Time Series Clustering with Topological and Geometric Mixed Distance
Abstract
1. Introduction
- We propose a TGMD measure for time series clustering analysis by combining the local geometric and global topological features of time series. Topology is a generic representation that enables a qualitative description of global features of a time series, while geometry describes local quantitative differences of the raw time series.
- Our mixed distance measure is based on a tuning function that modulates the TGMD matrix according to the proximity of topological properties.
- Experiments confirm that the method proposed in this paper outperforms topological-only methods or geometric-only methods in clustering noisy biological data for oscillatory activity clustering identification. Additionally, our method achieves competitive results on real datasets compared with other standard time series clustering methods. The visualization of TGMD metric space also demonstrate its effectiveness.
2. Background and Previous Work
2.1. Time Series Similarity Measures
2.2. Topological Time Series Analysis
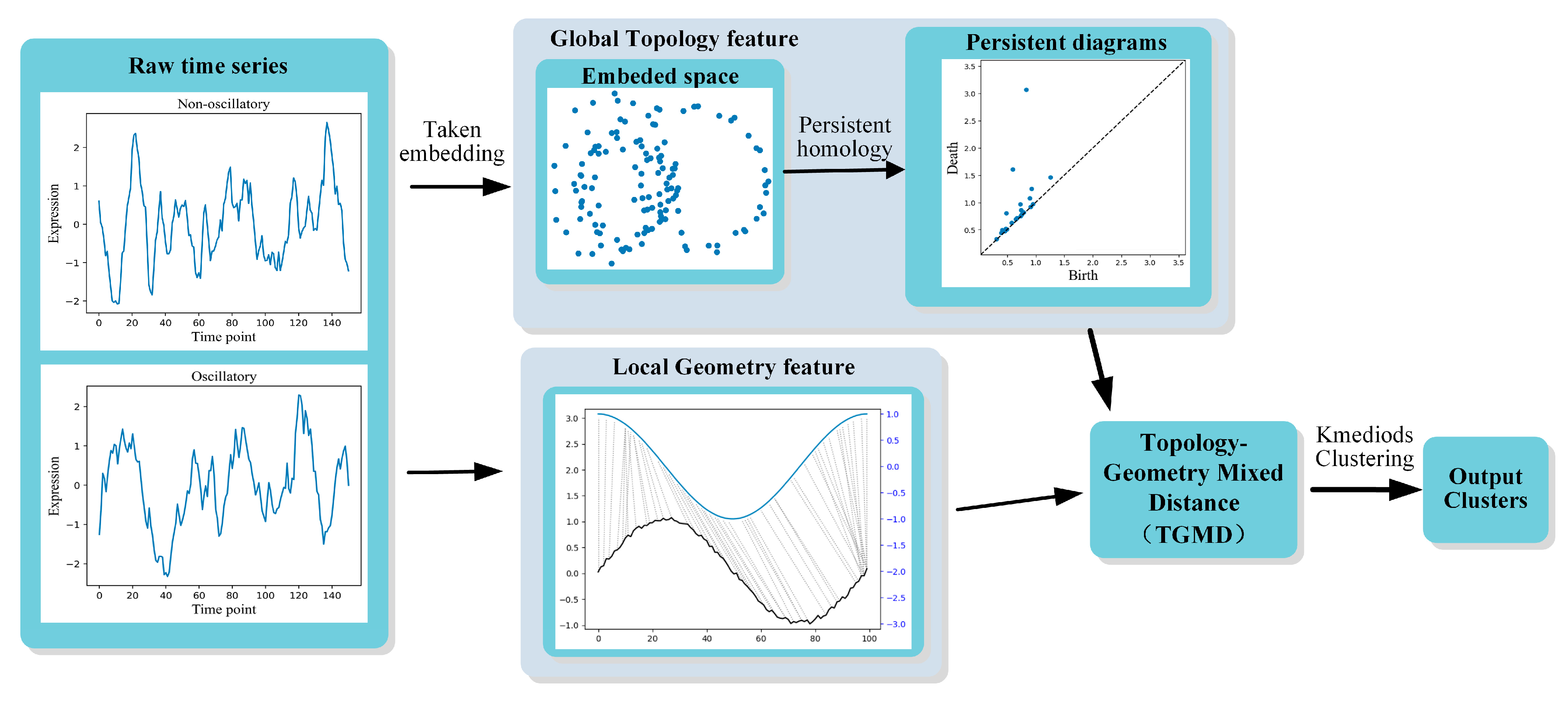
3. Methods
3.1. Global and Local Features Extraction
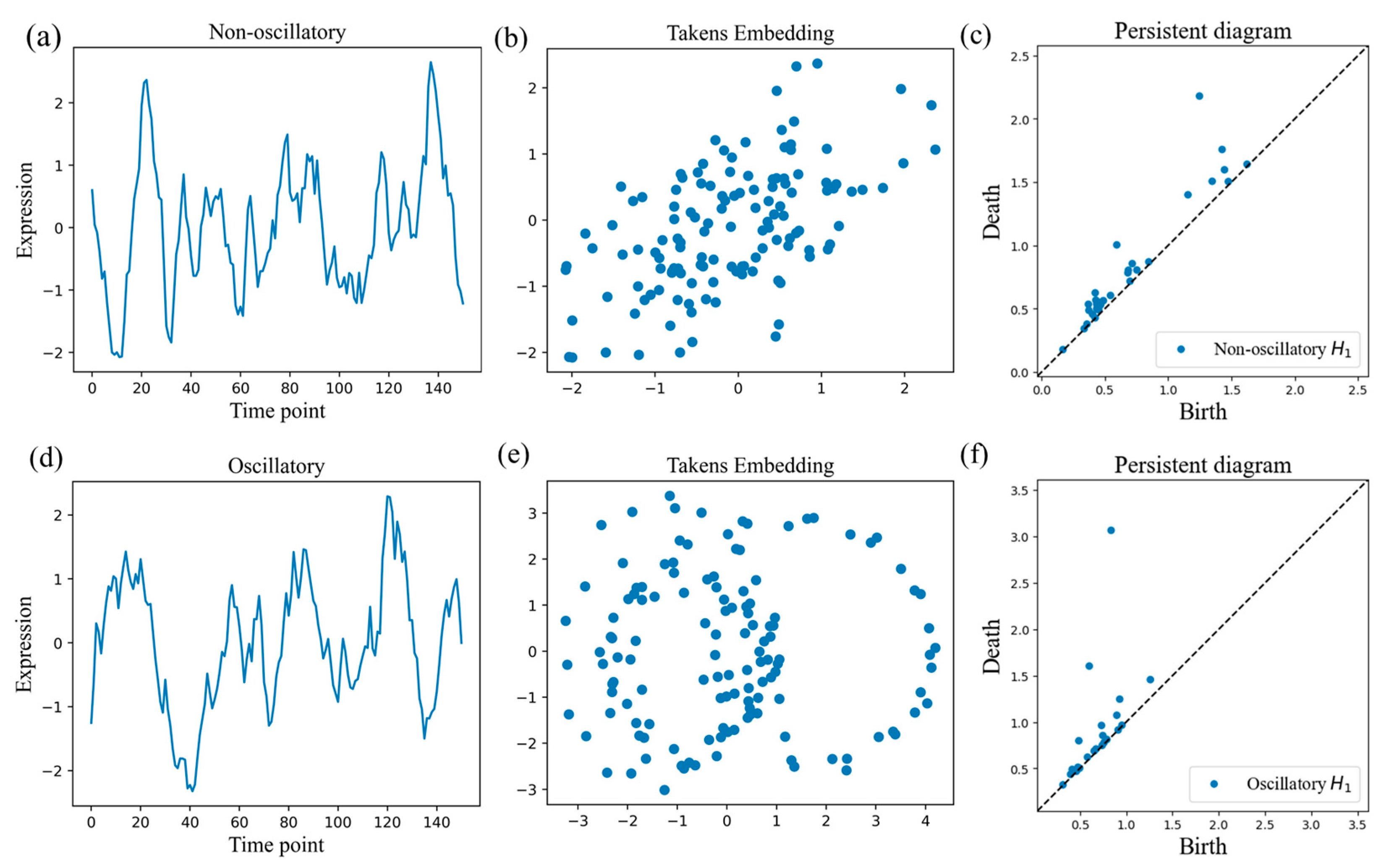
3.1.1. Global Topological Properties of Time Series
3.1.2. Local Properties of Time Series
3.2. Topological-Geometric Mixed Distance
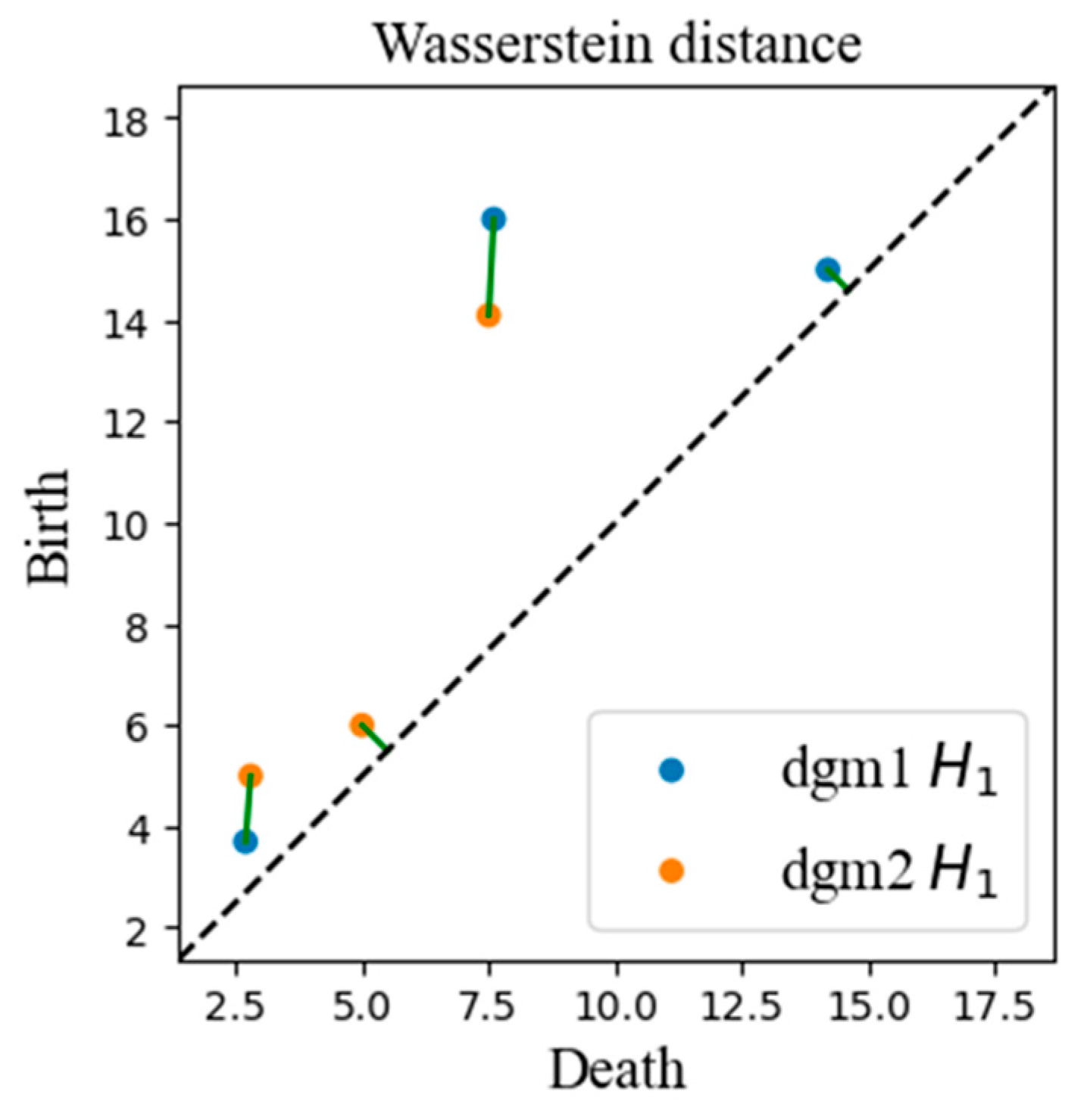
3.2.1. Topological Similarity
3.2.2. Geometric Similarity of Time Series
3.2.3. Topological-Geometric Mixed Distance
3.2.4. Clustering Algorithms
4. Experiment
4.1. Experiment Datasets
4.1.1. Synthetic Single-Cell Data
4.1.2. UCR Time Series Archive
4.2. Baselines and Metrics
4.2.1. Baselines
4.2.2. Metrics
4.3. Experimental Setup
4.4. Clustering Analysis of the Synthetic Single-Cell Data
4.5. Clustering Analysis of the Real Time Series Data
4.6. Visualization Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aghabozorgi, S.; Shirkhorshidi, A.S.; Wah, T.Y. Time-series clustering—A decade review. Inf. Syst. 2015, 53, 16–38. [Google Scholar] [CrossRef]
