Customized Deep Learning Classifier for Detection of Acute Lymphoblastic Leukemia Using Blood Smear Images
Abstract
:1. Introduction
2. Materials and Methods
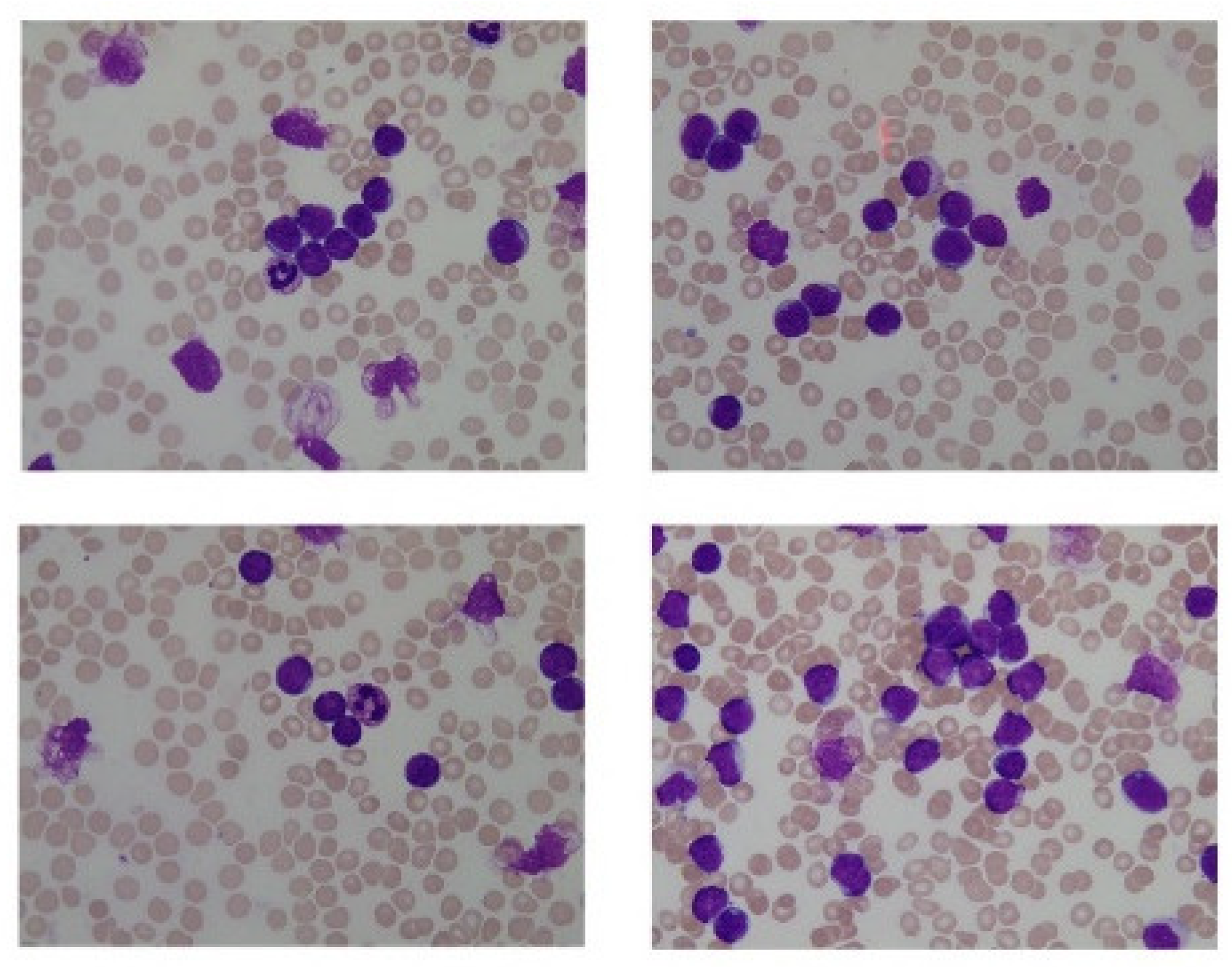
2.1. Input Data
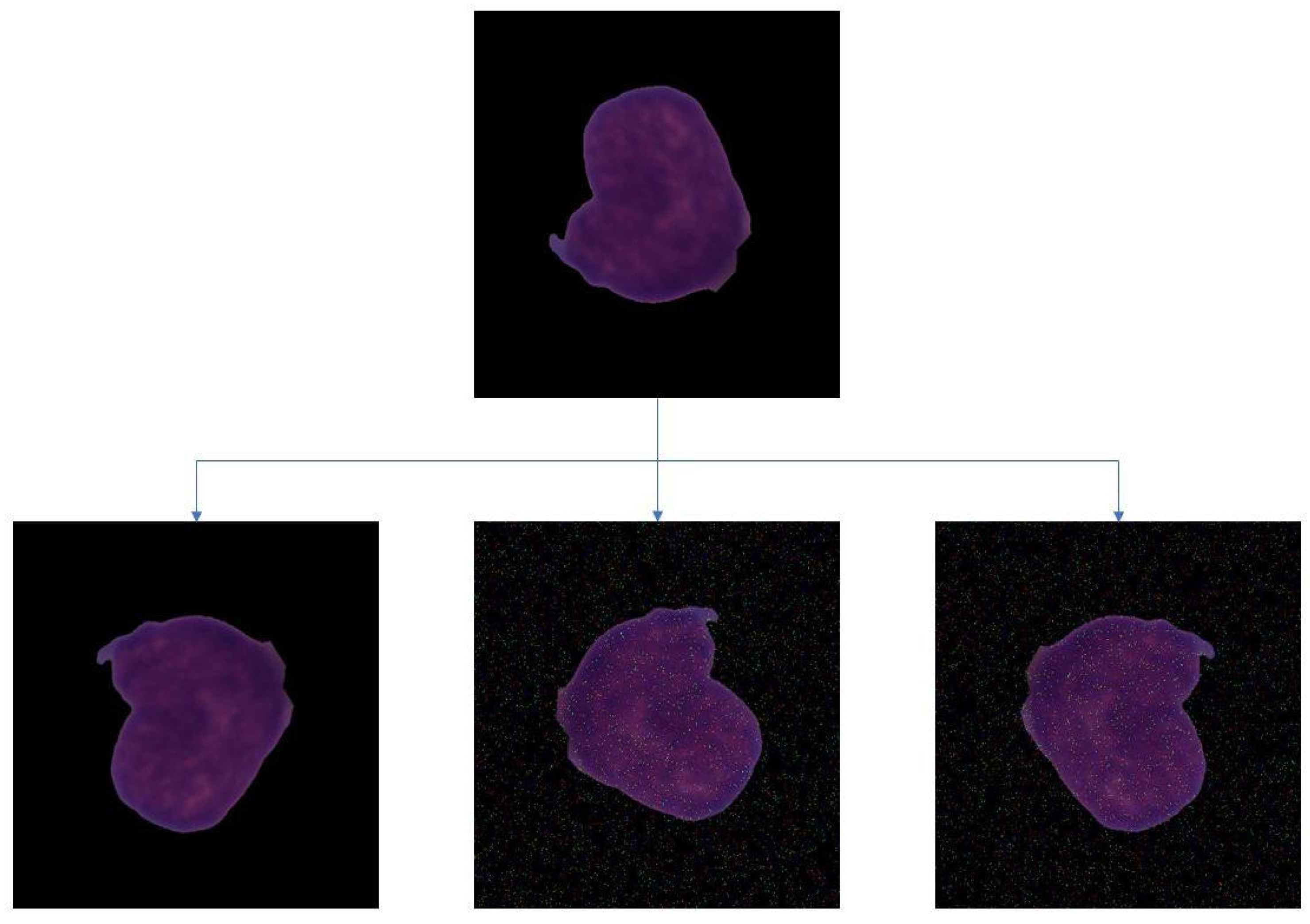
2.2. Data Augmentation
2.3. CNN
- Convolution (CONV) LayersThe convolution layers are the main building blocks of CNN. They comprise a set of independent filters, which are convolved with the input volume to compute an activation map made of neurons. The useful features from the input images are extracted by having multilayered architecture. Each of the filters can be of a different type, and they extract different features, such as vertical lines, horizontal lines, and edges. The CNN layers help in extracting features through convolution. The extracted deep features play a major role in the decision support system.
- m, n = no. of rows, no. of columns, respectively
- i, j = row index and column index
- n = convolution kernel size
- p = padding
- s = stride
- Pooling (POOL) LayerConvolutional neural networks often use pooling layers to decrease the representation size and increase the speed of computation. A pooling layer summarizes the activities of local patches of nodes in the convolutional layers. Pooling can be done in two ways: max pooling and average pooling. In max pooling, the maximum value for each patch of the feature map is stored, while others are discarded. The intuition behind using max pooling is that the maximum value indicates that it has the most impact on that patch of the image. Hence other patches can be discarded. Average pooling follows a similar procedure, except in the place of the maximum value, the average value of the patch is taken, and all other values are discarded.
