Validation of a Natural Language Processing Algorithm for the Extraction of the Sleep Parameters from the Polysomnography Reports
Abstract
:1. Introduction
2. Methods
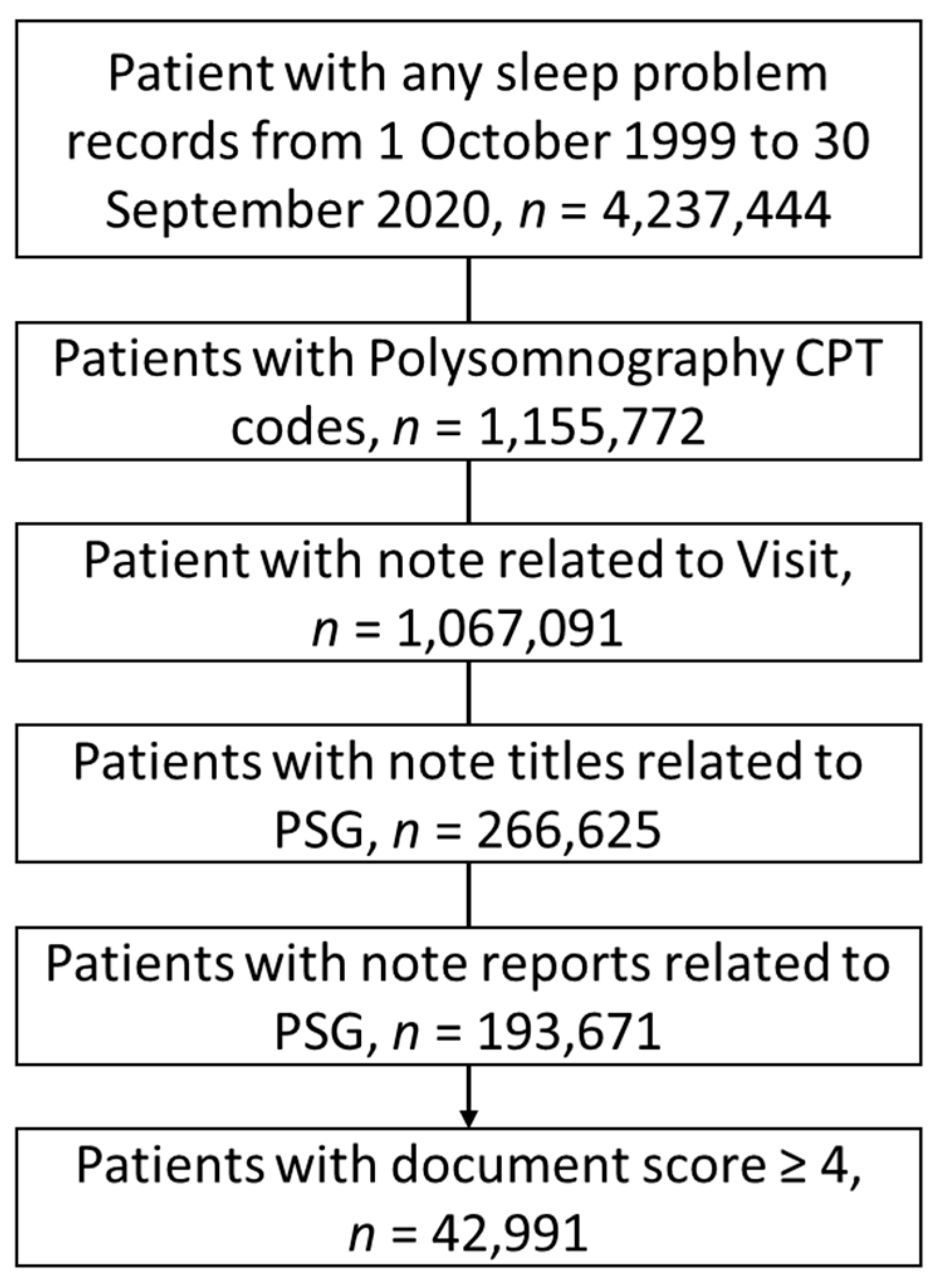
2.1. Cohort
2.2. Database
2.3. Sampling Strategy for the NLP Development and Reference Standard
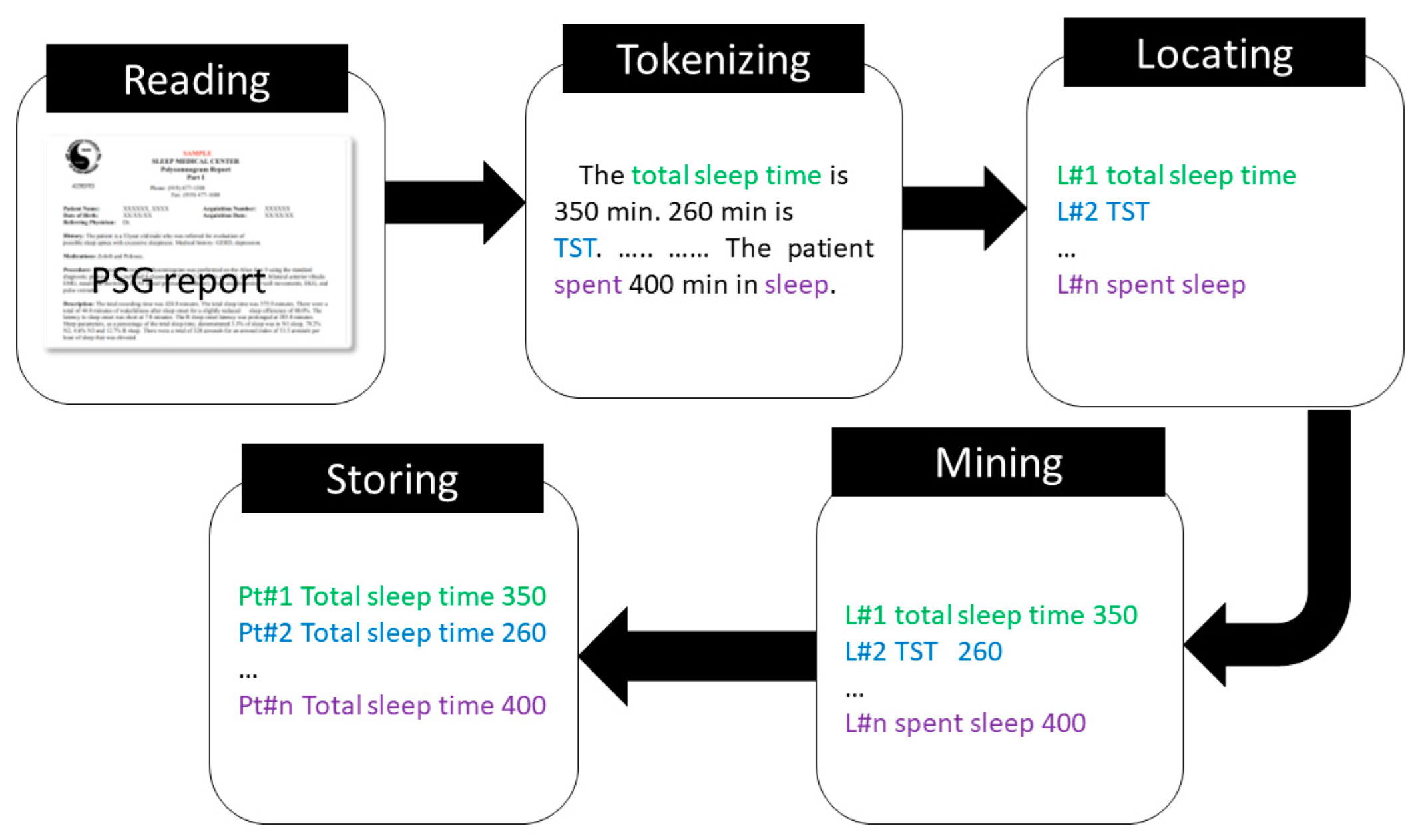
2.4. NLP Algorithm Development for the Extraction of the Sleep Parameters
- Reading: Read the PSG reports and store in the memory.
- Tokenizing: Tokenize the PSG reports.
- Locating: Identify the location of the sleep parameter of interest.
- Mining: Identify the related quantity associated with the sleep parameter of interest.
- Storing: Store the sleep parameter and associated quantity to a csv file.