- Subhani, N.; Rueda, L.; Ngom, A.; Burden, C.J. Multiple gene expression profile alignment for microarray time-series data clustering. Bioinformatics 2010, 26, 2281–2288. [Google Scholar] [CrossRef]
- Liu, X.; Tian, Y.; Zhang, X.; Wan, Z. Identification of Urban Functional Regions in Chengdu Based on Taxi Trajectory Time Series Data. ISPRS Int. J. Geo-Inf. 2020, 9, 158. [Google Scholar] [CrossRef]
- Gajowniczek, K.; Bator, M.; Ząbkowski, T. Whole Time Series Data Streams Clustering: Dynamic Profiling of the Electricity Consumption. Entropy 2020, 22, 1414. [Google Scholar] [CrossRef]
- Hsu, Y.-C.; Chen, A.-P. A clustering time series model for the optimal hedge ratio decision making. Neurocomputing 2014, 138, 358–370. [Google Scholar] [CrossRef]
- Chu, S. Iterative deepening dynamic time warping for time series. In Proceedings of the 2002 SIAM International Conference on Data Mining. Society for Industrial and Applied Mathematics, Arlington, VA, USA, 11–13 April 2002; pp. 148–156. [Google Scholar]
- Faloutsos, C.; Ranganathan, M.; Manolopoulos, Y. Fast subsequence matching in time-series databases. ACM Sigmod Rec. 1994, 23, 419–429. [Google Scholar] [CrossRef]
- Vlachos, M.; Kollios, G.; Gunopulos, D. Discovering similar multidimensional trajectories. In Proceedings of the 18th International Conference on Data Engineering, San Jose, CA, USA, 26 February–1 March 2002; pp. 673–684. [Google Scholar]
- Li, Y.; Liu, R.W.; Liu, Z.; Liu, J. Similarity Grouping-Guided Neural Network Modeling for Maritime Time Series Prediction. IEEE Access 2019, 7, 72647–72659. [Google Scholar] [CrossRef]
- Pereira, C.M.M.; de Mello, R.F. Persistent homology for time series and spatial data clustering. Expert Syst. Appl. 2015, 42, 6026–6038. [Google Scholar] [CrossRef]
- Ferreira, L.N.; Zhao, L. Time series clustering via community detection in networks. Inf. Sci. 2016, 326, 227–242. [Google Scholar] [CrossRef]
