Application of a Deep Learning Approach to Analyze Large-Scale MRI Data of the Spine
Abstract
1. Introduction
2. Materials and Methods
2.1. German National Cohort
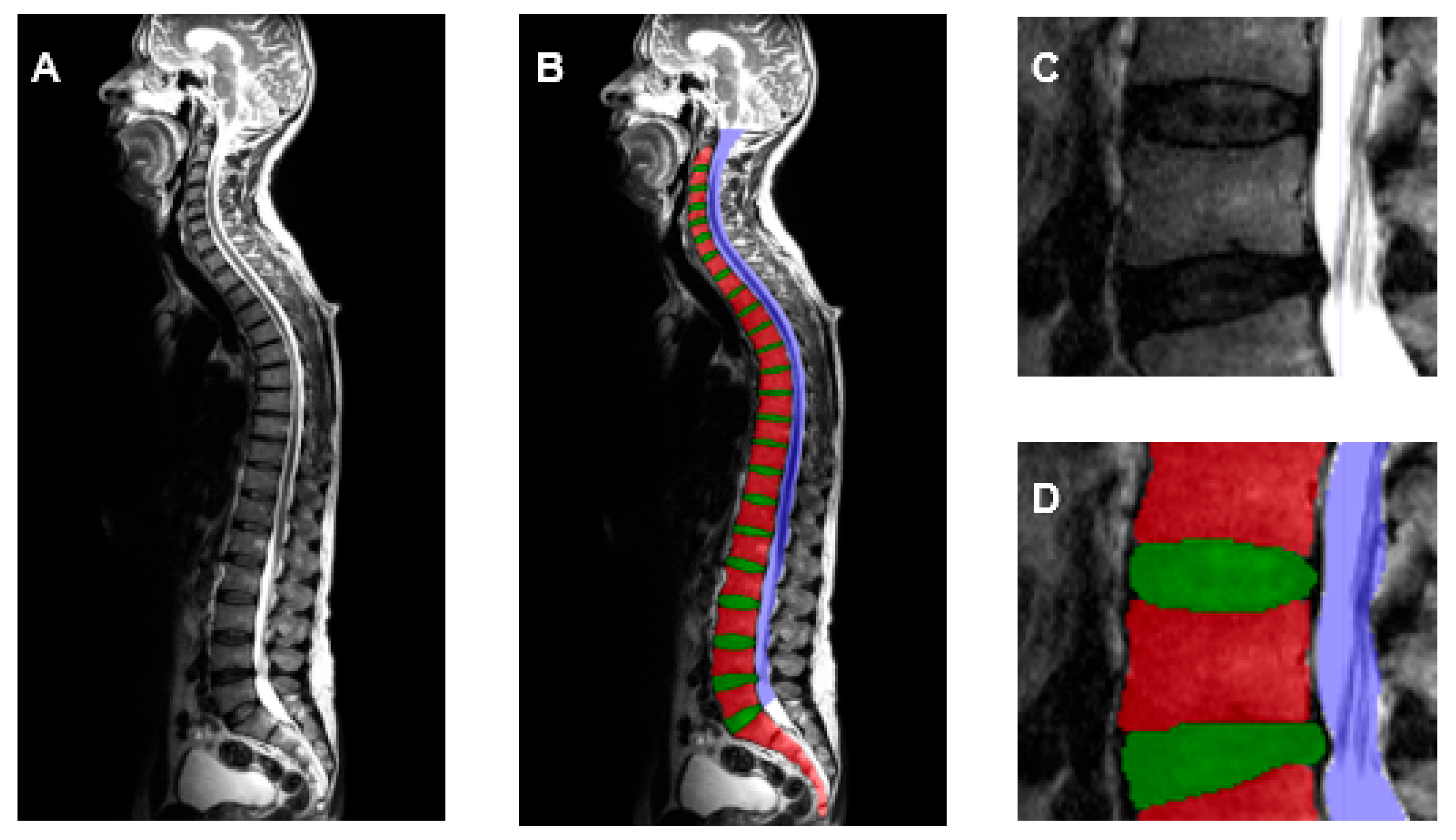
2.2. Generation of Training Dataset
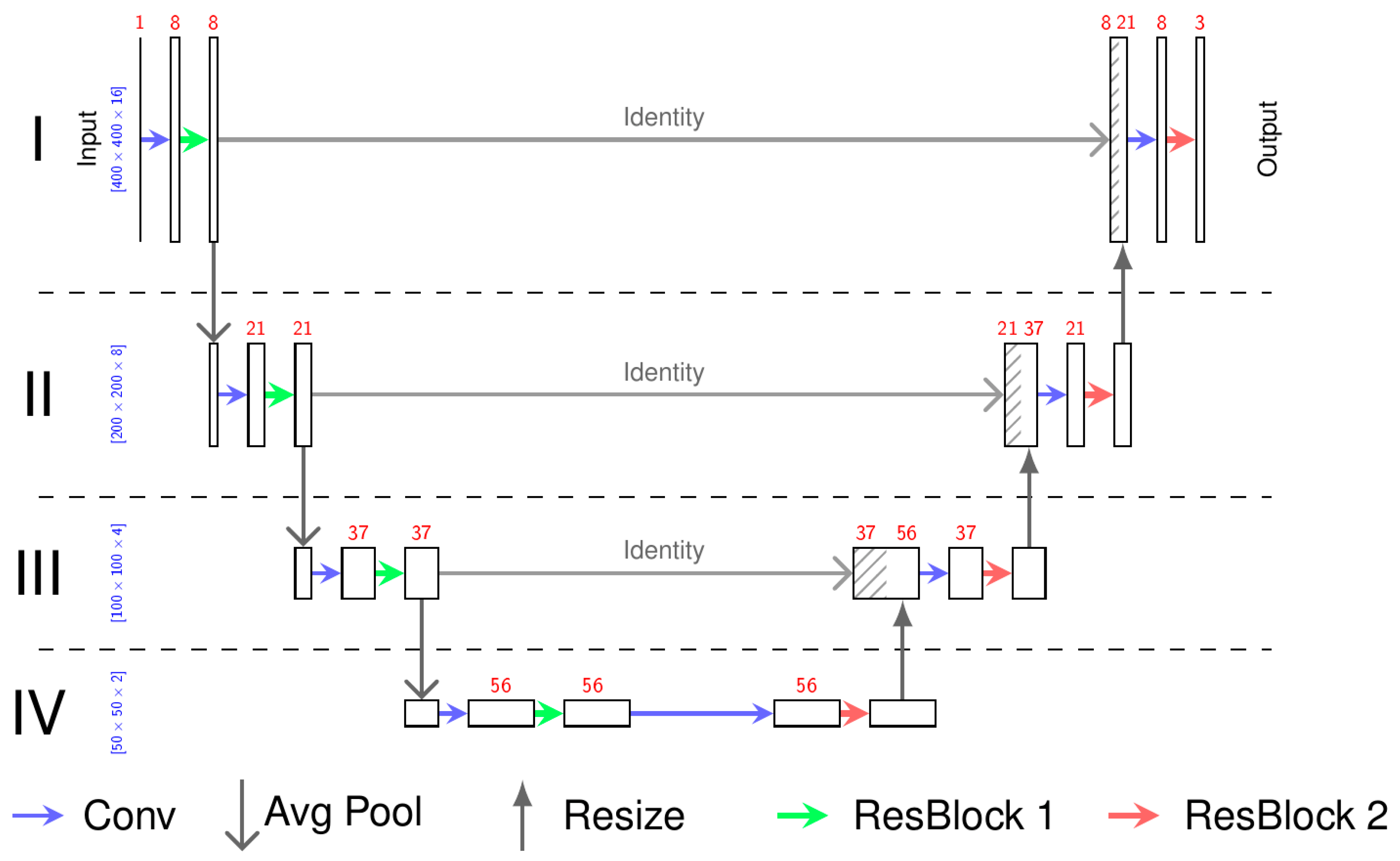
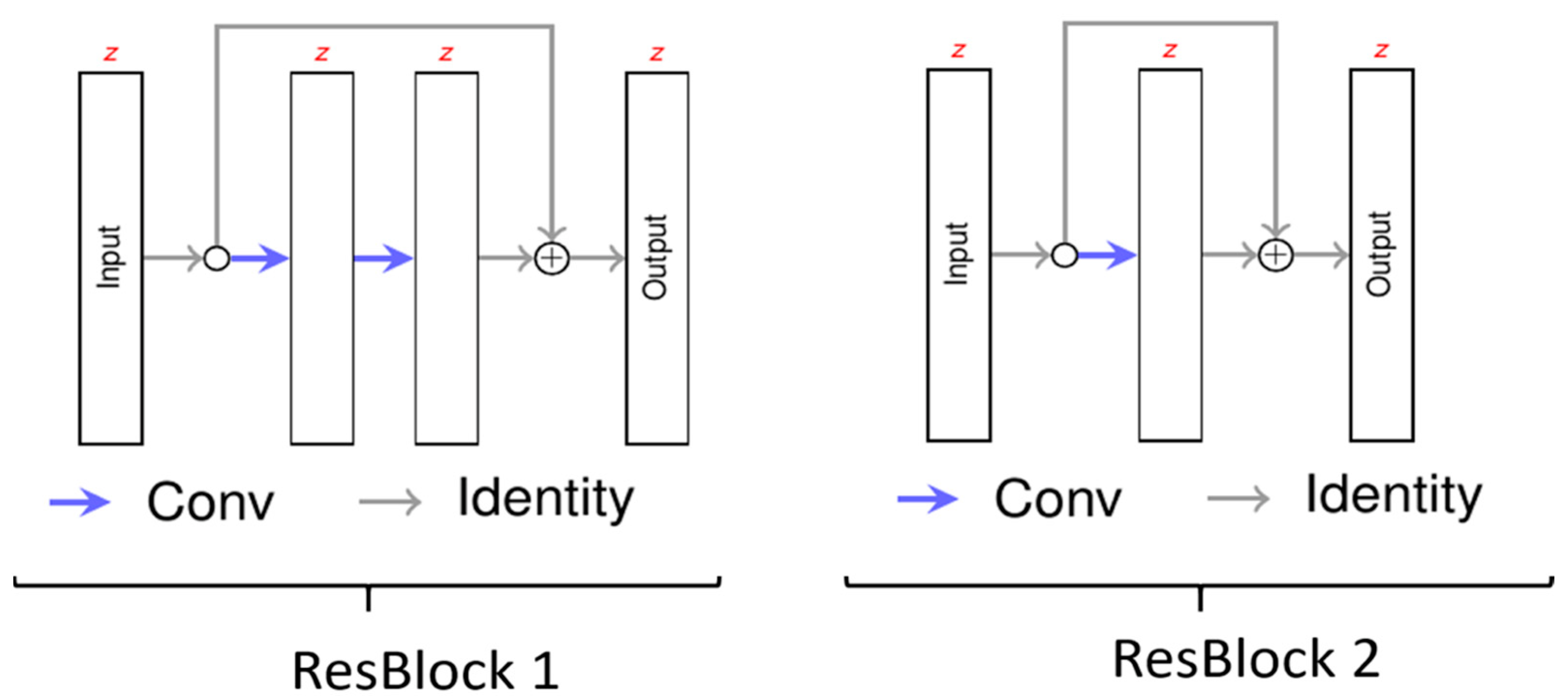
2.3. Neural Network
2.4. Training of the Deep Learning Algorithm
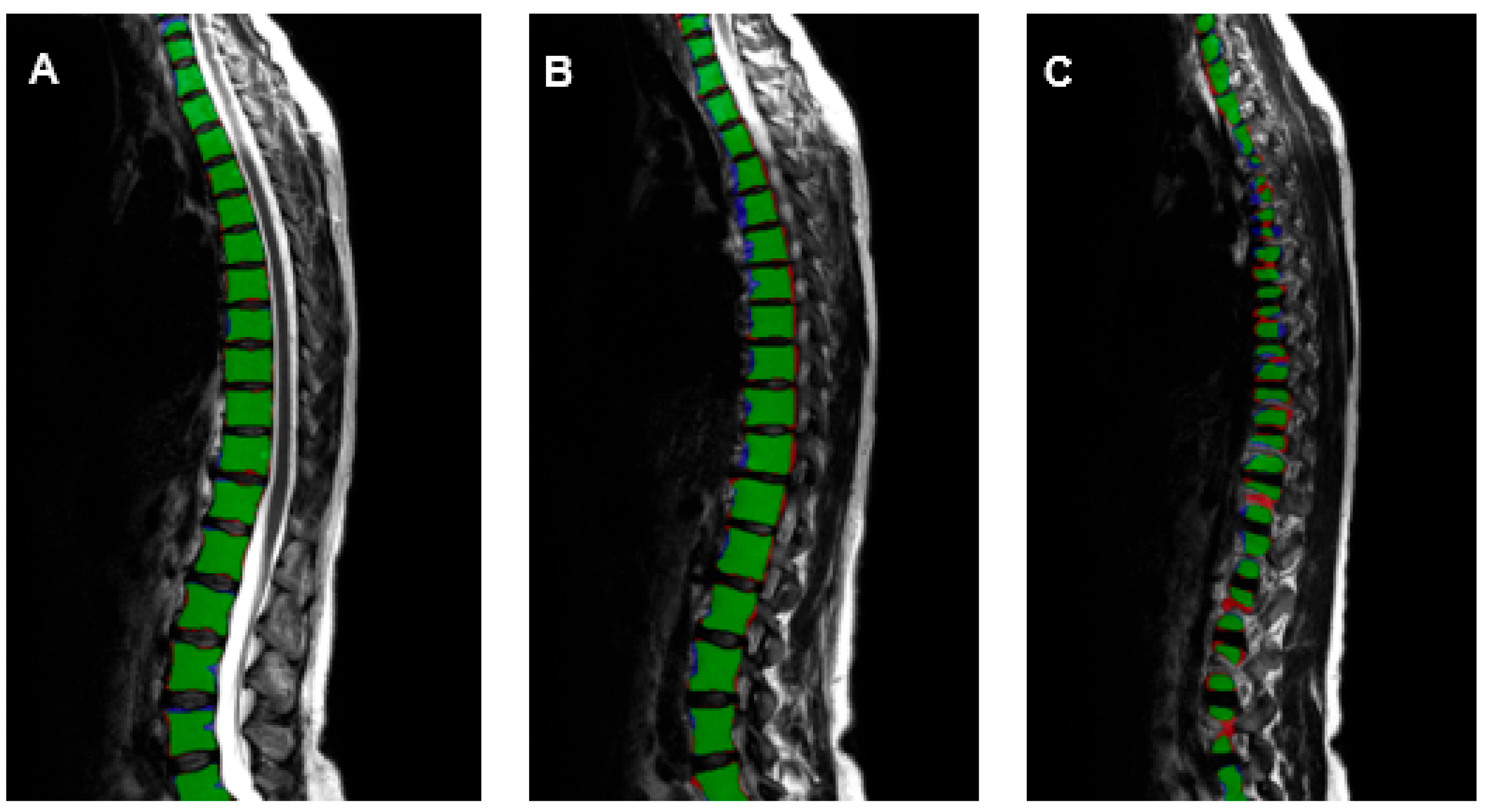
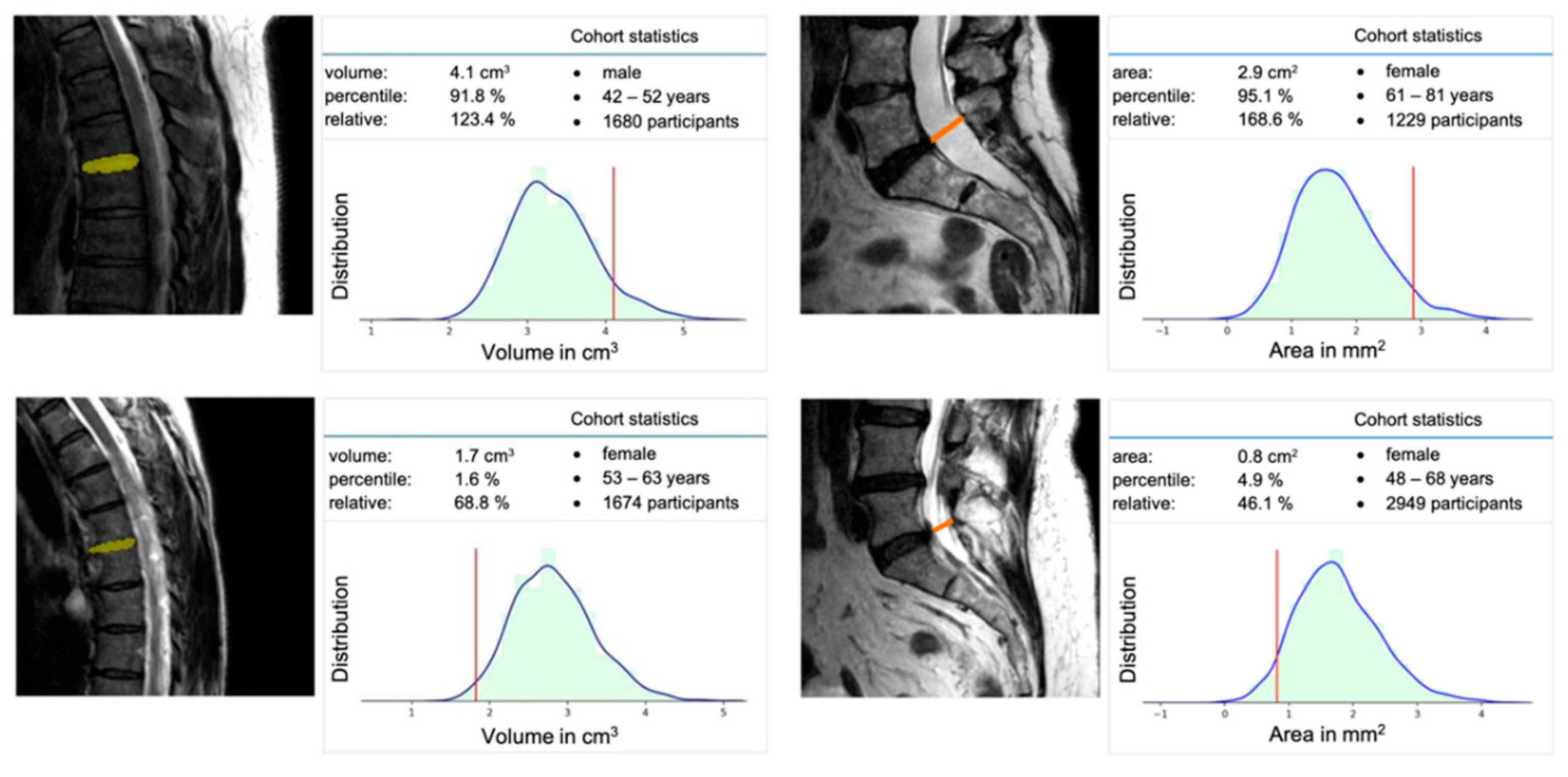
2.5. Extraction of Population-Based Data