2.5. Performance of the NLP Algorithm
3. Results
3.1. Reliability Analysis
3.2. Performance Analysis
4. Discussion
4.1. Methods and Results of the NLP Algorithm
4.2. Strength and Weakness
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gerstenslager, B.; Slowik, J.M. Sleep Study; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Mayo Clinic. Polysomnography (Sleep Study). 2022. Available online: https://www.mayoclinic.org/tests-procedures/polysomnography/about/pac-20394877 (accessed on 10 January 2022).
- Shrivastava, D.; Jung, S.; Saadat, M.; Sirohi, R.; Crewson, K. How to interpret the results of a sleep study. J. Community Hosp. Intern. Med. Perspect. 2014, 4, 24983. [Google Scholar] [CrossRef] [PubMed]
- Bajeh, A.O.; Abikoye, O.A.; Mojeed, H.A.; Saliku, S.A.; Oladipo, I.D.; Abdulraheem, M.; Awotunde, J.B.; Sangaiah, A.K.; Adewole, K.S. Application of computational intelligence models in IoMT big data for heart disease diagnosis in personalized health care. In Intelligent IoT Systems in Personalized Health Care; Elsevier: Amsterdam, The Netherlands, 2021; pp. 177–206. [Google Scholar]
- Luo, L.; Li, L.; Hu, J.; Wang, X.; Hou, B.; Zhang, T.; Zhao, L. A hybrid solution for extracting structured medical information from unstructured data in medical records via a double-reading/entry system. BMC Med. Inform. Decis. 2016, 16, 114. [Google Scholar] [CrossRef] [PubMed]
- Elbattah, M.; Arnaud, E.; Gignon, M.; Dequen, G. The Role of Text Analytics in Healthcare: A Review of Recent Developments and Applications. In Proceedings of the 14th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC), Vienna, Austria, 11–13 February 2021; pp. 825–832. [Google Scholar]
- Su, Y.-H.; Chao, C.-P.; Hung, L.-C.; Sung, S.-F.; Lee, P.-J. A Natural Language Processing Approach to Automated Highlighting of New Information in Clinical Notes. Appl. Sci. 2020, 10, 2824. [Google Scholar] [CrossRef]
- Murtaugh, M.A.; Gibson, B.S.; Redd, D.; Zeng-Treitler, Q. Regular expression-based learning to extract bodyweight values from clinical notes. J. Biomed. Inform. 2015, 54, 186–190. [Google Scholar] [CrossRef] [PubMed]
- Garvin, J.H.; DuVall, S.L.; South, B.R.; Bray, B.E.; Bolton, D.; Heavirland, J.; Pickard, S.; Heidenreich, P.; Shen, S.; Weir, C.; et al. Automated extraction of ejection fraction for quality measurement using regular expressions in Unstructured Information Management Architecture (UIMA) for heart failure. J. Am. Med. Inform. Assoc. 2012, 19, 859–866. [Google Scholar] [CrossRef]
- Garvin, J.H.; Kim, Y.; Temple Gobbel, G.; Matheny, M.E.; Redd, A.; Bray, B.E.; Heidenreich, P.; Bolton, D.; Heavirland, J.; Kelly, N.; et al. Automating quality measures for heart failure using natural language processing: A descriptive study in the department of veterans affairs. JMIR Med. Inform. 2018, 6, e9150. [Google Scholar] [CrossRef] [PubMed]
- Veena, G.; Hemanth, R.; Hareesh, J. Relation extraction in clinical text using NLP based regular expressions. In Proceedings of the 2019 2nd International Conference on Intelligent Computing, Instrumentation and Control Technologies (ICICICT), Kannur, India, 5–6 July 2019. [Google Scholar]
- Sada, Y.; Hou, J.; Richardson, P.; El-Serag, H.; Davila, J. Validation of case finding algorithms for hepatocellular cancer from administrative data and electronic health records using natural language processing. Med. Care 2016, 54, e9. [Google Scholar] [CrossRef] [PubMed]
