Abstract
(1) Background: Water is necessary for the preparation of some medicines found in pharmacies where the local water source does not meet the required purity. This study aimed to investigate the presence of coliform contamination in water used for drug reconstitution in community pharmacies in Jordan. (2) Methods: Two water samples from 50 randomly selected community pharmacies representing all Jordanian governorates were filtered and then cultured in plate count agars to determine total microbial count, and in m-Endo Agar Les and Eosin Methylene Blue (EMB) agar to cultivate Escherichia coli (E. coli). The presence of E. coli was further characterized with gram stains, biochemical tests, and Polymerase chain reaction (PCR). Antibiotic susceptibility of isolated E. coli was tested against a variety of standard antibiotics. (3) Results: Community pharmacies used droppers filled with water from coolers (62%), bottled water (20%), boiled tap water (16%) and tap water (2%). The majority of the sampled water contained coliform bacteria (88%), and E. coli was isolated from 26% of all samples. Statistical analysis showed no significant difference in the percentage of contaminated water samples based on its source location. Nonetheless, the results showed a tendency for higher proportions of contamination in droppers filled from boiled tap water (37.5%; SE: 17.1), followed by water from water coolers (25.8%; SE: 7.9), and then from bottled water (20%; SE: 12.7). All of the isolated E. coli were sensitive to gentamycin, ciprofloxacin and levofloxacin. The susceptibility of the isolates to ceftazidime, doxycycline, tetracycline, azithromycin and amoxicillin/clavulanic acid were 92%, 61%, 46%, 23% and 15%, respectively. (4) Conclusions: This study confirms the widespread presence of multidrug-resistant bacteria in water intended for reconstituting drugs in local pharmacies. These findings expose an alarming situation that needs special attention by the acting pharmacists and competent authorities. Higher levels of personal hygiene in the pharmacies coupled with regular inspection of water quality may reduce the risk of microbial contamination in compounded products, especially multidrug-resistant strains of E. coli and other index microorganisms.
1. Introduction
Water is considered a carrier of faecal-borne disease. The consumption of such contaminated water can lead to infection with many bacterial, viral and protozoal diseases [1]. However, water is one of the primary requirements for most pharmaceutical endevors as a raw material, ingredient and solvent in the processing, formulation and manufacturing of pharmaceutical products, active pharmaceutical ingredients (APIs) and intermediates [2,3]. Accordingly, water used in pharmaceutical preparations must be of higher purity than that usually available in domestic water sources [4]. Therefore, water used to produce pharmaceutical products must fulfil quality requirements as dictated in published standards. According to those standards, pharmacies and pharmaceutical companies set up special systems for water purification, which constitute an important part of the infrastructure of their institutions [5,6,7].
Bacteria, viruses and protozoa are microbes that may be present in water as pollutants. Coliform bacteria are microbes that emerge from the intestinal tracts of warm-blooded animals and exist in soil and vegetation. The existence of coliform bacteria in water indicates the possibility that disease-causing bacteria are also present in the water [8]. Hence, applying bioburden testing in water samples establishes the number of existing microorganisms, ensuring that bacterial loads do not exceed mandated pharmacopeia indices. Microbial testing of water includes the estimation of the number of specific index bacteria present in a given quality of water, which includes Escherichia coli (E. coli), Staphylococcus aureus, Pseudomonas aeruginosa and Aspergillus niger [9].
E. coli belongs to the Enterobacteriaceae family and is described as a facultative anaerobic, gram-negative, non-spore-forming, rod-shaped bacterium that contains the enzyme β-glucuronidase [10]. E. coli is an extremely versatile bacteria with over 250 serotypes, ranging from innocuous gut commensals to intra- or extra-intestinal pathogens that can colonize common medical devices, and is considered the primary cause of recurrent urogenital infections [11,12,13]. E. coli is commonly found in faeces, and its presence in pharmaceutical preparations is considered a direct indicator of faecal contamination in those products. Products contaminated with E. coli are discarded due to the possible presence of enteric pathogens, which include some pathogenic strains of E. coli, such as 0157: H7 [14].