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rezazade Mehrizi, M.H.; van Ooijen, P.; Homan, M. Applications of artificial intelligence (AI) in diagnostic radiology: A technography study. Eur. Radiol. 2021, 31, 1805–1811. [Google Scholar] [CrossRef] [PubMed]
- Bonacchi, R.; Filippi, M.; Rocca, M.A. Role of artificial intelligence in MS clinical practice. Neuroimage Clin. 2022, 35, 103065. [Google Scholar] [CrossRef] [PubMed]
- Argentiero, A.; Muscogiuri, G.; Rabbat, M.G.; Martini, C.; Soldato, N.; Basile, P.; Baggiano, A.; Mushtaq, S.; Fusini, L.; Mancini, M.E.; et al. The Applications of Artificial Intelligence in Cardiovascular Magnetic Resonance—A Comprehensive Review. J. Clin. Med. 2022, 11, 2866. [Google Scholar] [CrossRef]
- Zhou, B.; Yang, X.; Curran, W.J.; Liu, T. Artificial Intelligence in Quantitative Ultrasound Imaging: A Survey. J. Ultrasound Med. 2022, 41, 1329–1342. [Google Scholar] [CrossRef] [PubMed]
- Joseph, G.B.; McCulloch, C.E.; Sohn, J.H.; Pedoia, V.; Majumdar, S.; Link, T.M. AI MSK clinical applications: Cartilage and osteoarthritis. Skeletal Radiol. 2022, 51, 331–343. [Google Scholar] [CrossRef]