- Umeda, Y. Time Series Classification via Topological Data Analysis. Inf. Media Technol. 2017, 12, 228–239. [Google Scholar] [CrossRef]
- Tran, Q.H.; Hasegawa, Y. Topological time-series analysis with delay-variant embedding. Phys. Rev. E 2019, 99, 032209. [Google Scholar] [CrossRef]
- Majumdar, S.; Laha, A.K. Clustering and classification of time series using topological data analysis with applications to finance. Expert Syst. Appl. 2020, 162, 113868. [Google Scholar] [CrossRef]
- Seversky, L.M.; Davis, S.; Berger, M. On Time-Series Topological Data Analysis: New Data and Opportunities. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Las Vegas, NV, USA, 26 June–1 July 2016; pp. 1014–1022. [Google Scholar]
- Takens, F. Detecting strange attractors in turbulence. In Dynamical Systems and Turbulence, Warwick 1980; Springer: Berlin/Heidelberg, Germany, 1981; pp. 366–381. [Google Scholar]
- Edelsbrunner, H.; Letscher, D.; Zomorodian, A. Topological persistence and simplification. In Proceedings of the 41st Annual Symposium on Foundations of Computer Science, Redondo Beach, CA, USA, 12–14 November 2000; pp. 454–463. [Google Scholar]
- Zomorodian, A.J. Topology for Computing; Cambridge University Press: Cambridge, UK, 2005; Volume 16. [Google Scholar]
- Carlsson, G. Topology and data. Bull. Am. Math. Soc. 2009, 46, 255–308. [Google Scholar] [CrossRef]
- Cohen-Steiner, D.; Edelsbrunner, H.; Harer, J. Stability of Persistence Diagrams. Discret. Comput. Geom. 2007, 37, 103–120. [Google Scholar] [CrossRef]
- Carrière, M.; Cuturi, M.; Oudot, S. Sliced Wasserstein Kernel for Persistence Diagrams. In Proceedings of the 34th International Conference on Machine Learning, Proceedings of Machine Learning Research, Sydney, Australia, 6–11 August 2017; pp. 664–673. [Google Scholar]
- Yin, J.; Wang, R.; Zheng, H.; Yang, Y.; Li, Y.; Xu, M. A new time series similarity measurement method based on the morphological pattern and symbolic aggregate approximation. IEEE Access 2019, 7, 109751–109762. [Google Scholar] [CrossRef]