- Reeves, R.M.; Christensen, L.; Brown, J.R.; Conway, M.; Levis, M.; Gobbel, G.T.; Shah, R.U.; Goodrich, C.; Ricket, I.; Minter, F.; et al. Adaptation of an NLP system to a new healthcare environment to identify social determinants of health. J. Biomed. Inform. 2021, 120, 103851. [Google Scholar] [CrossRef]
- Ehrenfeld, J.M.; Gottlieb, K.G.; Beach, L.B.; Monahan, S.E.; Fabbri, D. Development of a natural language processing algorithm to identify and evaluate transgender patients in electronic health record systems. Ethn. Dis. 2019, 29 (Suppl. 2), 441. [Google Scholar] [CrossRef]
- Gundlapalli, A.V.; South, B.R.; Phansalkar, S.; Kinney, A.Y.; Shen, S.; Delisle, S.; Perl, T.; Samore, M.H. Application of natural language processing to VA electronic health records to identify phenotypic characteristics for clinical and research purposes. Summit Transl. Bioinform. 2008, 2008, 36–40. [Google Scholar]
- Nowakowski, S.; Razjouyan, J.; Sharafkhaneh, A.; Kunik, M.; Naik, A. Polysomnographic Sleep Is Associated with Time to Develop Dementia: A Study Using 19-Year VA National EHR Data. Innov. Aging 2020, 4 (Suppl. 1), 469. [Google Scholar] [CrossRef]
- Nowakowski, S.; Razjouyan, J.; Naik, A.; Agrawal, R.; Velamuri, K.; Singh, S.; Sharafkhaneh, A. 1180 The Use of Natural Language Processing to Extract Data from Psg Sleep Study Reports Using National Vha Electronic Medical Record Data. Sleep 2020, 43, A450–A451. [Google Scholar] [CrossRef]
- Khurshid, S.; Reeder, C.; Harrington, L.X.; Singh, P.; Sarma, G.; Friedman, S.F.; Di Achille, P.; Diamant, N.; Cunningham, J.W.; Turner, A.C.; et al. Cohort design and natural language processing to reduce bias in electronic health records research. NPJ Digit. Med. 2022, 5, 47. [Google Scholar] [CrossRef] [PubMed]
- The Department of Veterans Affairs. VHA Corporate Data Warehouse (CDW). 2022. Available online: https://www.hsrd.research.va.gov/for_researchers/cdw.cfm (accessed on 12 January 2022).
- Razjouyan, J.; Helmer, D.A.; Li, A.; Naik, A.D.; Amos, C.I.; Bandi, V.; Sharafkhaneh, A. Differences in COVID-19-related testing and healthcare utilization by race and ethnicity in the veterans health administration. J. Racial Ethn. Health Disparities 2022, 9, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Kang, H. Sample size determination and power analysis using the G*Power software. J. Educ. Eval. Health Prof. 2021, 18, 1149215. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.; Lim, H.K. Reliability Study of the Items of the Alberta Infant Motor Scale (AIMS) Using Kappa Analysis. Int. J. Environ. Res. Public Health 2022, 19, 1767. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Taylor, P.A.; Haller, S.P.; Kircanski, K.; Stoddard, J.; Pine, D.S.; Leibenluft, E.; Brotman, M.A.; Cox, R.W. Intraclass correlation: Improved modeling approaches and applications for neuroimaging. Hum. Brain Mapp. 2018, 39, 1187–1206. [Google Scholar] [CrossRef]
- Python Software Foundation. 2022. Available online: https://www.python.org/psf-landing/ (accessed on 16 January 2022).
- NLTK. Natural Language Processing Toolkit. 2022. Available online: https://www.nltk.org/ (accessed on 15 January 2022).