- Gandhi, D.; Garg, T.; Patel, L.; Elkassem, A.A.; Bansal, V.; Smith, A. Artificial intelligence in gastrointestinal and hepatic imaging: Past, present and future scopes. Clin. Imaging 2022, 87, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Nijiati, M.; Ma, J.; Hu, C.; Tuersun, A.; Abulizi, A.; Kelimu, A.; Zhang, D.; Li, G.; Zou, X. Artificial Intelligence Assisting the Early Detection of Active Pulmonary Tuberculosis From Chest X-rays: A Population-Based Study. Front. Mol. Biosci. 2022, 9, 874475. [Google Scholar] [CrossRef]
- Jairam, M.P.; Ha, R. A review of artificial intelligence in mammography. Clin. Imaging 2022, 88, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Smith-Bindman, R.; Kwan, M.L.; Marlow, E.C.; Theis, M.K.; Bolch, W.; Cheng, S.Y.; Bowles, E.J.A.; Duncan, J.R.; Greenlee, R.T.; Kushi, L.H.; et al. Trends in Use of Medical Imaging in US Health Care Systems and in Ontario, Canada, 2000–2016. JAMA 2019, 322, 843–856. [Google Scholar] [CrossRef]
- Bundesärztekammer 2021. Statistik der BÄK und KBV 2021. Available online: https://www.bundesaerztekammer.de/fileadmin/user_upload/BAEK/Ueber_uns/Statistik/Statistik_2021/2021_Statistik.pdf (accessed on 30 August 2022).