- Berndt, D.J.; Clifford, J. Using dynamic time warping to find patterns in time series. In Proceedings of the KDD Workshop, Seattle, WA, USA, 31 July 1994; pp. 359–370. [Google Scholar]
- Petitjean, F.; Ketterlin, A.; Gançarski, P. A global averaging method for dynamic time warping, with applications to clustering. Pattern Recognit. 2011, 44, 678–693. [Google Scholar] [CrossRef]
- Paparrizos, J.; Gravano, L. k-shape: Efficient and accurate clustering of time series. In Proceedings of the 2015 ACM SIGMOD International Conference on Management of Data, New York, NY, USA, 31 May–4 June 2015; pp. 1855–1870. [Google Scholar]
- Yang, J.; Leskovec, J. Patterns of temporal variation in online media. In Proceedings of the fourth ACM International Conference on Web Search and Data Mining, Hong Kong, China, 9–12 February 2011; pp. 177–186. [Google Scholar]
- Zulkepli, N.F.S.; Noorani, M.S.M.; Razak, F.A.; Ismail, M.; Alias, M.A. Cluster Analysis of Haze Episodes Based on Topological Features. Sustainability 2020, 12, 3985. [Google Scholar] [CrossRef]
- Perea, J.A. SW1PerS: Sliding windows and 1-persistence scoring; discovering periodicity in gene expression time series data. BMC Bioinform. 2015, 16, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Frahi, T.; Chinesta, F.; Falcó, A.; Badias, A.; Cueto, E.; Choi, H.Y.; Han, M.; Duval, J.-L. Empowering Advanced Driver-Assistance Systems from Topological Data Analysis. Mathematics 2021, 9, 634. [Google Scholar] [CrossRef]
- Kim, K.; Kim, J.; Rinaldo, A. Time series featurization via topological data analysis. arXiv 2018, arXiv:1812.02987. [Google Scholar]
- Gidea, M.; Goldsmith, D.; Katz, Y.; Roldan, P.; Shmalo, Y. Topological recognition of critical transitions in time series of cryptocurrencies. Phys. A Stat. Mech. Appl. 2020, 548, 123843. [Google Scholar] [CrossRef]