Globally, antimicrobial resistance is a growing concern that poses a threat to public health. Antibiotic resistance is expected to cause increased morbidity and mortality rates that may reach as high as ten million extra deaths by 2050, in addition to increases in healthcare expenditure [15,16]. One of the major potential reservoirs for antimicrobial resistance genes has been identified in the commensal E. coli of humans and animals [17]. Moreover, these genes can transfer to E. coli or to other bacteria found in people, animals, and in the environment [18], which complicates the effort to control the spread of antibiotic resistance.
To the best of our knowledge, no work has been published on the occurrence of E. coli as an indicator of coliform contamination in water used for the reconstitution of drugs in Jordanian community pharmacies. Therefore, this work was conducted to fill this knowledge gap.
2. Materials and Methods
2.1. Sampling Plan
This microbial study was conducted in the microbiology laboratory at the Faculty of Pharmacy at Al-Zaytoonah University of Jordan in July 2020. Sampling of water used for drug compounding was conducted on water samples collected from 50 randomly selected community pharmacies representing all governorates of Jordan. In Jordan, water used for drug compounding in the community pharmacies is directly taken from water droppers, with the source of that water being either bottled water, tap water, boiled tap water, or water from coolers. Therefore, water was sampled from the droppers that are continuously refilled without prior or subsequent sterilization or treatment.
2.2. Collection of Water Samples
Two water samples were collected aseptically from the sampled pharmacies in a sterile bottle with a cap, each of 100 mL capacity. The bottles were labelled with a code number corresponding to the pharmacy and the date of collection and were stored immediately in an icebox for transportation. The volume of each water sample was around 100 mL. Samples were collected between 10 a.m. and 2 p.m. [19] and were immediately transported to the microbiology laboratory. Microbiological examination was started promptly to avoid unpredictable changes (preferably within 2 hrs of arrival).
2.3. Bacterial Propagation and Identification
The sampled water was tested for microbial contamination using the total plate count (TPC) and total coliform count (TCC) methods. Four different types of media were used for the propagation, isolation and detection of Enterobacteriaceae. These media included: plate count agar medium (Biolab, Budapest, Hungary), for enumeration of total microorganisms in water samples, m-Endo Agar Les medium (Liofilchem, Italy) for the detection and enumeration of coliforms in water samples, and MacConkey agar and Eosin Methylene Blue medium (Biolab, Budapest, Hungary) for the isolation, enumeration and differentiation of E. coli. The above media were prepared according to manufacturers’ instructions. Sterility of the four prepared media types was achieved by autoclaving at 121 °C for 15 min.
2.4. Detection and Enumeration of Total Viable Microorganisms, Total Coliforms and E. coli by Membrane Filtration
The recommended techniques used to determine the faecal contamination of water by E. coli are the multiple tube fermentation method and the membrane filtration method [20], the latter of which was implemented in this study. For all samples, two volumes of about 100 mL were filtered through 0.45 μm pore-sized nylon membrane filters (Agela Technologies, China) using a sterile filtration unit (Thermo Scientific Nalgene Filtration Products, Nalgene, Mexico) and vacuum pump (Vp 280′2831). The membranes were aseptically removed using sterile forceps, rotated upside down and placed on plates of plate count agar and m-Endo Agar Les, ensuring that no air bubbles were trapped. The plates were then incubated at 35 ± 0.5 °C for 22 to 24 h. Colonies of E. coli exhibited a distinctive pink-to-dark red colour with a metallic green sheen in the EMB agar.
Coliform density was reported as the number of colonies per 100 mL of sample. Samples of sterile distilled water were used as negative controls. One strain of E. coli (ATCC 8739) was mixed with 100 mL sterile distilled water, filtered as above, and used to produce 20–80 coliform colonies per filter. This sample was used as a positive control to enumerate coliform density according to the following equation:
If growth covered the entire filtration area of the membrane, or a portion of it without discrete colonies, the results were then reported as “Confluent Growth With or Without Coliforms.” If the total number of colonies (coliforms plus non-coliforms) exceeded 200 per membrane, or the colonies were too indistinct for accurate counting, the results were reported as “Too Numerous to Count (TNTC)” [20].