- Fully Connected (FC) LayerThe FC input layer takes the output of the previous layers and turns them into a single vector that can be connected to the input layer of the next stage. This layer contains a softmax layer at the end, which predicts the correct label (0, 1). The output layer gives the final probability for each layer. The fully connected part of the CNN determines the most accurate weights by going through its backpropagation. The weights that each node receives are used to determine their respective labels. Since this project is of binary classification, the nodes will be prioritized to either 1 or 0.
- Batch NormalizationBatch normalization decreases the covariance shift, i.e., the amount by which the hidden unit values shift. If the algorithm is trained to map some input x to some output y, and if the distribution of x changes, the prediction will not work as well, and retraining might be required. Batch normalization allows the learning of each layer in an independent manner. An advantage of using batch normalization is that learning rates can be set higher as it makes sure that no activation goes high or low. Batch normalization also reduces overfitting as it has regularization effects.To improve the stability of the neural network, batch normalization normalizes the previous activation layers’ outputs. This process adds two parameters to each layer, so the normalized output gets multiplied by gamma (standard deviation) and beta (mean).Mathematically, the mini-batch mean is given by Equation (4):Mini-batch variance is shown in Equation (5):Normalization is given by Equation (6):
- DropoutDeep neural networks are likely to overfit early on any given dataset. Dropout is a method of regularization that approximates training a large number of neural networks with different architectures in parallel. While training, some dropout layers are randomly ignored. This simulates the effect of a new layer, making the neural network treat it in such a way. In effect, each update is performed with a new outlook on the layer. This method makes the network more robust as additional noise is introduced.
- Loss functionIn the model, the categorical cross-entropy loss function has been implemented. The performance of this binary classification model, i.e., whose output lies between 0 and 1, is measured by the mentioned loss function. Categorical cross-entropy compares the distribution of the predictions with the true distribution, where the probability of the class in consideration is set to 1 and the probability of the other classes is set to 0.Categorical cross-entropy is shown in Equation (7):where y-hat is the predicted expected value and y is the observed value.
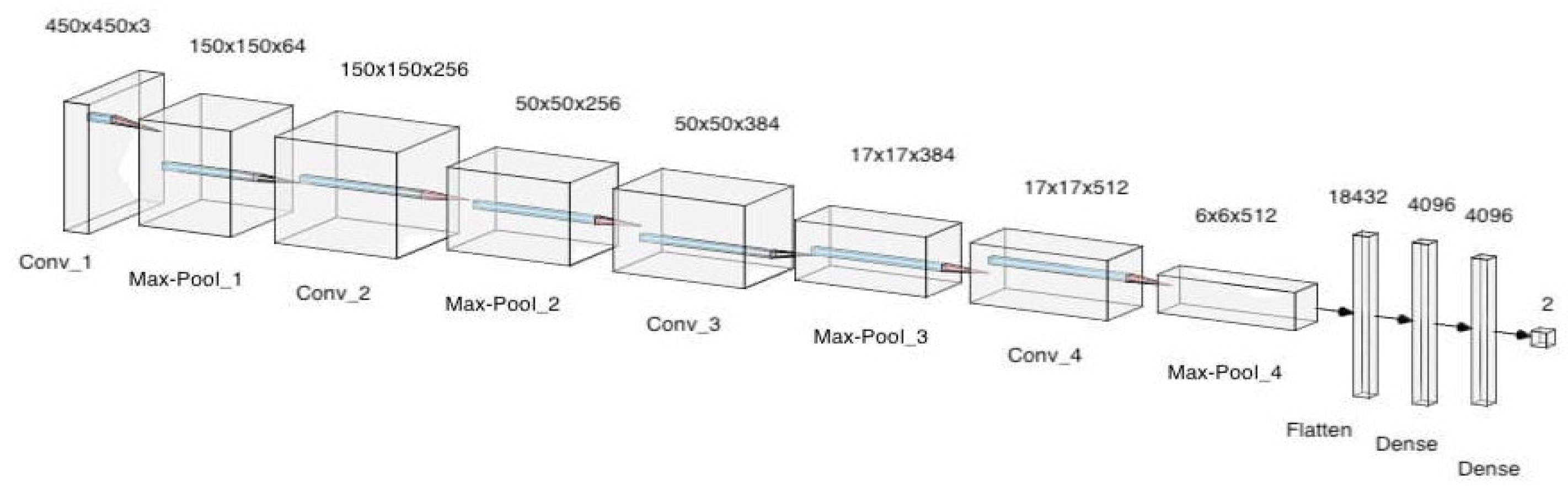
- OptimizerOptimizers are algorithms used to change the attributes of the neural network such as learning rates and weights to reduce the loss. In the model, adaptive movement estimation (Adam) has been incorporated. Adam is a combination of the root mean square propagation (RMSProp) and adaptive gradient algorithm (AdaGrad). The proposed convolution neural network architecture is shown in Figure 2, which makes use of pooling layers, fully connected layers, convolutional layers, dropout and batch normalization. Features were automatically extracted from the input images by the CNN. Feature extraction is then performed by the convolutional layers and the pooling layers. Four convolutional layers, four max-pooling layers, and 3 fully connected layers were utilized. Batch normalization and Dropout were applied to account for overfitting, vanishing, and exploding of gradients. This model consisted of 95,099,266 parameters in total. The architecture for the designed model can be seen in Figure 9. A more detailed description of the model is described in Table 2.