- Chen, R.; Zhang, J.; Ravishanker, N.; Konduri, K.J. Clustering Activity–Travel Behavior Time Series using Topological Data Analysis. J. Big Data Anal. Transp. 2019, 1, 109–121. [Google Scholar] [CrossRef]
- Rousseeuw, P.J. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef]
- Majumdar, K.; Jayachandran, S. A geometric analysis of time series leading to information encoding and a new entropy measure. J. Comput. Appl. Math. 2018, 328, 469–484. [Google Scholar] [CrossRef]
- Mileyko, Y.; Mukherjee, S.; Harer, J. Probability measures on the space of persistence diagrams. Inverse Probl. 2011, 27, 124007. [Google Scholar] [CrossRef]
- Monk, N.A.M. Oscillatory Expression of Hes1, p53, and NF-κB Driven by Transcriptional Time Delays. Curr. Biol. 2003, 13, 1409–1413. [Google Scholar] [CrossRef]
- Dau, H.A.; Bagnall, A.; Kamgar, K.; Yeh, C.M.; Zhu, Y.; Gharghabi, S.; Ratanamahatana, C.A.; Keogh, E. The UCR time series archive. IEEE/CAA J. Autom. Sin. 2019, 6, 1293–1305. [Google Scholar] [CrossRef]
- Huang, X.; Ye, Y.; Xiong, L.; Lau, R.Y.K.; Jiang, N.; Wang, S. Time series k-means: A new k-means type smooth subspace clustering for time series data. Inf. Sci. 2016, 367–368, 1–13. [Google Scholar] [CrossRef]
- Tavenard, R.; Faouzi, J.; Vandewiele, G.; Divo, F.; Androz, G.; Holtz, C.; Payne, M.; Yurchak, R.; Rußwurm, M.; Kolar, K. Tslearn, a machine learning toolkit for time series data. J. Mach. Learn. Res. 2020, 21, 1–6. [Google Scholar]
- Hubert, L.; Arabie, P. Comparing partitions. J. Classif. 1985, 2, 193–218. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Melville, J. Umap: Uniform manifold approximation and projection for dimension reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]







| Dataset | Data Size | Length | No. of Classes | Type |
|---|---|---|---|---|
| Synthetic Control | 600 | 60 | 6 | SIMULATED |
| ECG200 | 200 | 96 | 2 | ECG |
| ECGfiveday | 884 | 136 | 2 | ECG |
| ECG5000 | 500 | 140 | 5 | ECG |
| TwoLeadECG | 1162 | 82 | 2 | ECG |
| Fish | 350 | 463 | 7 | IMAGE |
| Face four | 112 | 350 | 4 | IMAGE |