2.5. Identification of E. coli
E. coli isolates were identified based on colonial morphology and with gram staining. Under light microscopy, E. coli cells are typically gram negative and are short and rod-like in appearance. Conventional biochemical tests, which included catalase and oxidase tests, were used for further characterization of the bacteria, according to methods described elsewhere [20]. Further identification of the isolates was performed using the HB010 Hi E. coli™ Identification Kit (HiMedia Laboratories, India), which is a standardized colorimetric identification system that uses eight conventional biochemical tests and four carbohydrate utilization tests.
2.6. Identification of E. coli by PCR
2.6.1. DNA Extraction
Extraction of DNA from propagated bacterial cells was performed using the G-spin Total DNA Extraction Kit according to manufacturer’s instructions (iNtRON Biotechnology, Seoul, Korea). Briefly, a pure single colony of E. coli was transferred from a nutrient agar plate (Biolab, Budapest, Hungary) into 5 mL nutrient broth media (Biolab, Budapest, Hungary) and then incubated overnight at 37 ℃ until it obtained an OD600 value of 0.8~1.0 on a spectrophotometer (UV-M51, BelEngineering, Monza, Italy). Then, 1–2 mL of the bacteria suspension was transferred to a 2 mL tube and centrifuged for 1 min at 13,000 rpm. Next, 200 μL Buffer CL, 20 μL Proteinase K and 5 μL RNase A solution were added to the sample tube and mixed using a vortex. The lysate was then incubated at 56 ℃ for 10–30 min in a water bath. Then, 200 μL of Buffer BL was added to the sample tube, mixed thoroughly, and incubated at 70 °C for 5 min. Finally, the suspension was centrifuged at 13,000 rpm for 5 min and the supernatant was collected and used as a DNA template for PCR.
2.6.2. PCR for Isolates Identification
PCR was performed to confirm the identity of the propagated isolates by detecting the β-galactosidase gene using the E. coli-specific lacZ3 primers (Table 1), as previously described [21]. All PCR reactions were done using a MultiGene Conventional PCR machine (Labnet, Edison, New Jersey, USA). The gene was amplified using 5 μL of PCR Master Mix 5× (FIREPol® Master Mix Ready to Load, Solis BioDyne, Estonia). The volume was made up to 25 μL using nuclease free water (Integrated DNA Technologies, Coralville, IA, USA). Amplification was done by initial denaturation at 95 °C for 3 min, followed by denaturation at 95 °C for 30 s; annealing temperature of primers was 58 °C for 30 s and extension at 72 °C for 1 min. The final extension was conducted at 72 °C for 10 min and the total reaction was performed for 37 cycles. The amplified PCR products were analysed by electrophoresis in 1.5% agarose gel at 100 v (NanoPAC Power supply, Cleaver Scientific, Rugby, UK) for 45 min, stained with ethidium bromide, and finally, visualized with UV Transilluminator.

Table 1.
The targeted gene, primer sequence and product size.
2.7. Antibiotic Susceptibility Test
To measure the antimicrobial drug susceptibility of all E. coli isolates, the Kirby–Bauer disk diffusion method was used according to the Clinical and Laboratory Standards Institute guidelines [22]. Susceptibility patterns of the isolates were determined against doxycycline (DO, 30 mcg), ceftazidime (CAZ, 30 mcg), gentamicin (CN, 10 mcg), ciprofloxacin (CIP5, 5 mcg), azithromycin (AZM, 15 mcg), amoxicillin/clavulanic acid (AMC, 30 mcg), levofloxacin (LVX5, 5 mcg) and tetracycline (TE, 30 mcg). All standard antibiotic discs were obtained from Oxoid (Basingstoke, Hampshire, UK). The results of antimicrobial testing were recorded as sensitive, intermediate sensitive and resistant according to zone diameter interpretative standards [22]. E. coli ATCC 8739 strain was included as a positive control.
2.8. Statistics
Data were transferred to Microsoft ExcelTM to generate statistical summaries. Statistically significant difference between the sources of contaminated water samples was based on p-values of >0.05, inferred from the Z-score for the differences between the means or proportions of infections.
3. Results
3.1. Detection and Enumeration of Total Coliforms and E. coli
The bacteria were cultured in plate count agar to determine total microbial count; m-Endo Agar LES was used, which encourages E. coli growth as red-coloured colonies with a metallic green sheen (Figure 1).

Figure 1.
Colonies of Escherichia coli grown on m-Endo Agar LES, producing a dark-red metallic sheen.
