen, 2020 [12] | China | 9 ICC+ | 68 NILM-, 78 NILM+ | NR | 9 (100) | 78 (53.4) | vaginal (swab) | V3-V4 16S rRNA | III: 1 (11.1), IV: 8 (88.9) | NILM-: I: 14 (20.6), II: 2 (2.9), III: 32 (47.1), IV: 20 (29.4); NILM+: I: 14 (17.9), II (2.6), III: 28 (35.9), IV: 32 (41.0), V: 2 (2.6) | NR | NR | ↑ cases: 367.76 ± 208.63 vs. NILM-: 84.02 ± 73.88 (q ≤ 0.001) vs. NILM+: 272.26 ± 191.62 (Chao index); cases: 2.47 ± 0.98 vs. NILM-: 0.94 ± 0.95 (q ≤ 0.001) vs. NILM+: 1.49 ± 1.01 (q < 0.05) (Shannon index) | cases vs. NILM-: R = 0.284, p = 0.001; cases vs. NILM+: R = −0.0359, p = 0.656 (Unweighted Unifrac) |
| Fan, 2021 [13] | China | 65 ICC | 54 NILM | 16, 18, 31, 33, 52, 58, 35, 39, 45, 51, 56, 59, 68 | 63 (96.9) | 47 (87) | vaginal (swab) | V3-V4 16S rRNA | NR | NR | NR | NR | ↑ p < 0.0001 (Chao1, Shannon, Simpson, OTUs) | p < 0.05 |
| Han, 2024 [14] | China | 84 ICC | 180 NILM | NR | NR | NR | vaginal (swab) | V3-V4 16S rRNA | NR | NR | NR | NR | ↔ p > 0.05 (Chao, Shannon, Simpson) | p = 0.001 (Bray–Curtis) |
| Ivanov, 2023 [15] | Russia | 17 ICC | 77 NILM | 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 26, 53, 66, 68, 73, 82 | 16 (94.1) | 19 (24.7) | cervical (swab) | V3-V4 16S rRNA | NR | NR | NR | NR | p ≤ 0.001 (Shannon, OTUs), ↑ p = 0.000344 (Faith’s) | NR |
| Kang, 2021 [16] | Korea | 8 ICC | 7 NILM | 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68, 69, 73, 82, 6, 11, 40, 42, 44, 53, 54, 70 | 8 (100) | 0 (0) | vaginal (swab) | V3 16S rRNA | NR | NR | NR | NR | ↑ p = 0.0012 (Richness index), ↔ p > 0.05 (Shannon index), ↔ p > 0.05 (Simpson index) | p = 0.001 (Bray–Curtis) |
| Kwom, 2018 [17] | Korea | 12 ICC | 18 NILM | NR | NR | NR | cervical (swab) | Whole-genome sequencing | NR | NR | NR | NR | p = 0.1218 (Shannon), ↔ p = 0.0863 (Simpson) | p = 0.087 (Bray–Curtis), p = 0.094 (Jaccard) |
| Łaniewski, 2018 [18] | USA | 10 ICC | 51 NILM | 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68 | 9 (90) | 31 (60.8) | cervical (swab, lavage) | V4 16S rRNA | NR | NR | LDo: 2 (20), LDe: 8 (80) | NILM-: LDo: (60), LDe: (40); NILM+: LDo: (68), LDe: (32) | NR | NR |
| Li C, 2022 [19] | China | 6 ICC+ | 25 NILM- | NR | 6 (100) | 0 (0) | cervical (swab) | V3-V4 16S rRNA | I: (40.9), II: (4.6), III: (31.8), IV: (18.2), V: (4.5) | II: (50), IV: (50) | NR | NR | NR | p = 0.044 (unweighted Unifrac) |
| Li X, 2023 [20] | China | 79 ICC | 79 NILM-, 80 NILM+ | 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68, and 13 LR | NR | NR | vaginal (swab) | V3-V4 16S rRNA | NR | NR | NR | NR | ↑ p < 0.01 (Chao, Shannon, Simpson, OTUs, Pielou) | NR |
| Li Y, 2023 [21] | China | 26 ICC | 53 NILM | 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 68, 26, 53, 66, 73, 82, 6, 11, 81 | NR | NR | vaginal (swab) | V4 16S rRNA | III: 7 (26.9), IV: 19 (73.1) | I: 15 (28.3), III: 18 (34), IV: 18 (34), V: 1 (1.9) | LDo: 7 (26.9), LDe: 19 (73.1) | LDo: 33 (62.3), LDe: 19 (35.8) | ↑ p < 0.05 (Chao), ↔ p = 0.065 (Shannon) | p = 0.002 (Bray–Curtis) |
| Liu, 2022 [22] | China | 41 ICC+ | 34 NILM+ | 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, and 82 | 41 (100) | 34 (100) | cervical (swab) | 16S rRNA | III: 4 (9.8), IV: 37 (90.2) | I: 9 (26.5), III: 14 (41.2), IV: 11 (32.3) | LDo: 4 (9.8), LDe: 37 (90.2) | LDo: 23 (67.7); LDe: 11 (32.3) | ↑ p < 0.05 (Chao index), ↑ p < 0.001 (Shannon index) | R = 0.109, p = 0.001 (Bray–Curtis) |