| DiatonSizeReduction | 322 | 345 | 4 | IMAGE |
| ItalyPowerDemand | 1096 | 24 | 2 | SENSOR |
| Trace | 200 | 275 | 4 | SENSOR |
| GunPoint | 200 | 150 | 2 | MOTION |
| Datasets | TS Kmeans | PCA-Kmeans | Kshape | k-med-DTW | k-med-ED | k-med-TGMD (DTW + TS) | k-med-TGMD (ED + TS) |
|---|---|---|---|---|---|---|---|
| Synthetic Controls | 0.5902 | 0.6157 | 0.6080 | 0.6502 | 0.3765 | 0.8238 | 0.5048 |
| ECG200 | 0.0783 | 0.2194 | 0.1802 | 0.0981 | 0.2640 | 0.3096 | 0.3135 |
| ECGfiveday | −0.0275 | −0.0434 | 0.6680 | 0.0515 | 0.0042 | 0.3993 | 0.2398 |
| Two lead ECG | 0.1136 | 0.0535 | 0.0502 | −0.0254 | 0.1129 | 0.2877 | 0.1129 |
| ECG5000 | 0.4253 | 0.4849 | 0.3859 | 0.4014 | 0.4943 | 0.4448 | 0.5726 |
| Face four | 0.3464 | 0.3377 | 0.3833 | 0.3548 | 0.2376 | 0.7283 | 0.2844 |
| fish | 0.3653 | 0.1796 | 0.1515 | 0.2061 | 0.1801 | 0.3230 | 0.2362 |
| Italy power | 0.0025 | 0.0025 | 0.5277 | 0.4427 | 0.8263 | 0.4843 | 0.8263 |
| Trace | 0.6160 | 0.3266 | 0.3319 | 0.6288 | 0.3649 | 0.6810 | 0.6730 |
| GunPoint | 0.1148 | −0.0053 | −0.0053 | −0.0050 | −0.0050 | 0.2775 | 0.2470 |
| DiatomSizeReduction | 0.8607 | 0.5221 | 0.4348 | 0.6450 | 0.6669 | 0.7857 | 0.8267 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Shi, Q.; Zhu, J.; Peng, J.; Li, H. Time Series Clustering with Topological and Geometric Mixed Distance. Mathematics 2021, 9, 1046. https://doi.org/10.3390/math9091046
Zhang Y, Shi Q, Zhu J, Peng J, Li H. Time Series Clustering with Topological and Geometric Mixed Distance. Mathematics. 2021; 9(9):1046. https://doi.org/10.3390/math9091046
Chicago/Turabian StyleZhang, Yunsheng, Qingzhang Shi, Jiawei Zhu, Jian Peng, and Haifeng Li. 2021. "Time Series Clustering with Topological and Geometric Mixed Distance" Mathematics 9, no. 9: 1046. https://doi.org/10.3390/math9091046
APA StyleZhang, Y., Shi, Q., Zhu, J., Peng, J., & Li, H. (2021). Time Series Clustering with Topological and Geometric Mixed Distance. Mathematics, 9(9), 1046. https://doi.org/10.3390/math9091046