- Bundesärztekammer 2011. Statistik der BÄK und der KBV 2011. Available online: https://www.bundesaerztekammer.de/fileadmin/user_upload/_old-files/downloads/Stat11Abbildungsteil1.pdf (accessed on 30 August 2022).
- Hunold, P.; Bucher, A.M.; Sandstede, J.; Janka, R.; Fritz, L.B.; Regier, M.; Loose, R.; Barkhausen, J.; Mentzel, H.-J.; Zimmer, C.; et al. Positionspapier der Deutschen Röntgengesellschaft (DRG), der Deutschen Gesellschaft für Neuroradiologie (DGNR) und der Gesellschaft für Pädiatrische Radiologie (GPR) zu den fachlichen Anforderungen an Durchführung und Befundung von MRT-Untersuchungen außerhalb des Fachgebietes Radiologie. Rofo 2021, 193, 1050–1061. [Google Scholar] [CrossRef] [PubMed]
- Bamberg, F.; Kauczor, H.-U.; Weckbach, S.; Schlett, C.L.; Forsting, M.; Ladd, S.C.; Greiser, K.H.; Weber, M.-A.; Schulz-Menger, J.; Niendorf, T.; et al. Whole-Body MR Imaging in the German National Cohort: Rationale, Design, and Technical Background. Radiology 2015, 277, 206–220. [Google Scholar] [CrossRef] [PubMed]
- Sollmann, N.; Fields, A.J.; O’Neill, C.; Nardo, L.; Majumdar, S.; Chin, C.T.; Tosun, D.; Han, M.; Vu, A.T.; Ozhinsky, E.; et al. Magnetic resonance imaging of the lumbar spine-recommendations for acquisition and image evaluation from the BACPAC Spine Imaging Working Group. Pain Med. 2022, 20, 130. [Google Scholar] [CrossRef] [PubMed]
- Hosten, N.; Bülow, R.; Völzke, H.; Domin, M.; Schmidt, C.O.; Teumer, A.; Ittermann, T.; Nauck, M.; Felix, S.; Dörr, M.; et al. SHIP-MR and Radiology: 12 Years of Whole-Body Magnetic Resonance Imaging in a Single Center. Healthcare 2021, 10, 33. [Google Scholar] [CrossRef] [PubMed]
- Nell, C.; Bülow, R.; Hosten, N.; Schmidt, C.O.; Hegenscheid, K. Reference values for the cervical spinal canal and the vertebral bodies by MRI in a general population. PLoS ONE 2019, 14, e0222682. [Google Scholar] [CrossRef]
- Gaonkar, B.; Villaroman, D.; Beckett, J.; Ahn, C.; Attiah, M.; Babayan, D.; Villablanca, J.P.; Salamon, N.; Bui, A.; Macyszyn, L. Quantitative Analysis of Spinal Canal Areas in the Lumbar Spine: An Imaging Informatics and Machine Learning Study. AJNR Am. J. Neuroradiol. 2019, 40, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Huber, F.A.; Stutz, S.; Vittoria de Martini, I.; Mannil, M.; Becker, A.S.; Winklhofer, S.; Burgstaller, J.M.; Guggenberger, R. Qualitative versus quantitative lumbar spinal stenosis grading by machine learning supported texture analysis-Experience from the LSOS study cohort. Eur. J. Radiol. 2019, 114, 45–50. [Google Scholar] [CrossRef]
- Avanzo, M.; Wei, L.; Stancanello, J.; Vallières, M.; Rao, A.; Morin, O.; Mattonen, S.A.; El Naqa, I. Machine and deep learning methods for radiomics. Med. Phys. 2020, 47, e185–e202. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar]
- Kuss, O.; Becher, H.; Wienke, A.; Ittermann, T.; Ostrzinski, S.; Schipf, S.; Schmidt, C.O.; Leitzmann, M.; Pischon, T.; Krist, L.; et al. Statistical Analysis in the German National Cohort (NAKO)—Specific Aspects and General Recommendations. Eur. J. Epidemiol. 2022, 37, 429–436. [Google Scholar] [CrossRef]
- German National Cohort (GNC) Consortium. The German National Cohort: Aims, study design and organization. Eur. J. Epidemiol. 2014, 29, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Yushkevich, P.A.; Piven, J.; Hazlett, H.C.; Smith, R.G.; Ho, S.; Gee, J.C.; Gerig, G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 2006, 31, 1116–1128. [Google Scholar] [CrossRef]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016; Springer: Cham, Switzerland, 2016; Volume 9901, pp. 424–432. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. Available online: https://arxiv.org/abs/1512.03385 (accessed on 10 December 2015).