| Ma, 2023 [23] | China | 27 ICC | 30 NILM-, 22 NILM+ | 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 26, 53, 66, 73, 82, 6, 11, and 81 | 21 (77.8) | 22 (42.3) | vaginal (swab) | V4 16S rRNA | III: 7 (25.9), IV: 20 (74.1) | HPV-: I: 9 (30), III: 10 (33.3), IV: 10 (33.3), V: 1 (3.3); HPV+: 6 (27.3), III: 8 (36.4), IV: 12 (26.7), V: 2 (4.4) | LDo: 7 (25.9), LDe: 20 (74.1) | NILM-: LDo: 19 (63.3), LDe: 11 (36.7); NILM+: LDo: 14 (63.6), 8 (36.4) | ↔ NILM- vs. NILM+ (p > 0.05) vs. ↑ cases (p < 0.01) (Shannon index); ↔ NILM- vs. NILM+ (p > 0.05) vs. ↑ cases (p < 0.05) (Simpson index); ↔ NILM- vs. NILM+ (p > 0.05) vs. ↑ cases (p < 0.001) (Sobs) | NR |
| Mitra, 2015 [24] | England | 20 ICC | 5 NILM | 16, 18, 12, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68 | NR | NR | vaginal (swab) | V1-V2 16S rRNA | I: 1 (20), II: 1 (20), IV: 2 (40), V: 1 (20) | I: 10 (50), III 8 (40), IV: 2 (10) | NR | NR | NR | NR |
| Musa, 2023 [25] | Nigeria | 30 ICC | 19 NILM | 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 69, 73, 82, 6, 11, 40, 42, 43, 44, 54, 61, 70 | 27 (90) | 8 (42.1) | cervico- vaginal (lavage) | V3-V4 16S rRNA | I: 2 (0.7), III: 3 (10), IV: 25 (83.3) | I: 2 (10.5), III: 8 (42.1), IV: 9 (47.4) | NR | NR | NR | NR |
| Ou, 2024 [26] | China | 25 ICC | 10 NILM | NR | 9 (90) | 22 (88) | vaginal (swab), cervico- vaginal (lavage) | V3-V4 or V4-V5 16S rRNA | NR | NR | NR | NR | NR | p < 0.001 (Bray–Curtis) |
| Sekaran, 2023 [27] | India | 65 ICC | 54 NILM | NR | NR | NR | vaginal (swab), cervico- vaginal (lavage) | 16S rRNA | NR | NR | NR | NR | NR | p = 0.001 (Bray–Curtis) |
| Stoian, 2023 [28] | Romania | 9 ICC+ | 20 NILM-, 9 NILM+ | 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68 | 9 (100) | 9 (31) | cervical (swab) | V3-V4 16S rRNA | NR | NR | NR | NR | ↑ p = 0.0019 (Shannon) | NR |
| Teka, 2023 [29] | Ethiopia | 60 ICC | 35 NILM | 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 69, 73, 82, 6, 11, 40, 42, 43, 44, 54, 61, 70 | NR | NR | cervical (swab, brush) | V4 16S rRNA | NR | NR | NR | NR | ↑ p = 0.00000054 (Shannon), p = 0.000005 (Simpson) | p = 0.001 (weighted UniFrac) |
| Wang, 2022 [30] | China | 26 ICC | 40 NILM | 16, 18, 11, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68, | 18 (69.2) | 1 (2.5) | vaginal (swab) | V3-V4 16S rRNA | NR | NR | NR | NR | ↑ p < 0.001 (Shannon), ↓ p < 0.001 (Simpson) | R = 0.464, p = 0.001 (Bray–Curtis) |
| Wei, 2022 [31] | China | 11 ICC | 10 NILM-, 13 NILM+ | 12, 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68 | 11 (100) | 13 (56.5) | cervical (biopsy) | V3-V4 16S rRNA | I: 1 (9.1), II: 7 (63.6), III: 3 (27.3) | NILM-: I: 6 (60), II: 4 (40); NILM+: I: 5 (38.5), II: 5 (38.5), III: 3 (23.1) | NR | NR | NILM- vs. ↑ NILM+: p = 0.03971 vs. ↑ cases: p = 0.004151 (Shannon); NILM- vs. ↑ NILM+: p = 0.01851 vs. ↑ cases: p = 0.000894 (Simpson) | NR |
| Wu, 2021 [32] | China | 13 ICC | 28 NILM-, 12 NILM+ | 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68, 53, 6, 11, 42, 43, 44, CP8304(81) | 10 (76.9) | 12 (30) | cervical (swab) | V4 16S rRNA | II: 11 (85), III: 2 (15) | NILM-: NR; NILM+: II: 10 (83), 2 (17) | NR | NR | ↑ p < 0.05 (Shannon, Simpson) | p < 0.01 (weighted Unifrac) |