3. Results and Discussion
3.1. Performance Metrics
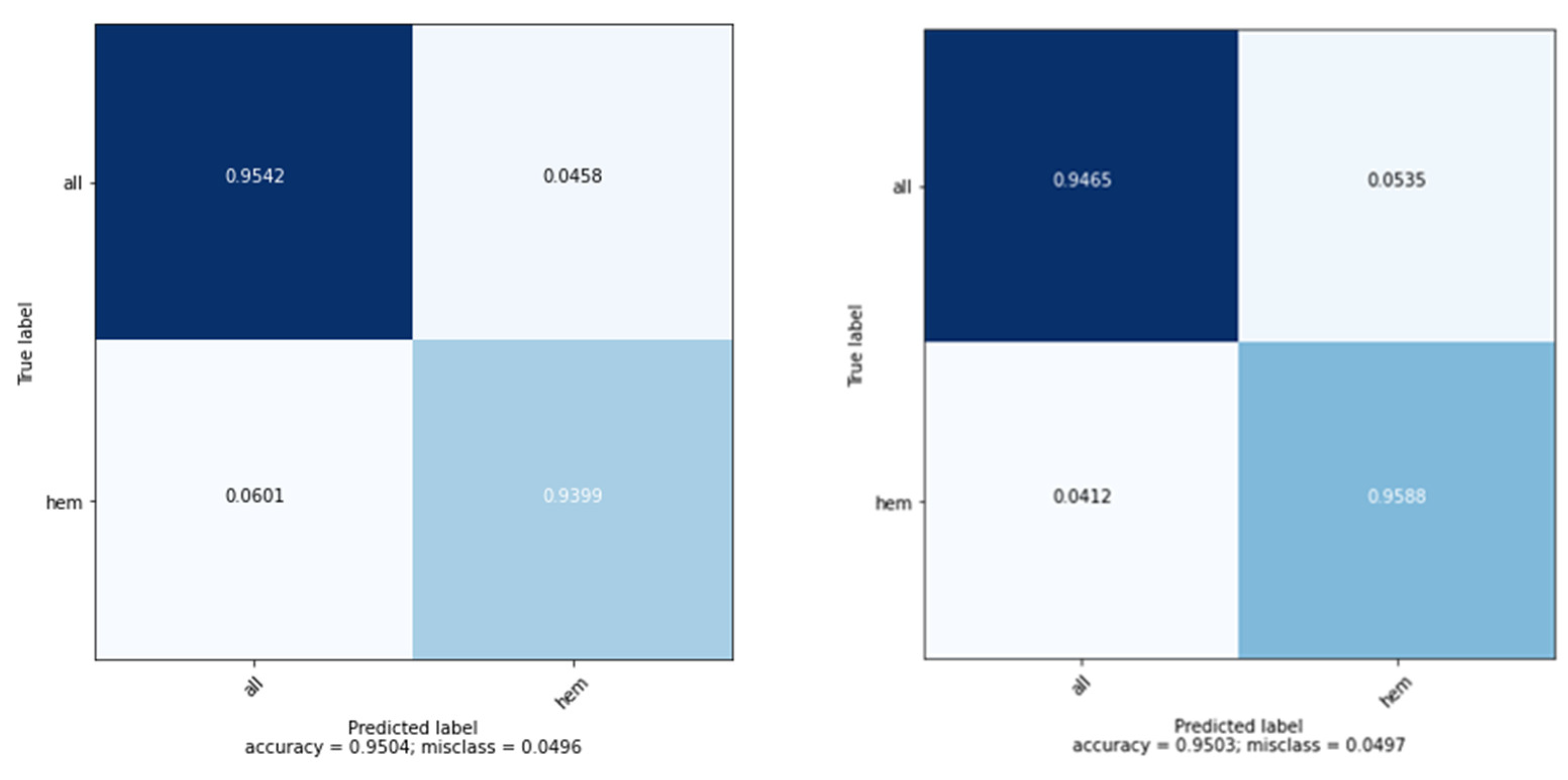
3.2. Model Evaluation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cho, S.; Tromburg, C.; Forbes, C.; Tran, A.; Allapitan, E.; Fay-McClymont, T.; Reynolds, K.; Schulte, F. Social adjustment across the lifespan in survivors of pediatric acute lymphoblastic leukemia (ALL): A systematic review. J. Cancer Surviv. 2022, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Sheykhhasan, M.; Manoochehri, H.; Dama, P. Use of CAR T-cell for acute lymphoblastic leukemia (ALL) treatment: A review study. Cancer Gene Ther. 2022, 29, 1080–1096. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, H.; Luan, Y.; Liu, T.; Yang, W.; Roberts, K.G.; Qian, M.X.; Zhang, B.; Yang, W.; Perez-Andreu, V.; et al. Noncoding ge-netic variation in GATA3 increases acute lymphoblastic leukemia risk through local and global changes in chromatin con-formation. Nat. Genet. 2022, 54, 170–179. [Google Scholar] [CrossRef]
- Khullar, K.; Plascak, J.J.; Parikh, R.R. Acute lymphoblastic leukemia (ALL) in adults: Disparities in treatment intervention based on access to treatment facility. Leuk. Lymphoma 2021, 63, 170–178. [Google Scholar] [CrossRef]
- Chadaga, K.; Prabhu, S.; Vivekananda, B.K.; Niranjana, S.; Umakanth, S. Battling COVID-19 using machine learning: A review. Cogent Eng. 2021, 8, 1958666. [Google Scholar] [CrossRef]
- Nono Djotsa, A.; Helmer, D.A.; Park, C.; Lynch, K.E.; Sharafkhaneh, A.; Naik, A.D.; Razjouyan, J.; Amos, C.I. Assessing Smoking Status and Risk of SARS-CoV-2 Infection: A Machine Learning Approach among Veterans. Healthcare 2022, 10, 1244. [Google Scholar] [CrossRef]
- Chadaga, K.; Prabhu, S.; Umakanth, S.; Bhat, V.K.; Sampathila, N.; Chadaga, R.P.; Prakasha, K.K. COVID-19 Mortality Prediction among Patients Using Epidemiological Parameters: An Ensemble Machine Learning Approach. Eng. Sci. 2021, 16, 221–233. [Google Scholar] [CrossRef]
- Absar, N.; Das, E.K.; Shoma, S.N.; Khandaker, M.U.; Miraz, M.H.; Faruque, M.R.I.; Tamam, N.; Sulieman, A.; Pathan, R.K. The Efficacy of Machine-Learning-Supported Smart System for Heart Disease Prediction. Healthcare 2022, 10, 1137. [Google Scholar] [CrossRef]
- Chadaga, K.; Prabhu, S.; Bhat, K.V.; Umakanth, S.; Sampathila, N. Medical diagnosis of COVID-19 using blood tests and machine learning. In Journal of Physics: Conference Series, Proceedings of the 1st International Conference on Artificial Intelligence, Computational Electronics and Communication System (AICECS 2021), Manipal, India, 28–30 October 2021; IOP Publishing: Bristol, UK, 2022; Volume 2161. [Google Scholar] [CrossRef]