Coliform contamination was detected in 44 of the 50 water samples (88%) collected from community pharmacies in Jordan (Table 2). Only 12% (6 of 50) of water samples were free of coliform microbial contamination. Total microbial colonies were too numerous to count (TNTC) in most of the sampled water (58%; 29 of 50), while 30% of the samples (15 of 50) had countable total microbial colonies with a range of 5–30 CFU/100 mL. The presence of E. coli was confirmed in 13 samples of the collected 50 samples (26%; 95% confidence interval: 14.6–40.4%). The total coliform count in the water samples contaminated with E. coli had a range of 1–10 CFU/100 mL, except for one sample that produced TNTC.

Table 2.
Total microbial and total coliform counts found in water samples used for reconstitution of drugs in Jordanian community pharmacies.
The majority of the community pharmacies filled the droppers with water from water coolers (n = 31), followed by bottled water (n = 10), boiled tap water (n = 8), and then tap water (n = 1, Table 3). With the exception of the latter group, statistical analysis showed no significant difference in the proportion of contaminated water samples based on its source. Nonetheless, the results showed a tendency for higher proportions of contamination in droppers filled from boiled tap water (37.5%; SE: 17.1), followed by droppers filled from cooler water (25.8%; SE: 7.9), then droppers filled from bottled water (20%; SE: 12.7).

Table 3.
Summary statistics on source of water in droppers used for pharmaceutical compounding in Jordanian community pharmacies. The table shows number of samples contaminated with Escherichia coli, mean (%) and standard error (SE). Statistical significance between source groups was inferred using Z-score and the p-values between the groups was reported.
3.2. Identification of E. coli
In this study, 13 isolates of coliform bacteria were identified as E. coli (26%). The identification of E. coli was based on gram staining, biochemical testing and culturing of each isolate on MacConkey agar and Eosin Methylene Blue agar (Figure 2), and PCR.
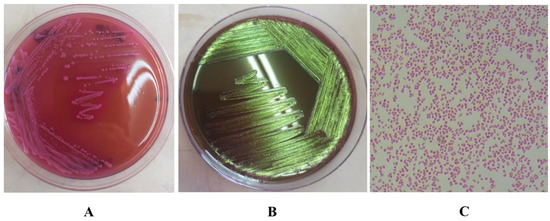
Figure 2.
(A). The growth of Escherichia coli on MacConkey agar show colonies that are dry, donut shaped and dark pink in colour, surrounded with dark pink area of precipitated bile salts. (B). Growth of E. coli on EMB agar shows the characteristic green metallic sheen. (C). Morphology of E. coli cells by microscopy after gram staining, showing gram negative, short rods.
Biochemical tests used for the characterization of E.coli isolates from the water samples included: Methyl red (positive), Voges Proskauer’s (negative), Citrate utilization (negative), Indole (positive), Glucuronidase (positive), Nitrate reduction (positive), O-nitrophenyl-beta-D-galactopyranoside (ONPG) (positive), Lysine utilization (positive), Lactose (positive), Glucose (positive), Sucrose (the bacteria were variable for sucrose testing from 11%–89%), Sorbitol (positive), Catalase (positive) and Oxidase (negative). The positive control group was E. coli ATCC 8739. The identification of E. coli was confirmed by PCR amplification of the β-galactosidase gene, which gave a fragment of 243 bp.
3.3. Antibiotic Susceptibility
The 13 isolates of E. coli were analysed for antibiotic susceptibility. The results of antimicrobial susceptibility testing showed that all E. coli isolates (13 of 13; 100%) were sensitive to gentamycin, ciprofloxacin and levofloxacin. The susceptibility of the isolates to ceftazidime, doxycycline, tetracycline, azithromycin and amoxicillin/clavulanic acid were 92%, 61%, 46%, 23% and 15%, respectively. Out of 13 E. coli isolates, 10 (77%), 6 (46%) and 1 (8%) were resistant to azithromycin, amoxicillin/clavulanic acid and ceftazidime, respectively (Figure 3).
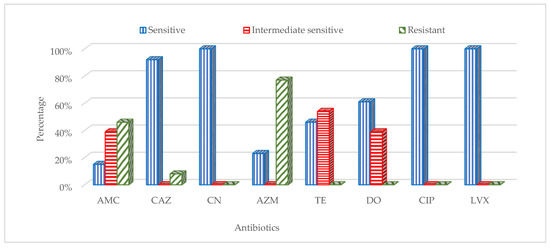
Figure 3.