| Xie, 2020 [33] | China | 18 ICC+ | 25 NILM- | 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 73, 82, 6, 11, 40, 42, 43, 44, 54, 61, 81, 83 | 18 (100) | 0 (0) | vaginal (swab) | V4 16S rRNA | NR | NR | LDo: 3 (18.4), LDe: 15 (81.6) | LDo: 9 (35.6), LDe: 16 (64.4) | ↔ p = 0.2609 (Shannon), p = 0.2245 (Simpson) | NR |
| Xu, 2022 [34] | China | 10 ICC | 10 NILM | 16, 18, 31, 33, 35, 39, 42, 43, 44, 45, 51, 52, 56, 58, 59, 68 | NR | NR | cervico- vaginal (swab) | V3-V4 16S rRNA | NR | NR | NR | NR | ↑ p = 0.04 (Shannon), p = 0.02 (Simpson) | F = 1.8557, R2 = 0.1407, p = 0.008 (Bray–Curtis) |
| Zeber-Lubecka, 2022 [35] | Poland | 16 ICC | 30 NILM- | NR | NR | 0 (0) | cervical (swab) | V2-V3-V4-V6-V7-V8-V9 16S rRNA | NR | NR | NR | NR | premenopause: ↔ p = 0.055 (Chao), ↑ p = 0.0025 (Shannon); postmenopause: ↔ p = 0.7 (Chao), ↑ p = 0.026 (Shannon) | NR |
| Zeng, 2023 [36] | China | 15 ICC | 15 NILM | NR | NR | NR | vaginal (swab) | V3-V4 16S rRNA | NR | NR | NR | NR | ↑ p = 0.0023 (Chao1), p = 0.0023 (Shannon), p = 0.0043 (Simpson), p = 0.0012 (OTUs), p = 0.0010 (PD whole tree), p = 0.0007 (goods coverage) | NR |
| Zhai, 2021 [37] | China | 38 ICC | 29 NILM-, 29 NILM+ | NR | NR | NR | cervical (swab) | V3-V4 16S rRNA | NR | NR | NR | NR | ↔ p > 0.05(Chao1, Shannon, Simpson, PD whole tree, ACE), ↓ cases vs. NILM-: p ≤ 0.05 (OTUs) | p ≤ 0.05 (weighted UniFrac) |
| Zhang, 2024 [38] | China | 22 ICC+ | 22 NILM-, 21 NILM + | 16, 18, 33, 51, 52, 53, 58 | 22 (100) | 21 (48.8) | vaginal (swab) | 16s rDNA | NR | NR | NR | NR | NILM- vs. ↑ NILM+: 0.013 vs. cases: ↑ 0.00055 (Chao1), NILM- vs. ↑ NILM+: p=0.005 vs. ↑ cases: 6.7 × 10−7 (Shannon), NILM- vs. ↑ NILM+: 0.0039 vs. ↑ cases: 1.3 × 10−6 (Simpson) | R2 = 0.189, p = 0.001 (unweighted UniFrac), R2 = 0.05, p = 0.017 (weighted UniFrac) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Incognito, G.G.; Ronsini, C.; Palmara, V.; Romeo, P.; Vizzielli, G.; Restaino, S.; La Verde, M.; De Tommasi, O.; Palumbo, M.; Cianci, S. The Interplay Between Cervicovaginal Microbiota Diversity, Lactobacillus Profiles and Human Papillomavirus in Cervical Cancer: A Systematic Review. Healthcare 2025, 13, 599. https://doi.org/10.3390/healthcare13060599
Incognito GG, Ronsini C, Palmara V, Romeo P, Vizzielli G, Restaino S, La Verde M, De Tommasi O, Palumbo M, Cianci S. The Interplay Between Cervicovaginal Microbiota Diversity, Lactobacillus Profiles and Human Papillomavirus in Cervical Cancer: A Systematic Review. Healthcare. 2025; 13(6):599. https://doi.org/10.3390/healthcare13060599
Chicago/Turabian StyleIncognito, Giosuè Giordano, Carlo Ronsini, Vittorio Palmara, Paola Romeo, Giuseppe Vizzielli, Stefano Restaino, Marco La Verde, Orazio De Tommasi, Marco Palumbo, and Stefano Cianci. 2025. "The Interplay Between Cervicovaginal Microbiota Diversity, Lactobacillus Profiles and Human Papillomavirus in Cervical Cancer: A Systematic Review" Healthcare 13, no. 6: 599. https://doi.org/10.3390/healthcare13060599
APA StyleIncognito, G. G., Ronsini, C., Palmara, V., Romeo, P., Vizzielli, G., Restaino, S., La Verde, M., De Tommasi, O., Palumbo, M., & Cianci, S. (2025). The Interplay Between Cervicovaginal Microbiota Diversity, Lactobacillus Profiles and Human Papillomavirus in Cervical Cancer: A Systematic Review. Healthcare, 13(6), 599. https://doi.org/10.3390/healthcare13060599