- Lukić, I.; Ranković, N.; Savić, N.; Ranković, D.; Popov, Ž.; Vujić, A.; Folić, N. A Novel Approach of Determining the Risks for the Development of Hyperinsulinemia in the Children and Adolescent Population Using Radial Basis Function and Support Vec-tor Machine Learning Algorithm. Healthcare 2022, 10, 921. [Google Scholar] [CrossRef]
- Yamamoto, N.; Sukegawa, S.; Watari, T. Impact of System and Diagnostic Errors on Medical Litigation Outcomes: Machine Learning-Based Prediction Models. Healthcare 2022, 10, 892. [Google Scholar] [CrossRef]
- Li, L.; Han, C.; Yu, X.; Shen, J.; Cao, Y. Targeting AraC-Resistant Acute Myeloid Leukemia by Dual Inhibition of CDK9 and Bcl-2: A Systematic Review and Meta-Analysis. J. Healthc. Eng. 2022, 25, 2022. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, P.; Khanna, K.; Singh, V. LeuFeatx: Deep learning–based feature extractor for the diagnosis of acute leukemia from mi-croscopic images of peripheral blood smear. Comput. Biol. Med. 2022, 19, 105236. [Google Scholar] [CrossRef] [PubMed]
- Alqudah, R.; Suen, C.Y. Intensive Survey on Peripheral Blood Smear Analysis Using Deep Learning. In Advances in Pattern Recognition and Artificial Intelligence; World Scientific Publishing: Singapore, 2022; pp. 23–45. [Google Scholar] [CrossRef]
- Toret, E.; Demir-Kolsuz, O.; Ozdemir, Z.C.; Bor, O. A Case Report of Congenital Thrombotic Thrombocytopenic Purpura: The Peripheral Blood Smear Lights the Diagnosis. J. Pediatr. Hematol. 2022, 44, e243–e245. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.M. An efficient multi-scale CNN model with intrinsic feature integration for motor imagery EEG subject classification in brain-machine interfaces. Biomed. Signal Process. Control 2022, 74, 103496. [Google Scholar] [CrossRef]
- Anagha, V.; Disha, A.; Aishwarya, B.Y.; Nikkita, R.; Biradar, V.G. Detection of Leukemia Using Convolutional Neural Network. In Emerging Research in Computing, Information, Communication and Applications; Springer: Singapore, 2021; pp. 229–242. [Google Scholar] [CrossRef]
- Jiang, Z.; Dong, Z.; Wang, L.; Jiang, W. Method for Diagnosis of Acute Lymphoblastic Leukemia Based on ViT-CNN Ensemble Model. Comput. Intell. Neurosci. 2021, 23, 2021. [Google Scholar] [CrossRef]
- Ahmed, N.; Yigit, A.; Isik, Z.; Alpkocak, A. Identification of leukemia subtypes from microscopic images using convolutional neu-ral network. Diagnostics 2019, 9, 104. [Google Scholar] [CrossRef]
- Ghaderzadeh, M.; Aria, M.; Hosseini, A.; Asadi, F.; Bashash, D.; Abolghasemi, H. A fast and efficient CNN model for B-ALL diag-nosis and its subtypes classification using peripheral blood smear images. Int. J. Intell. Syst. 2021, 37, 5113–5133. [Google Scholar] [CrossRef]