Resistance patterns of Escherichia coli isolates against the following antibiotics: doxycycline (DO, 30 mcg), ceftazidime (CAZ, 30 mcg), gentamicin (CN, 10 mcg), ciprofloxacin (CIP5, 5 mcg), azithromycin (AZM, 15 mcg), amoxicillin/clavulanic acid (AMC, 30 mcg), levofloxacin (LVX5, 5 mcg) and tetracycline (TE, 30 mcg).
4. Discussion
This study was conducted to investigate the presence of microbial contamination of coliform bacteria, particularly E. coli, in water used for drug compounding in community pharmacies in Jordan. Unfortunately, in Jordan there are no official guidelines or published resolutions establishing any standards for the quality of water or practices for drug reconstitution in local pharmacies. Additionally, there is a lack of information on the prevalence of pathogenic E. coli in water used for pharmaceutical compounding in community pharmacies in Jordan. To the best of our knowledge, we believe that this study is the first one conducted on screening water used in drug compounding in community pharmacies in Jordan.
The results of this study clearly indicate the presence of microbial contamination in the majority of water samples collected from community pharmacies (88%). The total microbial count was too numerous to count in most of the contaminated samples (58%) regardless of the source water, while only 12% of the water samples were free of microbial contamination. Such high levels of microbial growth may indicate accumulation of contaminations, mainly in the droppers themselves. However, the presence of contamination in the main water sources has not been tested, and therefore cannot be ruled out. A similar study was conducted in Brazil [23], where there are national regulations ensuring the quality of water used for drug compounding with quality control testing every month. That study included testing for coliform bacteria and E. coli in 744 samples from 30 pharmacies collected over a 4 year period and showed lack of evidence of coliform bacteria in all of the tested samples. Unfortunately, no other similar studies could be found in the literature. Nonetheless, the results of that study clearly indicate the potential effect of implementing regulations to ensure the quality of water used in reconstitution of medications, especially if such activities are practiced in hospitals or in reconstituting drugs for parenteral use.
In our study, most of the water samples from droppers filled with bottled water were contaminated with coliform bacteria. The chance of isolating coliform bacteria from bottled water was once tested in Tanzania, with 3.6% of the 131 bottled water samples testing positive for coliform bacteria [24]. None of the visited community pharmacies in our study had procedures for decontamination or instruments for sterilizing water droppers. Accordingly, and to overcome the chance of using contaminated bottled water, we recommend discarding used water droppers, or alternatively, using disposable bottled water from companies with known quality control status.
This study has shown that coliform bacteria were present in all water droppers sampled, regardless of the source of water. This may be directly related to the hygiene practices of the workers in the pharmacies rather than the level of contamination of the water source. Alternatively, the continuous filling of the same droppers without periodic decontamination or sterilization of the droppers may have resulted in accumulation of bacterial contaminants over time.
Due to the small size of sampled water from droppers filled with tap water, no conclusions could explain the lack of isolating coliform bacteria from such droppers. Nonetheless, the bacteriological quality of the tap water could be explained by the boiling step or the effectiveness of disinfection processes used for treating water before distribution.
In this study, 26% of the water samples collected from community pharmacies had E. coli, which poses a threat to the quality of compounded pharmaceutical products. The possible existence of coliform bacteria in treated drinking water may indicate a failure of treatment. The presence of antibiotic-resistant E. coli in water intended for drug compounding is a serious health risk to all consumers and particularly to immunocompromised individuals [25]. The risk of acquiring infections with such multidrug-resistant strains is further complicated by the fact that, in Jordan, several other sources of such strains can be obtained from surface water, drinking water and even from clinics [26,27,28].
The circulation of multidrug-resistant bacteria can be supported by the fact that the currently isolated E. coli were sensitive to gentamycin, ciprofloxacin and levofloxacin, while other isolates were resistant to azithromycin, amoxicillin/clavulanic acid and ceftazidime. This pattern is reflected in local clinical studies from environmental samples collected from hospitals and homes [28], as well as from patients in Jordan [29], which exhibited similar patterns of multidrug-resistant E. coli. The identification of similar resistance patterns in E. coli from these sites and from the sites sampled in this study might suggest the contribution of sub-optimal practices in local pharmacies in the dissemination of multidrug-resistant bacteria to human patients.