- Li, B.; Chen, Y.W.; Chen, Y.Q. The nearest neighbor algorithm of local probability centers. IEEE Trans. Syst. Man Cybern. Part B Cybern. 2008, 38, 141–154. [Google Scholar] [CrossRef]
- Akgün, K.M.; Sigel, K.; Cheung, K.-H.; Kidwai-Khan, F.; Bryant, A.K.; Brandt, C.; Justice, A.; Crothers, K. Extracting lung function measurements to enhance phenotyping of chronic obstructive pulmonary disease (COPD) in an electronic health record using automated tools. PLoS ONE 2020, 15, e0227730. [Google Scholar] [CrossRef]
- Velupillai, S.; Dalianis, H.; Hassel, M.; Nilsson, G.H. Developing a standard for de-identifying electronic patient records written in Swedish: Precision, recall and F-measure in a manual and computerized annotation trial. Int. J. Med. Inform. 2009, 78, e19–e26. [Google Scholar] [CrossRef]
- Hripcsak, G.; Rothschild, A.S. Agreement, the F-Measure, and Reliability in Information Retrieval. J. Am. Med. Inform. Assoc. 2005, 12, 296–298. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.J.; Powers, R.; Montelione, G.T. Protein NMR recall, precision, and F-measure scores (RPF scores): Structure quality assessment measures based on information retrieval statistics. J. Am. Chem. Soc. 2005, 127, 1665–1674. [Google Scholar] [CrossRef] [PubMed]
- Loewen, A.H.; Korngut, L.; Rimmer, K.; Damji, O.; Turin, T.C.; Hanly, P.J. Limitations of split-night polysomnography for the diagnosis of nocturnal hypoventilation and titration of non-invasive positive pressure ventilation in amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. Front. Degener. 2014, 15, 494–498. [Google Scholar] [CrossRef] [PubMed]
- Devlin, J.; Chang, M.-W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Haghayegh, S.; Khoshnevis, S.; Smolensky, M.H.; Diller, K.R.; Castriotta, R.J. Accuracy of wristband Fitbit models in assessing sleep: Systematic review and meta-analysis. J. Med. Internet Res. 2019, 21, e16273. [Google Scholar] [CrossRef]
| Sleep Parameter * | N (%) | ICC (95% CI) |
|---|---|---|
| TST | 195 (97.5) | 0.83 (0.78, 0.87) |
| SE | 196 (98.0) | 0.59 (0.49, 0.68) |
| SOL | 194 (97.0) | 0.89 (0.86, 0.92) |
| WASO | 184 (92.0) | 0.51 (0.40, 0.61) |
| AHI | 192 (96.0) | 0.62 (0.52, 0.70) |
| Sleep Parameter | Accuracy | Precision | Recall | F-1 Score |
|---|---|---|---|---|
| Training | ||||
| TST | 0.91 | 0.98 | 0.93 | 0.95 |
| SOL | 0.91 | 1.0 | 0.91 | 0.96 |
| SE | 0.98 | 1.0 | 1.0 | 0.99 |
| WASO | 0.98 | 1.0 | 0.98 | 0.99 |
| AHI | 0.96 | 0.99 | 0.96 | 0.98 |
| Validation | ||||
| TST | 0.95 | 1.0 | 0.95 | 0.97 |
| SOL | 0.90 | 1.0 | 0.90 | 0.95 |
| SE | 1.0 | 1.0 | 1.0 | 1.0 |
| WASO | 0.95 | 1.0 | 0.95 | 0.97 |
| AHI | 0.95 | 1.0 | 0.95 | 0.97 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, M.; Nowakowski, S.; Agrawal, R.; Naik, A.; Sharafkhaneh, A.; Razjouyan, J. Validation of a Natural Language Processing Algorithm for the Extraction of the Sleep Parameters from the Polysomnography Reports. Healthcare 2022, 10, 1837. https://doi.org/10.3390/healthcare10101837
Rahman M, Nowakowski S, Agrawal R, Naik A, Sharafkhaneh A, Razjouyan J. Validation of a Natural Language Processing Algorithm for the Extraction of the Sleep Parameters from the Polysomnography Reports. Healthcare. 2022; 10(10):1837. https://doi.org/10.3390/healthcare10101837
Chicago/Turabian StyleRahman, Mahbubur, Sara Nowakowski, Ritwick Agrawal, Aanand Naik, Amir Sharafkhaneh, and Javad Razjouyan. 2022. "Validation of a Natural Language Processing Algorithm for the Extraction of the Sleep Parameters from the Polysomnography Reports" Healthcare 10, no. 10: 1837. https://doi.org/10.3390/healthcare10101837
APA StyleRahman, M., Nowakowski, S., Agrawal, R., Naik, A., Sharafkhaneh, A., & Razjouyan, J. (2022). Validation of a Natural Language Processing Algorithm for the Extraction of the Sleep Parameters from the Polysomnography Reports. Healthcare, 10(10), 1837. https://doi.org/10.3390/healthcare10101837