- Qiao, Y.; Zhang, Y.; Liu, N.; Chen, P.; Liu, Y. An End-to-End Pipeline for Early Diagnosis of Acute Promyelocytic Leukemia Based on a Compact CNN Model. Diagnostics 2021, 11, 1237. [Google Scholar] [CrossRef]
- Jha, K.K.; Dutta, H.S. Mutual Information based hybrid model and deep learning for Acute Lymphocytic Leukemia detection in single cell blood smear images. Comput. Methods Programs Biomed. 2019, 179, 104987. [Google Scholar] [CrossRef]
- Vogado, L.H.; Veras, R.M.; Araujo, F.H.; Silva, R.R.; Aires, K.R. Leukemia diagnosis in blood slides using transfer learning in CNNs and SVM for classification. Eng. Appl. Artif. Intell. 2018, 72, 415–422. [Google Scholar] [CrossRef]
- Shafique, S.; Tehsin, S. Acute lymphoblastic leukemia detection and classification of its subtypes using pretrained deep convo-lutional neural networks. Technol. Cancer Res. Treat. 2018, 26, 1533033818802789. [Google Scholar]
- He, J.; Pu, X.; Li, M.; Li, C.; Guo, Y. Deep convolutional neural networks for predicting leukemia-related transcription factor bind-ing sites from DNA sequence data. Chemom. Intell. Lab. Syst. 2020, 199, 103976. [Google Scholar] [CrossRef]
- Sahlol, A.T.; Kollmannsberger, P.; Ewees, A.A. Efficient classification of white blood cell leukemia with improved swarm optimi-zation of deep features. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Banik, P.P.; Saha, R.; Kim, K.-D. An Automatic Nucleus Segmentation and CNN Model based Classification Method of White Blood Cell. Expert Syst. Appl. 2020, 149, 113211. [Google Scholar] [CrossRef]
- Naz, I.; Muhammad, N.; Yasmin, M.; Sharif, M.; Shah, J.H.; Fernandes, S.L. Robust Discrimination of Leukocytes Protuberant Types for Early Diagnosis Of Leukemia. J. Mech. Med. Biol. 2019, 19, 1950055. [Google Scholar] [CrossRef]
- Rehman, A.; Abbas, N.; Saba, T.; Rahman, S.I.U.; Mehmood, Z.; Kolivand, H. Classification of acute lymphoblastic leukemia using deep learning. Microsc. Res. Tech. 2018, 81, 1310–1317. [Google Scholar] [CrossRef]
- Gupta, A.; Gupta, R. ALL Challenge Dataset of ISBI 2019 [Data Set]. In The Cancer Imaging Archive; National Cancer Institute: Bethesda, ME, USA, 2019. [Google Scholar] [CrossRef]
- Gehlot, S.; Gupta, A.; Gupta, R. SDCT-AuxNet: DCT augmented stain deconvolutional CNN with auxiliary classifier for cancer diagnosis. Med. Image Anal. 2020, 61, 101661. [Google Scholar] [CrossRef]
- Goswami, S.; Mehta, S.; Sahrawat, D.; Gupta, A.; Gupta, R. Heterogeneity Loss to Handle Intersub-ject and Intrasubject Variability in Cancer, ICLR workshop on Affordable AI in healthcare. arXiv 2020, arXiv:2003.03295. [Google Scholar]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef]