5. Conclusions
This study provides basic information about the presence of E. coli in water used for reconstitution of drugs in Jordanian community pharmacies. The current isolation of multidrug-resistant bacteria in most sampled water used for drug compounding is an alarming situation that needs special attention by pharmacists and competent authorities. Stricter measures should be implemented by the concerned regulatory authorities, such as the Jordan Food and Drug Administration and the Pharmacists Syndicate, to ensure high-quality compounding practices in the pharmacies and/or better conformation to the regulations of international pharmacopoeias. Regular inspection of water quality is expected to reduce the risk of microbial contamination in compounded products, particularly the multidrug-resistant strains of E. coli and other index microorganisms.
Author Contributions
Conceptualization, M.N.S.A.-S. and M.K.A.-S.; methodology, R.A.M. and D.H.A.; software, M.N.S.A.-S.; validation, M.K.A.-S., R.A.M. and D.H.A.; formal analysis, M.N.S.A.-S.; investigation, R.A.M.; resources, M.K.A.-S.; data curation, M.K.A.-S.; writing—original draft preparation, M.K.A.-S.; writing—review and editing, M.N.S.A.-S.; visualization, M.K.A.-S.; supervision, M.K.A.-S.; project administration, M.K.A.-S.; funding acquisition, M.K.A.-S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Deanship of scientific research and innovation of Al-Zaytoonah University of Jordan, grant number 09/18/2018-2019.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors want to thank Aurum Biotech, Amman, Jordan, for their valuable technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kaper, J.; Nataro, J.; Mobley, H. Nature reviews. Microbiology. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- WHO. Who Drug Information; WHO: Geneva, Switzerland, 2021; pp. 606–853.
- Mistry, H.; Patel, T.; Patel, A. Microbial analysis of water system for veterinary vaccine manufacturing facility. Int. J. Vet. Sci. Anim. Husb. 2018, 3, 24–29. [Google Scholar]
- European Pharmacopoeia. EDQM.226; Council of Europe: Strasbourg, France, 2011. [Google Scholar]
- Tyagi, S.; Sharma, B. Water Quality Assessment in Terms of Water Quality Index. Am. J. Water Resour. 2013, 1, 34–38. [Google Scholar] [CrossRef]
- WHO. Guidelines for Drinking-Water Quality; WHO: Geneva, Switzerland, 2008; p. 668.
- Aziz, H.M. The investigated microbiological (coliform) among different drinking water sources in Kalar city. IOSR-JESTFT 2016, 10, 56–58. [Google Scholar] [CrossRef]
- Mishra, M.; Patel, A.K.; Behera, N. Prevalence of multidrug resistant E. coli in the river Mahanadi of Sambalpur. Curr. Res. Microbiol. Biotechnol. 2013, 1, 239–244. [Google Scholar] [CrossRef]
- Shield, K.F.; Bain, R.E.; Cronk, R. Association of supply type with fecal contamination of source water and household stored drinking water in developing countries: A bivariate meta-analysis. Environ. Health Perspect. 2015, 123, 1222–1231. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Sharma, S.; Sharma, P. Escherichia coli biofilm: Development and therapeutic strategies. J. Appl. Microbiol. 2016, 121, 309–319. [Google Scholar] [CrossRef]
- Tarawneh, O.; Alwahsh, W.; Abul-Futouh, H.; Al-Samad, L.A.; Hamadneh, L.; Abu Mahfouz, H.; Fadhil Abed, A. Determination of Antimicrobial and Antibiofilm Activity of Combined LVX and AMP Impregnated in p (HEMA) Hydrogel. Appl. Sci. 2021, 11, 8345. [Google Scholar] [CrossRef]
- Korajkic, A.; McMinn, B.R.; Harwood, V.J. Relationships between microbial indicators and pathogens in recreational water settings. Int. J. Environ. Res. Public Health 2018, 15, 2842. [Google Scholar] [CrossRef]
- de Kraker, M.E.; Stewardson, A.J.; Harbarth, S. Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef]