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. Available online: https://arxiv.org/abs/1412.6980 (accessed on 22 December 2014).
- Gautschi, O.P.; Stienen, M.N.; Joswig, H.; Smoll, N.R.; Schaller, K.; Corniola, M.V. The usefulness of radiological grading scales to predict pain intensity, functional impairment, and health-related quality of life after surgery for lumbar degenerative disc disease. Acta Neurochir. 2017, 159, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Baur, D.; Kroboth, K.; Heyde, C.-E.; Voelker, A. Convolutional Neural Networks in Spinal Magnetic Resonance Imaging: A Systematic Review. World Neurosurg. 2022, 166, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Ganbold, B.; Kim, K.G. Web-Based Spine Segmentation Using Deep Learning in Computed Tomography Images. Healthc. Inform. Res. 2020, 26, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Kou, Z.; Shang, L.; Zhang, Y.; Wang, D. HC-COVID. Proc. ACM Hum. Comput. Interact. 2022, 6, 1–25. [Google Scholar] [CrossRef]
- Abu-Salih, B. Domain-specific knowledge graphs: A survey. J. Netw. Comput. Appl. 2021, 185, 103076. [Google Scholar] [CrossRef]
| Training Parameter | |
|---|---|
| sample size | 400 × 400 × 16 |
| optimizer | ADAM with a decaying learning rate |
| loss | cross-entropy with focal loss (γ = 1.0) |
| samples per epoch | 1024 |
| number of epochs | 400 |
| VB | VD | SC | |
|---|---|---|---|
| Precision | 0.908 | 0.902 | 0.926 |
| Recall | 0.909 | 0.908 | 0.924 |
| Dice-score | 0.908 | 0.905 | 0.925 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Streckenbach, F.; Leifert, G.; Beyer, T.; Mesanovic, A.; Wäscher, H.; Cantré, D.; Langner, S.; Weber, M.-A.; Lindner, T. Application of a Deep Learning Approach to Analyze Large-Scale MRI Data of the Spine. Healthcare 2022, 10, 2132. https://doi.org/10.3390/healthcare10112132
Streckenbach F, Leifert G, Beyer T, Mesanovic A, Wäscher H, Cantré D, Langner S, Weber M-A, Lindner T. Application of a Deep Learning Approach to Analyze Large-Scale MRI Data of the Spine. Healthcare. 2022; 10(11):2132. https://doi.org/10.3390/healthcare10112132
Chicago/Turabian StyleStreckenbach, Felix, Gundram Leifert, Thomas Beyer, Anita Mesanovic, Hanna Wäscher, Daniel Cantré, Sönke Langner, Marc-André Weber, and Tobias Lindner. 2022. "Application of a Deep Learning Approach to Analyze Large-Scale MRI Data of the Spine" Healthcare 10, no. 11: 2132. https://doi.org/10.3390/healthcare10112132
APA StyleStreckenbach, F., Leifert, G., Beyer, T., Mesanovic, A., Wäscher, H., Cantré, D., Langner, S., Weber, M.-A., & Lindner, T. (2022). Application of a Deep Learning Approach to Analyze Large-Scale MRI Data of the Spine. Healthcare, 10(11), 2132. https://doi.org/10.3390/healthcare10112132