- Abunadi, I.; Senan, E.M. Multi-Method Diagnosis of Blood Microscopic Sample for Early Detection of Acute Lymphoblastic Leukemia Based on Deep Learning and Hybrid Techniques. Sensors 2022, 22, 1629. [Google Scholar] [CrossRef]
- Pan, Y.; Liu, M.; Xia, Y.; Shen, D. Neighborhood-Correction Algorithm for Classification of Normal and Malignant Cells. In ISBI 2019 C-NMC Challenge: Classification in Cancer Cell Imaging; Springer: Singapore, 2019; pp. 73–82. [Google Scholar]
- Khandekar, R.; Shastry, P.; Jaishankar, S.; Faust, O.; Sampathila, N. Automated blast cell detection for Acute Lympho-blastic Leukemia diagnosis. Biomed. Signal Processing Control 2021, 68, 102690. [Google Scholar] [CrossRef]
- Marzahl, C.; Aubreville, M.; Voigt, J.; Maier, A. Classification of Leukemic B-Lymphoblast Cells from Blood Smear Microscopic Images with an Attention-Based Deep Learning Method and Advanced Augmentation Techniques. In ISBI 2019 C-NMC Challenge: Classification in Cancer Cell Imaging; Springer: Singapore, 2019; pp. 13–22. [Google Scholar]
- Mayrose, H.; Sampathila, N.; Bairy, G.M.; Belurkar, S.; Saravu, K.; Basu, A.; Khan, S. Intelligent algorithm for detection of dengue using mobilenetv2-based deep features with lymphocyte nucleus. Expert Syst. 2021, e12904. [Google Scholar] [CrossRef]
- Krishnadas, P.; Sampathila, N. Automated Detection of Malaria implemented by Deep Learning in Pytorch. In Proceedings of the 2021 IEEE International Conference on Electronics, Computing and Communication Technologies (CONECCT), Bangalore, India, 9–11 July 2021; pp. 01–05. [Google Scholar] [CrossRef]
- Mayrose, H.; Niranjana, S.; Bairy, G.M.; Edwankar, H.; Belurkar, S.; Saravu, K. Computer Vision Approach for the detection of Thrombocytopenia from Microscopic Blood Smear Images. In Proceedings of the 2021 IEEE International Conference on Electronics, Computing and Communication Technologies (CONECCT), Bangalore, India, 9–11 July 2021; pp. 1–5. [Google Scholar] [CrossRef]
- Upadya, P.S.; Sampathila, N.; Hebbar, H.; Pai, B.S. Machine learning approach for classification of maculopapular and vesicular rashes using the textural features of the skin images. Cogent Eng. 2022, 9, 2009093. [Google Scholar] [CrossRef]
- Cornish, T.C.; McClintock, D.S. Whole Slide Imaging and Telepathology. In Whole Slide Imaging; Springer: Cham, Switzerland, 2021; pp. 117–152. [Google Scholar] [CrossRef]
- Díaz, D.; Corredor, G.; Romero, E.; Cruz-Roa, A. A web-based telepathology framework for collabora-tive work of pathologists to support teaching and research in Latin America. In Sipaim–Miccai Biomedical Workshop; Springer: Cham, Switzerland; pp. 105–112.