- Roberts, R.R.; Hota, B.; Ahmad, I.; Scott, R.D.; Foster, S.D.; Abbasi, F.; Schabowski, S.; Kampe, L.M.; Ciavarella, G.G.; Supino, M.; et al. Hospital and societal costs of antimicrobial-resistant infections in a Chicago teaching hospital: Implications for antibiotic stewardship. Arch. Clin. Infect. Dis. 2009, 49, 1175–1184. [Google Scholar] [CrossRef]
- Kinge, C.N.W.; Ateba, C.N.; Kawadza, D.T. Antibiotic resistance profiles of Escherichia coli isolated from different water sources in the Mmabatho locality, North-West Province, South Africa. S. Afr. J. Sci. 2010, 106, 44–49. [Google Scholar] [CrossRef]
- Balasubramaniam, A.; Eswaran, M.A.; Suresh, P.; Sukumar, K. Detection of tetracycline resistance determinant tet A gene and antimicrobial resistance pattern in Escherichia coli isolates recovered from healthy layer chickens. Vet. World 2014, 7, 635–638. [Google Scholar] [CrossRef]
- Cotruvo, J.A. WHO Guidelines for Drinking Water Quality: First Addendum to the Fourth Edition. J.-Am. Water Work. Assoc. 2017, 109, 44–51. [Google Scholar] [CrossRef]
- Lin, S.; Evans, R.L. An Analysis of Coliform Bacteria In The Upper Illinois Waterway. JAWRA J. Am. Water Resour. Assoc. 1974, 10, 1198–1217. [Google Scholar] [CrossRef]
- APHA. Standard Methods for the Examination of Water and Wastewater, 21st ed.; American Public Health Association, American Water Works Association, Water Environment Federation: Washington, DC, USA, 2005. [Google Scholar]
- Tille, P.M. Bailey & Scott’s Diagnostic Microbiology, 14th ed.; Elsevier: St. Louis, MO, USA, 2017. [Google Scholar]
- Molina, F.; Lopez-Acedo, E.; Tabla, R.; Roe, I.; Gomez, A.; Rebollo, J.E. Improved detection of Escherichia coli and coliform bacteria by multiplex PCR. BMC Biotechnol. 2015, 15, 48. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). M100-ED30:2020 Performance Standards for Antimicrobial Susceptibility Testing; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2020. [Google Scholar]
- Santos, N.V.; Moura, A.C.; Baptista, J.G.; Filho, A.F. Avaliação da qualidade de águas purificadas utilizadas em farmácias de manipulação (Assessing the quality of purified water used in compounding pharmacies). Rev. Ciênc. Farm. Básica Apl. 2014, 35, 419–423. [Google Scholar]
- Kassenga, G.R. The health-related microbiological quality of bottled drinking water sold in Dar es Salaam, Tanzania. J. Water Health 2007, 5, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Webster, L.F.; Thompson, B.C.; Fulton, M.H.; Chestnut, D.E.; Van Dolah, R.F.; Leight, A.K.; Scott, G.I. Identification of sources of Escherichia coli in South Carolina estuaries using antibiotic resistance analysis. J. Exp. Mar. Biol. Ecol. 2004, 298, 179–195. [Google Scholar] [CrossRef]
- Burjaq, S.Z.; Abu-Romman, S.M.; Haddad, M.A. Molecular characterization of virulence genes and antibiotic resistance among fecal Escherichia coli isolated from surface water of Wadi Shueib-Jordan. Int. Arab. J. Antimicrob. Agents 2017, 7, 1–7. [Google Scholar] [CrossRef]
- Swedan, S.; Abu Alrub, H. Antimicrobial resistance, virulence factors, and pathotypes of Escherichia coli isolated from drinking water sources in Jordan. Pathogens 2019, 8, 86. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, B.; Haddadin, R.N. Multiple drug resistance and biocide resistance in Escherichia coli environmental isolates from hospital and household settings. Antimicrob. Resist. Infect. Control 2018, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Abu Salah, M.A.; Badran, E.F.; Shehabi, A.A. High incidence of multidrug resistant Escherichia coli producing CTX-M-type ESBLs colonizing the intestine of Jordanian infants. Int. Arab. J. Antimicrob. Agents 2017, 7, 5. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).