| Research | Dataset | Algorithm Used | Accuracy | Precision | Recall |
|---|---|---|---|---|---|
| [23] | Three hybrid image databases | CNN and SVM | 99% | 99% | 99% |
| [24] | IDB dataset | AlexNet | 96% | - | 96.74% |
| [25] | DNA Sequence images | CNN and other ML models | 75% | - | - |
| [26] | GRTD dataset | VCGNet | 96% | 93% | 93% |
| [27] | BCCD ALL-IDB2 JTSC CellaVision databases | CNN | 97% | 80% | 94% |
| [28] | LISC and Dhruv dataset | CNN | 97% | 80% | 94% |
| [29] | Amreek Clinical Laboratory | CNN | 97.75% | - | - |
| Layer (Type) | Layer Shape | Number of Parameters |
|---|---|---|
| Conv2D | (450, 450, 3) | 1792 |
| Max_Pooling_2D | (150, 150, 64) | - |
| Conv2D | (150, 150, 256) | 147,712 |
| Max_Pooling_2D | (50, 50, 256) | - |
| Conv2D | (50, 50, 384) | 885,120 |
| Batch_Normalization | (50, 50, 384) | 1536 |
| Max_Pooling | (17, 17, 384) | - |
| Dropout | (17, 17, 384) | - |
| Conv2D | (17, 17, 512) | 1,769,984 |
| Batch Normalization | (17, 17, 512) | 2048 |
| Max_Pooling | (6, 6, 512) | - |
| Dropout | (6, 6, 512) | - |
| Flatten | 18,232 | - |
| Dense | 4096 | 75,501,568 |
| Dropout | 4096 | - |
| Dense | 4096 | 16,781,312 |
| Dropout | 4096 | - |
| Output | 2 | 8194 |
| Instance | Accuracy (%) | Specificity (%) | Recall (%) | F1-Score (%) | Precision (%) | Matthews Correlation Coefficient (MCC) |
|---|---|---|---|---|---|---|
| Instance 1 | 94.94 | 94.87 | 94 | 94.96 | 95.95 | 89.42 |
| Instance 2 | 94.5 | 95.8 | 93.2 | 93.2 | 96 | 89.53 |
| Instance 3 | 94.72 | 95.8 | 93.2 | 93.2 | 96 | 88.76 |
| Instance 4 | 95 | 95.91 | 94 | 94.96 | 95.95 | 88 |
| Instance 5 | 95.45 | 95 | 95.91 | 95.43 | 94.94 | 89.5 |
| Reference | Method | Salient Features | Performance Measure |
|---|---|---|---|
| [34] | Bagging Ensemble with Deep Learning | Bagging ensembling was utilized | F1-score of 88% |
| [35] | Neighborhood Correction Algorithm | Fine-tuning a pre-trained residual network, constructing a Fisher vector based on feature maps, and correction using weighted majority. | F1-score of 92% |
| [37] | CNN | Regional proposal subnetwork. | F1-score of 83% |
| Proposed | CNN | Improved accuracy | F1-score of 96% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sampathila, N.; Chadaga, K.; Goswami, N.; Chadaga, R.P.; Pandya, M.; Prabhu, S.; Bairy, M.G.; Katta, S.S.; Bhat, D.; Upadya, S.P. Customized Deep Learning Classifier for Detection of Acute Lymphoblastic Leukemia Using Blood Smear Images. Healthcare 2022, 10, 1812. https://doi.org/10.3390/healthcare10101812
Sampathila N, Chadaga K, Goswami N, Chadaga RP, Pandya M, Prabhu S, Bairy MG, Katta SS, Bhat D, Upadya SP. Customized Deep Learning Classifier for Detection of Acute Lymphoblastic Leukemia Using Blood Smear Images. Healthcare. 2022; 10(10):1812. https://doi.org/10.3390/healthcare10101812
Chicago/Turabian StyleSampathila, Niranjana, Krishnaraj Chadaga, Neelankit Goswami, Rajagopala P. Chadaga, Mayur Pandya, Srikanth Prabhu, Muralidhar G. Bairy, Swathi S. Katta, Devadas Bhat, and Sudhakara P. Upadya. 2022. "Customized Deep Learning Classifier for Detection of Acute Lymphoblastic Leukemia Using Blood Smear Images" Healthcare 10, no. 10: 1812. https://doi.org/10.3390/healthcare10101812
APA StyleSampathila, N., Chadaga, K., Goswami, N., Chadaga, R. P., Pandya, M., Prabhu, S., Bairy, M. G., Katta, S. S., Bhat, D., & Upadya, S. P. (2022). Customized Deep Learning Classifier for Detection of Acute Lymphoblastic Leukemia Using Blood Smear Images. Healthcare, 10(10), 1812. https://doi.org/10.3390/healthcare10101812







